Abstract
Insulin-like growth factor II mRNA-binding protein 1 (IMP1) belongs to a family of RNA-binding proteins implicated in mRNA localization, turnover, and translational control. Mouse IMP1 is expressed during early development, and an increase in expression occurs around embryonic day 12.5 (E12.5). To characterize the physiological role of IMP1, we generated IMP1-deficient mice carrying a gene trap insertion in the Imp1 gene. Imp1−/− mice were on average 40% smaller than wild-type and heterozygous sex-matched littermates. Growth retardation was apparent from E17.5 and remained permanent into adult life. Moreover, Imp1−/− mice exhibited high perinatal mortality, and only 50% were alive 3 days after birth. In contrast to most other organs, intestinal epithelial cells continue to express IMP1 postnatally, and Imp1−/− mice exhibited impaired development of the intestine, with small and misshapen villi and twisted colon crypts. Analysis of target mRNAs and global expression profiling at E12.5 indicated that Igf2 translation was downregulated, whereas the postnatal intestine showed reduced expression of transcripts encoding extracellular matrix components, such as galectin- 1, lumican, tenascin-C, procollagen transcripts, and the Hsp47 procollagen chaperone. Taken together, the results demonstrate that IMP1 is essential for normal growth and development. Moreover, IMP1 may facilitate intestinal morphogenesis via regulation of extracellular matrix formation.
Insulin-like growth factor II (IGF-II) mRNA-binding protein 1 (IMP1) and the closely related IMP2 and IMP3 proteins belong to a conserved family of RNA-binding proteins consisting of two RNA recognition motifs (RRM) and four K-homology (KH) domains (for review, see references 36 and 56). The latter are phylogenetically conserved in Drosophila melanogaster and Caenorhabditis elegans (GenBank T23837) and constitute a functionally independent entity (37). IMP1 is orthologous to the chicken zipcode-binding protein 1 (ZBP1) and the mouse c-Myc coding region determinant-binding protein (CRD-BP) (10, 45), whereas IMP3 is orthologous to the Xenopus laevis Vg1 mRNA-binding protein (Vg1-RBP/Vera) (8, 16). IMP2 has diverged phylogenetically from IMP1 and IMP3 and has no known orthologues, but a splice variant p62 has been isolated from hepatic carcinoma (59).
So far only a handful of mRNAs, including Igf2 leader 3, H19, c-myc, β-actin, and Vg1 mRNAs, have been identified as targets for the proteins (16, 25, 39, 45, 48). Binding sites are located in the 5′ untranslated region (UTR), the coding region, and the 3′ UTR of the mRNAs. Similar to other RNA-binding proteins such as Bruno, which mediates translational repression of oskar mRNA in D. melanogaster (24), multiple IMP molecules may associate with the RNA target. So far, there is no absolute consensus about the nature of the IMP attachment site. Therefore, it is not possible to predict putative target mRNAs solely on the basis of their sequences.
The IMPs have been implicated in posttranscriptional processes such as mRNA localization, turnover, and translational control (reviewed in references 36 and 56). IMP1 and the other members of the family are mainly cytoplasmic, but the presence of both putative import and export signals indicates that they also enter the nucleus (38, 43). This is further supported by the recent identification of IMP3 in spliceosomes (61) and of IMP1 in nucleolar preribosomal complexes (17). In the cytoplasm, IMP1 is present in large RNP granules, distributed along microtubules and filaments, and in motile cells it localizes to the leading edge (37).
An important lead to the function of the IMPs in RNA localization comes from studies of Xenopus laevis Vegetal 1 (Vg1) mRNA, which encodes a transforming growth factor beta-like growth factor that stimulates the formation of mesoderm at the vegetal cortex. In the early stages of oocyte development, Vg1 mRNA is evenly distributed in the ooplasm, but during stages III and IV it becomes localized to the vegetal pole via a Vg1-RBP/Vera-dependent mechanism (9). Later in development, Vg1-RBP/Vera promotes the migration of neural crest cells (55). Chicken ZBP1 was originally isolated from a protein complex associated with the β-actin mRNA zipcode, indicating that ZBP1 is implicated in localization of β-actin mRNA to the leading edge (45). In neurons, ZBP1 has recently been shown to be required for the localization of β-actin mRNA to dendrites, and reduced levels of ZBP1 and β-actin mRNA are associated with impaired growth of dendritic filopodia in response to growth factors (12). However, the biological function of IMPs may not be restricted to RNA trafficking. Coexpression of IMP1 and Igf2 leader 3 mRNA indicates that IMP1 is involved in translational control of the transcript (39). Finally, mouse CRD-BP, which binds to a 250-nucleotide element in the 3′ end of the coding region of c-myc mRNA stabilizes the messenger (18, 54), and it has been proposed that CRD-BP protects the transcript from an endonuclease.
Mouse IMPs are expressed in a biphasic fashion, with early expression in the oocytes and in the zygotes (36, 56). Later, a sharp increase in the expression of all three IMPs takes place around embryonic day 12.5 (E12.5), followed by a decline towards birth (39, 48). The tissue-specific distribution of the individual members has been partially characterized. At E12.5, Imp3/KOC mRNA is mainly found in the fore- and hindbrain, the snout, the branchial archs, the gut, the tail, the vertebrae, and the skin (31, 34). Moreover, high levels of Imp3/KOC mRNA are expressed in the placenta. Towards the end of embryogenesis, Imp3/KOC mRNA expression has almost disappeared except in the gut, thymus, extremities, and snout (31, 34). Vg1-RBP/Vera, zebra fish Vg1-RBP, and Drosophila Imp mRNAs are also expressed throughout the central nervous system during early neurogenesis (34, 40, 60). So far, the presence of IMPs in adult tissues has not been reported, but they are frequently found in transformed cells and are therefore considered oncofetal proteins. KOC (KH domain-containing protein overexpressed in cancer/IMP3) was originally isolated in a differential hybridization screen for genes overexpressed in pancreatic cancer (15, 33), and IMP1 is found in the majority of solid tumors from the breast, colon, and lung (11, 46; F. C. Nielsen, unpublished data). Moreover, many sarcomas in particular Ewing sarcoma exhibit high levels of Imp1 mRNA (20).
The restricted spatiotemporal expression and selective RNA binding are hallmarks of the IMP family. To study the physiological role of IMP1, we characterized IMP1-deficient mice generated by a gene trap insertion in intron 2 of the Imp1 gene. We show that IMP1-deficient mice become dwarfs and have increased perinatal mortality. Moreover, the animals exhibit imperfect development of the gut. Taken together, the results demonstrate that IMP1 is essential for normal embryonal development and perinatal survival.
MATERIALS AND METHODS
Gene trap insertion in ES cells and generation of Imp1−/− mice.
Embryonic stem (ES) cells with a retroviral gene trap vector inserted into the Imp1 locus (Omnibank no. OST33739) were generated as described previously (58) (Lexicon Genetics). The location of the gene trap insertion was determined by sequence analysis. The Imp1-targeted ES cells were microinjected into C57BL/6 blastocytes, followed by transfer to a foster mother to generate chimeric animals. Chimeric males were mated with C57BL/6 females, and F1 agouti pups were analyzed for the presence of the transgene by PCR analysis of tail genomic DNA with the following primers: Imp1, 5′-CCCTGCAGCTAGGGATGAAGA-3′ and 5′-GTATGAACCTGGAACATGACGCT-3′; and β-geo, 5′-CGTCGTGACTGGGAAAACCCT-3′ and 5′-GGCGGATTGACCGTAATGGGA-3′. Alternatively, the genotype was determined via addition of [α-32P]dCTP to the PCR mixture with the β-geo primers (se above) and H19 primers 5′-ACATGACATGGTCCGGTGTGAT-3′ and 5′-TCCCCGGATTCAAAGGCCCA-3′. Heterozygous Imp1+/− mice were crossed to obtain homozygous Imp1−/− mice. The strain was maintained on a 50% C57BL/6-50% 129 SvEvBrd background. The mice were bred on a 10-h light, 14-h dark cycle and had free access to drinking water and standard chow containing 0.9% calcium and 0.7% phosphorus (Altromin no. 1324; C. Petersen a/s, Ringsted, Denmark).
Measurement of mouse growth.
Wild-type, Imp1+/−, and Imp1−/− male and female mice were weighed to monitor growth. For developmental analysis of weights, embryos and placenta were dissected out of the yolk sac and weighed separately. For staging embryos, noon of the day that a vaginal plug was evident was designated E0.5.
Survival.
Wild-type, Imp1+/−, and Imp1−/− mice were monitored for 60 days for survival. Data were plotted into Kaplan-Meier survival curves with SPSS version 11.0.
Maternal behavior.
First-litter wild-type (n = 6), Imp1+/− (n = 3), and Imp1−/− (n = 4) mice (between 3 and 5 months old) were examined for nesting, nursing, and pup retrieval behaviors. The nesting behavior was scored 0 for no nesting, 1 for incomplete nesting (no enclosing walls), and 2 for complete nesting (enclosing walls). Nursing behavior was defined as crouching and providing milk to the pups and assigned a score of 0 for no nursing and 1 for nursing. Finally, pup retrieval behavior was determined as the total time required for the mother to retrieve all pups when they had been placed at the opposite end of the cage from the nesting area divided by the total number of pups. The mice were observed for 5 consecutive days after the pups were born. Data were calculated as means ± standard error of the mean.
β-Galactosidase expression pattern analysis.
Embryos were washed in 0.1 M phosphate buffer (pH 7.3) and fixed for approximately 45 min in 0.2% glutaraldehyde-5 mM EGTA (pH 7.3)-2 mM MgCl2-0.1 M phosphate buffer (pH 7.3). The embryos were then washed three times in 4 mM MgCl2-0.02% Nonidet-P40-0.1 M phosphate buffer (pH 7.3) and stained in the dark in 4 mM K3Fe(CN)6-4 mM K4Fe(CN)6-4 mM MgCl2-0.02% NP-40-0.1 M phosphate buffer (pH 7.3)-1 mg of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) per ml. Finally the embryos were washed, postfixed in 4% paraformaldehyde (PFA), and stored in 70% ethanol at 4°C. In all cases, Imp1+/− and Imp1−/− embryos were incubated with stage-matched wild-type embryos as controls for endogenous β-galactosidase activity. After whole-mount analysis, β-galactosidase-stained embryos were embedded in paraffin and cut in 4-μm sections.
Whole-mount RNA in situ hybridization.
Embryos were fixed overnight in 4% PFA at 4°C. After three washes in PBT (PBS plus 1% Tween 20), the embryos were rehydrated in a series of methanol-PBT, bleached in 6% hydrogen peroxide, treated with 10 μg of proteinase K (Roche) per ml for various times depending on the age of the embryo, and washed two times with 2 mg of glycine per ml in PBT for 10 min. The embryos were then postfixed in 4% PFA-0.2% glutaraldehyde for 20 min, washed three times in PBT for 10 min, and prehybridized in hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 5.0], 0.5% 3-[(cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 5 mM EDTA, 0.2% Tween 20, 1% sodium dodecyl sulfate [SDS], 100 μg of heparin per ml, and 50 μg of yeast tRNA per ml) at 65°C for 3 h. The embryos were then hybridized at 65°C overnight in hybridization buffer containing 1 μg of Imp1 digoxigenin-labeled antisense riboprobe per ml. After hybridization, the embryos were washed in hybridization buffer, in wash buffer I (50% formamide, 5× SSC [pH 5.0], and 1% SDS), and in RNase buffer (0.5 M NaCl, 10 mM Tris-HCl [pH 7.5], and 0.1% Tween 20) before they were treated with 20 μg of RNase per ml at 37°C and washed in wash buffer II (50% formamide, 2× SSC [pH 5.0], and 1% SDS). Finally, the embryos were blocked in blocking buffer (1% sheep serum, 1% blocking reagent [Roche], 1.4 M NaCl, 27 mM KCl, 0.25 M Tris-HCl [pH 7.5], and 1% Tween 20), and transcripts were detected by overnight incubation at 4°C with antidigoxigenin antibody (Roche) and nitro blue tetrazolium 5-bromo-4-chloro-3-indolyl phosphate (NBT-BCIP) (Roche) staining as recommended by the manufacturer′s instructions.
Organ analysis.
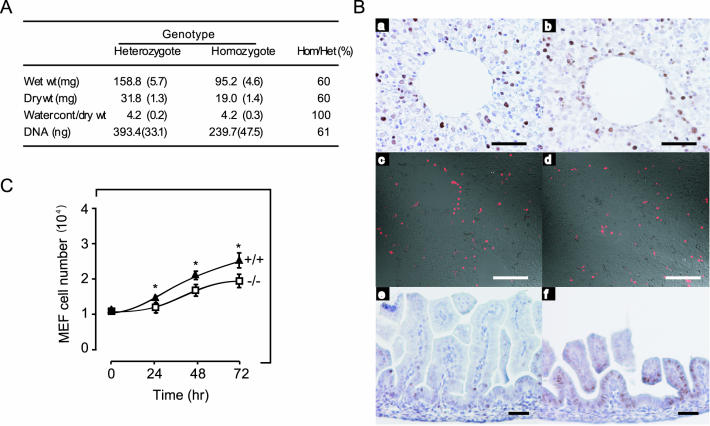
The weights of the organs from 2- to 3-week-old mice were determined and expressed relative to the mean weight of those of sex-matched, wild-type littermates. Moreover, the wet weight, dry weight, and DNA content of the kidneys from 3- to 4-month-old Imp1+/− (n = 3) and Imp1−/− (n = 3) mice were analyzed as recently described (57). Finally, tissue from 1-month-old wild-type (n = 2) and Imp1−/− (n = 2) mice was dissected, sectioned, and examined with a TE300 eclipse microscope (Nikon). The number of villi and the average height and width of villi, crypts, and mucosa were measured in three visual fields representing three different parts of the small intestine and colon, respectively. The results represent means and standard error of the mean of the individual measurements.
Histological staining.
Fresh tissue samples were placed in 4% PFA overnight and embedded in paraffin following standard procedures. The tissue was sectioned at 4 μm, deparaffinized by immersion in xylene for 10 and 5 min, respectively, and rehydrated in ethanol. After rehydration, sections were immersed in Mayer's hematoxylin solution (Sigma) for 30 s, followed by a 5-min rinse in running water. Then 0.5 ml of acetic acid was added to 100 ml of 0.5% eosin Y aqueous solution (Sigma), and sections were immersed for 5 min, followed by a 5-min wash in running water before dehydration in quick steps, and coverslipped with Pertex (Histolab). Slides were examined with a TE300 eclipse microscope (Nikon). For collagen staining, tissue sections were processed as described above. After rehydration, they were immersed in saturated picric acid solution containing 0.6 g of acid fuchsin/500 ml for 5 min. Sections were then washed, dehydrated, coverslipped with Pertex, and examined as described above.
Immunohistochemistry.
Tissue sections were processed as described above. After rehydration, antigens were retrieved by boiling in 10 mM citric acid (pH 6.0). Sections were preabsorbed with TBS (0.05 M Tris-HCl [pH 7.6], 0.15 M NaCl, 0.01% Triton X-100) containing 10% pig serum (Dako Cytomation) for 20 min before they were incubated for 1 h with IMP1 antibody diluted 1:2,000 in TBS containing 10% pig serum. For control sections the specific antibody was replaced with rabbit serum. Moreover, IMP1-specific antibodies could be preadsorbed with the peptide that was used for the immunizations. Following three rinses at 5-min intervals in TBS, the sections were incubated with EnVision horseradish peroxidase-conjugated secondary antibody (Dako Cytomation), and rinsed again, after which EnVision liquid diamidobensidine (DAB) solution (Dako Cytomation) was applied for 10 min. Sections were dehydrated in ethanol, coverslipped with Pertex, and viewed through a TE300 eclipse microscope (Nikon).
For collagen type XV α1 staining, primary antibody (Santa Cruz Biotechnology) was diluted 1:40 and applied as described above. Sections were incubated with fluorescein isothiocyanate-conjugated secondary antibody (Jackson Immuno Research) for 30 min before being coverslipped in nonfading glycerol medium (Dabco). Repeated rinses were performed between each step. Slides were examined with a Zeiss LSM 510 confocal laser scanning microscope.
Skeletal preparations.
Embryos or newborn mice were deskinned, eviscerated, and fixed in 100% ethanol for at least 2 days. Samples were then kept in acetone for 2 days, washed with water, and kept in staining solution (1 vol of 0.3% alcian blue in 70% ethanol, 1 vol of 0.1% alizarin red S in 95% ethanol, 1 vol of absolute acetic acid, and 17 vol of 100% ethanol) for 5 days at room temperature. The samples were washed with water, kept in clearing solution (1 vol of 1% KOH and 1 vol of 20% glycerol) at 37°C for 15 h, and finally stored in glycerol.
Isolation of MEFs, cell cultures, and cell proliferation assay.
E13.5 embryos were minced and incubated in 0.25% trypsin (Invitrogen) and 5 U of DNase I (Roche) at 37°C for 30 min. Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml was added to the cell suspension, and the cells were centrifuged at 1,000 rpm for 10 min. The pellet was resuspended in medium, and the cells were passaged by standard procedures. Cell proliferation was measured by plating mouse embryo fibroblasts (MEFs) from each genotype at a density of 2.0 × 104 cells per 35-mm petri dish. The cells were maintained and counted every day after plating. The cells were not used beyond passage 5. Counting was repeated with at least three different litters from crossed Imp1+/− mice. Statistical analysis was performed with a paired two-tailed t test.
PCNA and TUNEL assay.
Paraffin-embedded sections from wild-type and Imp1−/− E17.5 embryos were analyzed for apoptotic cells by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining with an In Situ Cell Death Detection kit (Roche) according to the manufacturer's instructions. Proliferating cell nuclear antigen (PCNA) was detected on paraffin-embedded sections with a PCNA staining kit (Zymed) as recommended by the supplier. Red-labeled DNA strand breaks and dark-brown-stained nuclei were examined in a Zeiss LSM 510 confocal laser scanning microscope. Positive cells were counted and expressed as positive cells per square arbitrary unit. Data were obtained from at least three sections from two different animals.
RNA analysis.
Total RNA was isolated from various tissues with Trizol reagent (Invitrogen). For reverse transcription-PCR (RT-PCR), cDNA was synthesized with the avian myeloblastosis virus reverse transcriptase (Promega) as described by the supplier. The cDNAs were amplified with primers 5′-TCTAACCGGGAGCAGACCAG-3′ and 5′-CTGCGTAGGCACCAGGAGCC-3′, resulting in a 237-bp Imp1 fragment, and primers 5′-ACTGGGACGACATGGAGAAG-3′ and 5′-GGGGTGTTGAAGGTCTCAAA-3′, resulting in a 157-bp β-actin fragment as a control. The samples were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
Northern blot analysis was performed as previously described (39). The RNA was denatured in glyoxal-dimethyl sulfoxide, separated in 1% agarose gels, transferred to Hybond-N membranes (Amersham Pharmacia Biotech), and hybridized with [α-32P]dCTP-labeled Igf2 (coding region), H19 (bp 103 to 979), β-actin (bp 1385 to 1481), c-myc (bp 565 to 1337) and Gapdh (coding region) cDNA fragments. The intensity of bands was quantified by Phosphorimager counting (Fuji).
To confirm the array data, quantitative RT-PCR was performed with the LightCycler-FastStart DNA Master SYBR Green I kit (Roche) according to the instructions of the manufacturer. Primer sequences for the examined genes were as follows: Col1α1, 5′-GTAACGATGGTGCTGTTGGTGCTGC-3′ and 5′-GTTGTCCATCAGCACCAGGGTTTCC-3′; Col1α2, 5′-GTTTCCAAGGACCTGCTGGTGAACC-3′ and 5′-TTGAAGCCAGGAAGTCCAGGAGTCC-3′; Col6α2, 5′-AAGAGCAGGAAGCTGTCTCACC-3′ and 5′-TGAGAACTCGTTCTGCAGCTGG-3′; Col6α3, 5′-CAGCTGACACGCTGAAGTTGTACC-3′ and 5′-CAGGTTAGCCACACGTTCAAGACC-3′ ; Col15α1, 5′-GTGGGCAATGCTGGAGCTATGG-3′ and 5′-GGGCTTGTGTGTAGGTGACAGC-3′; Hsp47, 5′-CAGGCAATGATCTCCCTCTGAGG-3′ and 5′-GTGTGGTCGCCTTGGAACTTAGC-3′; and Tff2, 5′-CTTGGATGCTGCTTTGACTCTAGC-3′ and 5′-CAGACTGTGGGAAGAAACACCAGG-3′. The amplification program consisted of one cycle of 95°C with a 600-s hold (hot start), followed by 35 cycles of 95°C with a 0-s hold, specific annealing temperature with an 8-s hold, 72°C with an 11-s hold, and finally an acquisition temperature of 84°C with a 0-s hold. Amplification was monitored by melting-curve analysis with one cycle of 95°C with a 0-s hold, 70°C with a 0-s hold, and 99°C with s 0-s hold at the acquisition mode. Each gene was examined in three independent reactions repeated once. The relative amount of each mRNA was normalized to the level of TATA-box binding protein (Tbp) in each sample.
Polysome isolation.
For polysome isolation, frozen E12.5 embryos were crushed and lysed in 500 μl of 20 mM Tris-HCl (pH 8.5)-1.5 mM MgCl2-140 mM KCl-0.5 mM dithiothreitol-0.5% NP-40-1,000 U of RNasin (Promega) per ml-0.1 mM cycloheximide. The lysates were centrifuged at 10,000 × g for 10 min, and the supernatant was applied to a linear 20 to 47% sucrose gradient in 20 mM Tris-HCl (pH 8.0)-140 mM KCl-5 mM MgCl2. Centrifugation was carried out at 40,000 rpm for 2.25 h in a Beckman SW41 rotor. Fractions of 1 ml were collected, followed by isolation of RNA and Northern blot analysis.
Western blot analysis.
Western blot analysis was performed as described before (39). Briefly, protein extracts from mouse embryos were separated in SDS-10% polyacrylamide gels and transferred to Hybond-P membranes (Amersham Pharmacia Biotech). After blocking, membranes were incubated overnight with anti-β-actin (Sigma), anti-c-Myc (Santa Cruz Biotechnology), or anti-TBP (Santa Cruz Biotechnology) antibodies in blocking solution at 4°C and with horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin G (Transduction Laboratories) for 1 h at room temperature. Immunoreactive proteins were detected with Supersignal chemiluminescence reagents (Pierce) according to the manufacturer's instructions.
DNA microarray analysis.
Total RNA was isolated from E12.5 embryos or postnatal organs from 12-day-old mice with Trizol reagent. Eight micrograms of purified RNA was used to synthesize double-stranded cDNA with the Superscript Choice system (Invitrogen) with an oligo(dT) primer containing a T7 RNA polymerase promoter (GenSet). The cDNA was used as the template for an in vitro transcription reaction to synthesize biotin-labeled antisense cRNA (BioArray high yield RNA transcript labeling kit; Enzo Diagnostics). After fragmentation at 94°C for 35 min in fragmentation buffer (40 mM Tris, 30 mM magnesium acetate, 10 mM potassium acetate), the labeled cRNA was hybridized for 16 h to Affymetrix U74av2 arrays (Affymetrix Inc.). The arrays were washed and stained with phycoerythrin-streptavidin (SAPE) with the Affymetrix Fluidics Station 400, and the arrays were scanned in the Affymetrix GeneArray 2500 scanner, as described in the Affymetrix GeneChip protocol, to generate fluorescent images. Low-level absolute analysis was done with MicroArray Suite version 5 software from Affymetrix. A detection call and an intensity value (expression level) of each gene represented on the array were calculated based on the hybridization signal from 11 perfect-match and mismatch probe pairs. Three independent experiments with RNAs from different age- and sex-matched embryos and postnatal organs were performed, and expression index computation and outlier detection were performed with the dChip algorithm, as described before (27). Briefly, the individual experiments were normalized before a hierarchical cluster of up- and downregulated transcripts was based on a perfect-match-mismatch model, and a P value of 0.05 (two-tailed, two-sample unequal variance t test). Only genes which were up- or downregulated more than twofold were included in the final list.
RESULTS
Disruption of the Imp1 gene.
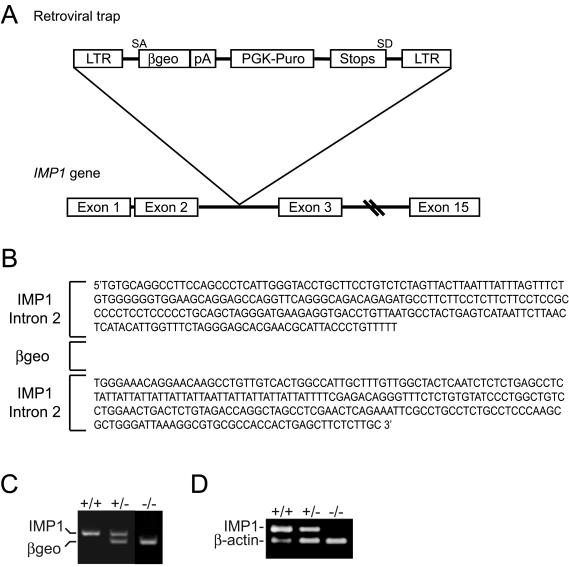
Imp1+/− mice were generated from ES cells containing a retroviral gene trap in the Imp1 locus as described previously (58). Sequence analysis showed that the gene trap was inserted in the ≈25-kb intron 2 of the Imp1 gene (Fig. 1A and B). The insertion led to the expression of an IMP1-β-galactosidase-neomycin (β-geo) fusion molecule containing the first RRM module encoded by exons 1 and 2 but lacking the second RRM motif and the four KH domains, which are essential for biological activity (37). The mice were genotyped by PCR with primers located at each side of the gene trap and in the β-geo cassette (Fig. 1C). To exclude that additional copies of the gene trap had erroneously been inserted into the genome, we initially also genotyped the animals with a competitive PCR amplifying the β-geo cassette and the H19 gene, located on chromosome 7. There were no discrepancies between the two methods of genotyping for 250 mice from heterozygous pairs, and the relative intensities of incorporated [α-32P]dCTP in the β-geo and H19 products demonstrated that only one copy of the insert had been introduced (data not shown).
FIG. 1.
Generation of IMP1-deficient mice. (A) Schematic representation of the insertion of the retroviral gene trap vector into intron 2 of the Imp1 gene. Exons are represented as boxes. LTR, long terminal repeat; β-geo, β-galactosidase-neomycin resistance cassette; PGK, phosphoglycerate kinase 1; Puro, puromycin; SA and SD, splice acceptor and splice donor sites, respectively. (B) Imp1 intron 2 sequences flanking the retroviral insert. Splicing of the gene trap produces a transcript containing an IMP1-β-geo fusion molecule that includes the first of the two RRM modules encoded by exons 1 and 2 but lacks the second RRM motif and the remaining four KH domains, which are encoded by exons 3 to 15. (C) Genotyping of wild-type, Imp1+/−, and Imp1−/− mice by multiplex RT-PCR. The reaction resulted in a 396-bp wild-type fragment and the 298-bp β-geo fragment with Imp1- and β-geo-specific primers and genomic DNA from mouse tails. The products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. (D) Expression of the Imp1 transcript in wild-type, Imp1+/−, and Imp1−/− mice. RNA from E12.5 embryos were examined for Imp1 expression by multiplex RT-PCR with specific primers for Imp1 and β-actin, resulting in 237-bp and 157-bp PCR fragments, respectively. The products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
The absence of Imp1 mRNA in Imp1−/− mice was confirmed with a multiplex RT-PCR, with β-actin as an internal control, on total mRNA from E12.5 embryos (Fig. 1D), in which Imp1 mRNA expression peaks (39, 48). Whereas no Imp1 mRNA was detected in the Imp1−/− mice, the transcript was present in both heterozygous and wild-type mice. Moreover, whole-mount RNA in situ hybridization with an antisense Imp1 riboprobe showed no expression in Imp1−/− embryos even after extended exposures (Fig. 2A, picture j). We conclude that Imp1 mRNA expression is absent in the Imp1−/− gene trap mice.
FIG.2.
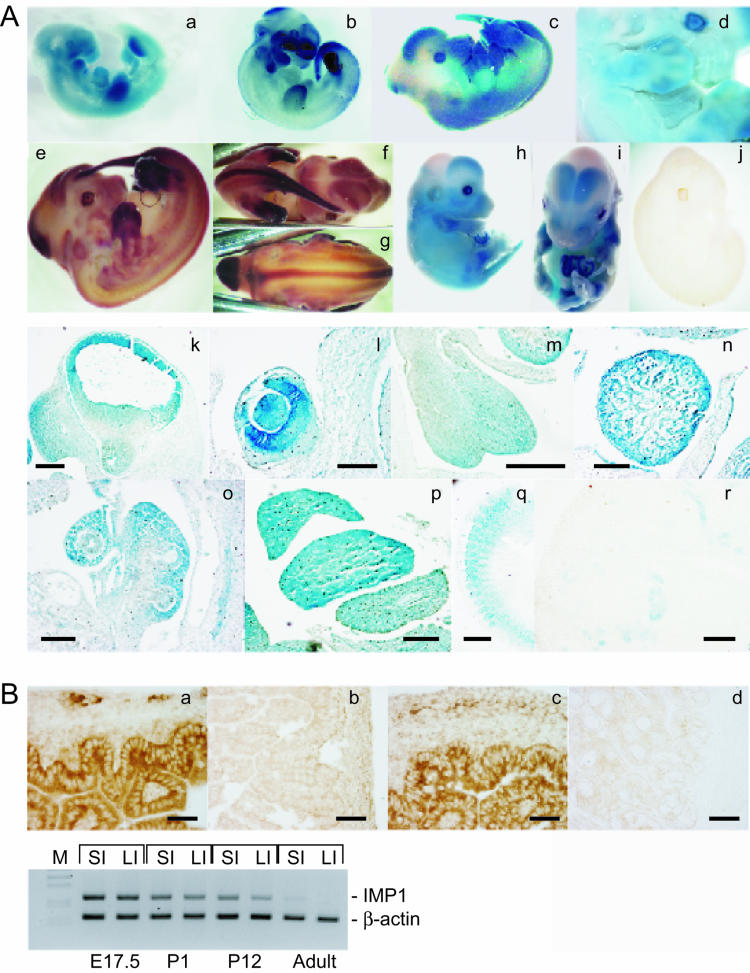
Expression of Imp1 during development and in adult tissues. (A) β-Galactosidase activity (a, c, d, h, i, and k to q) and Imp1 mRNA expression revealed by whole-mount in situ hybridization with an antisense Imp1 riboprobe (b, e, f, g, and j) was examined in E10.5 (a and b), E12.5 (c to g), E14.5 (h and i)m and E17.5 (r) embryos. No Imp1 mRNA expression was observed in Imp1−/− E12.5 embryos (j). E12.5 embryos were sectioned, and β-galactosidase activity was detected in snout and forebrain (k), eye (l), tongue (m), heart (n), lung (o), liver (p), and somites (q). Moreover IMP1 protein expression was detected in kidneys from E17.5 embryos (r). Bars: k, 350 μm; l, 125 μm; m, 250 μm; n, 250 μm; o, 140 μm; p, 250 μm; q, 250 μm; r, 120 μm. (B) Immunohistochemistry was performed to detect IMP1 in the small intestine (a) and large intestine (c) from E17.5 embryos. No staining was observed in adult large intestine (d). As a control IMP1-specific antibodies were preadsorbed with the peptide that was used for the immunizations (b). Bars: a to d, 60 μm. In the lower panel, the expression of Imp1 mRNA was examined in small and large intestines from E17.5, P1, P12, and adult mice by multiplex RT-PCR. Specific primers for Imp1 and β-actin, resulting in 237-bp and 157-bp PCR fragments, respectively, were used. The products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. SI, small intestine; LI, large intestine.
Imp1 expression during development and adult life.
The gene trap construct included a β-galactosidase cassette under control of the native Imp1 promoter, and we examined the spatial and temporal expression patterns of Imp1 in mouse fetuses by whole-mount β-galactosidase staining as well as whole-mount Imp1 RNA in situ hybridization. At E10.5, β-galactosidase and Imp1 mRNA expression was mainly observed in the fore- and hindbrain, the snout, the branchial arches, the developing limb buds, and the tail (Fig. 2A, a and b). At E12.5, β-galactosidase and Imp1 mRNA expression increased in the expanding fore- and hindbrain (Fig. 2A, c to g and k) as well as in the neural tract. Marked expression was also observed in the snout, the interdigital mesenchyme of the limb buds, the tail, the branchial arches and somites, and the developing eye, tongue, heart, lung, and liver (Fig. 2A, l to q). At E14.5, β-galactosidase expression was found in the same locations (Fig. 2A, h and i), but in limb buds β-galactosidase staining became restricted to the future tendons. Subsequently, at E17.5, Imp1 expression generally decreased but remained high in the intestine (Fig. 2B, a and c), in the developing tubules of the kidney (Fig. 2A, r), and in the liver (data not shown).
Multiplex RT-PCR of total mRNA from E17.5, postnatal day 1 (P1), P12, and adult small and large intestine showed that Imp1 mRNA was expressed until P12 and that low levels remained even in the intestines of adult mice (1 year old) (Fig. 2B, lower panel). Similar results were obtained for the kidney (data not shown). We infer that the Imp1 gene trap recapitulates the spatiotemporal expression of Imp1. Moreover, we demonstrate that Imp1 expression overlaps that of the previously described Imp3/KOC gene (31, 34).
Breeding data and postnatal lethality.
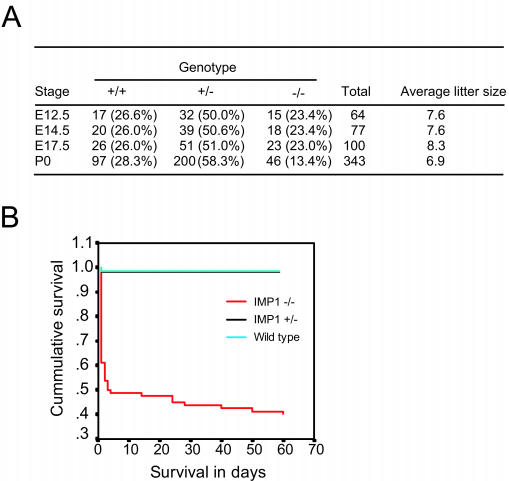
Breeding data were obtained from crosses between both heterozygous and homozygous mice, since both Imp1+/− and Imp1−/− males and females were fertile. In all cases, the sex ratio was approximately 1:1. The genotypes of the embryos from heterozygous pairs were present at approximately the expected ratio of 25:50:25 (Imp1+/+, Imp1+/−, and Imp1−/−), whereas the fraction of Imp1−/− mice was reduced to 13.4% (P < 0.05) after birth (P0 to P1), indicating that embryos were lost in the perinatal period (Fig. 3A). The survival curves for homozygous mice during the perinatal, postnatal, and adult periods up to 8 weeks after birth showed that only 50% of the pups were alive after 3 days, and after 8 weeks an additional 10% of the homozygous mice died (Fig. 3B). So in total only 40% of the Imp1−/− mice survive into adulthood. There was no difference in perinatal mortality among Imp1−/− female or male pups, but death was 3 times higher among pups from homozygous pairs. We therefore compared the maternal behavior of wild-type, Imp1+/−, and Imp1−/− mice. The nesting, nursing, and pup retrieval behaviors were scored as described in Materials and Methods and gave mean values ± standard error of the mean for wild-type, Imp1+/−, and Imp1−/− mice, respectively, as follows: nesting, 1.8 ± 0.1, 1.8 ± 0.2, and 1.9 ± 0.1; nursing, 1, 1, and 1 (scores were 0 or 1); and pup retrieval, 25 ± 1, 22 ± 6, and 27 ± 4. There were no significant differences in maternal behavior among the groups.
FIG. 3.
Breeding data and survival of IMP1-deficient mice. (A) The average genotypes and litter sizes of offspring from Imp1+/− matings at E12.5 to P0. (B) Kaplan-Meier survival curve for wild-type, Imp1+/−, and Imp1−/− mice. Statistically significant decreased survival (P < 0.05) was seen in Imp1−/− mice compared to wild-type and Imp1+/− mice.
Reduced growth of Imp1−/− mice.
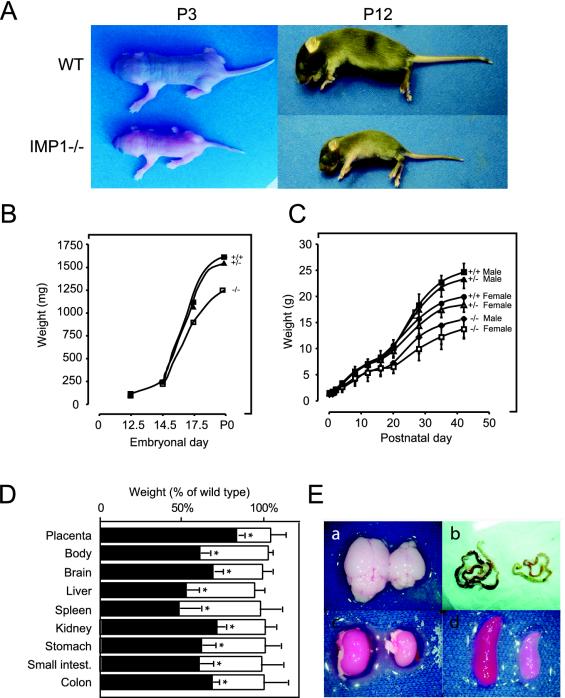
The most striking feature of adult Imp1−/− mice was their smaller body size (Fig. 4A). The mice also exhibited a slightly disproportionate body composition, with a relatively small trunk and a short snout, and in addition, some of the mice had tiny kinks on their tails. Since Imp1 expression peaks at E12.5 (39, 48), we monitored the growth kinetics from this day for up to 6 weeks after birth. At E12.5 and E14.5 there was no significant difference in body weight between wild-type, heterozygous, and homozygous mice. At E17.5, Imp1−/− embryos were 14% (P < 0.05) smaller than wild-type embryos (Fig. 4B), and the placenta was 18% (P < 0.05) smaller (Fig. 4D). The difference increased until birth, when Imp1−/− pups were 21% (P < 0.05) lighter than their littermates. Growth retardation continued progressively, and 1 week after birth, Imp1−/− mice were on average 45% (P < 0.05) smaller than their heterozygous and wild-type littermates (Fig. 4C). This difference remained stable for the following month, and adult Imp1−/− mice were about 40% (P < 0.05) smaller than wild-types and heterozygous mice and never reached the size of their normal littermates. The postnatal growth of females and males was proportionally reduced to the same extent. The weights of brain, liver, spleen, kidney, stomach, and intestine were also proportionally reduced in size to about 60% (P < 0.05) in the Imp1−/− mice compared to wild-type and heterozygous mice (Fig. 4D and E).
FIG. 4.
Pre- and postnatal growth retardation of IMP1-deficient mice. (A) Photographs of wild-type and Imp1−/− littermates at 3 and 12 days of age. (B) Embryonal growth. Mean body weight ± standard error of the mean of wild-type, Imp1+/−, and Imp1−/− embryos from E12.5, E14.5, E17.5, and P0 mice. Growth was significantly (P < 0.05) reduced in Imp1−/− E17.5 embryos and P0 mice. (C) Postnatal growth. Wild-type, Imp1+/−, and Imp1−/− male and female mice were weighed over a period of 6 weeks. Results are plotted as the mean body weight ± standard error of the mean. Growth was significantly (P < 0.05) reduced in Imp1−/− postnatal and adult mice. (D) Organs were proportionally reduced in size. Mean organ weights ± standard error of the mean for Imp1+/− (open columns) and Imp1−/− (solid columns) mice at 2 to 3 weeks of age (except placenta, which was from E17.5 embryos) are plotted as a percentage of the weights for the organs and placentas from wild-type mice. The size of all Imp1−/− organs was significantly (P < 0.05) reduced. (E) Macroscopic appearance of selected organs from IMP1-deficient mice. Hypoplasia was observed in various organs, including brain (a), small intestine (b), kidney (c), and spleen (d).
Macro- and microscopic appearance of organs.
To identify developmental defects and/or pathological features, we examined the macro- and microscopic appearance of a series of mouse embryos and organs from Imp1−/− mice. With the exception of the visible difference in size from day E17.5, Imp1−/− embryos were macroscopically indistinguishable from their littermates. In the first days after birth, however, Imp1−/− pups failed to thrive. They were dehydrated and inactive and had difficulties in competing with their littermates for breastfeeding. There was a substantial difference between the mice, and the disabilities ranged from complete exhaustion and early death to an almost normal appearance and activity. The only pathological feature that we observed in a few of the surviving animals was signs of neurological damage, including aggressive behavior, restlessness, and circular movements.
To determine the cause of the early mortality, we dissected a series of 1- to 12-day-old pups that had died within 1 h of inspection or whose death was imminent. In the animals with the most severe phenotype, we observed necrotic patches and adherences in the intestine. Moreover, anemia was a frequent finding, and hepatic steatosis was observed in some adult mice. The remaining organs looked normal, indicating that the early death was associated with intestinal dysfunction.
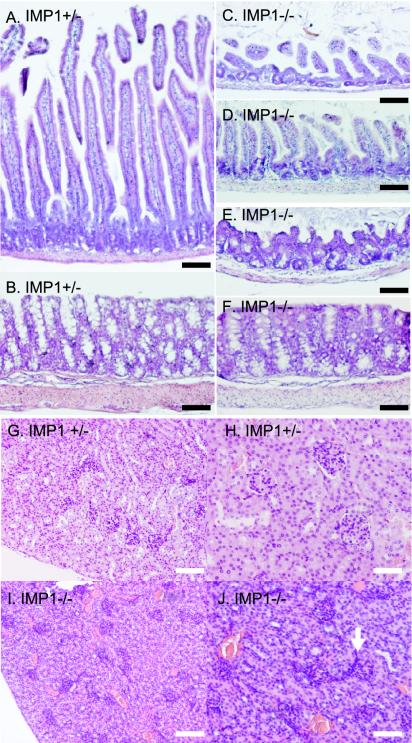
The examination of hematoxylin- and eosin-stained sections from organs revealed histological changes in gut and kidney and to a minor extent in liver. In the developing intestine of E17.5 embryos, reduced size and irregular shape of the villi was noted (see Fig. 7B, pictures e and f). In postnatal mice, the changes were even more prominent, and in an attempt to qualify the morphological observations, we compared the sizes of the small intestines and colons of two 1-month-old IMP1-deficient mice with those of these organs from two wild-type mice of the same age (Table 1). In the small intestine, the thickness of the muscular wall and the height and width of the villi were reduced. Moreover, the abnormal mucosal lining was characterized by irregular misshapen villi and loss of mesenchyme, but strikingly the number of villi was unchanged (compare Fig. 5A with C and D). In the colon, the thickness of the mucosal layer was reduced, and the crypts were deformed and twisted. Moreover, the muscular wall was thinned to about one third of the normal size, and the goblet cells were redistributed to the bottom of the crypts (compare Fig. 5B with E and F).
FIG. 7.
Reduced cellular proliferation in organs and MEFs from Imp1−/− mice. (A) The wet weight, dry weight, and DNA content of the kidney from 3- to 4-month-old Imp1+/− (n = 3) and Imp1−/− (n = 3) mice were analyzed as described in Materials and Methods. (B) Liver (a to d) and small intestine (e and f) sections from wild-type and Imp1−/− E17.5 embryos, respectively, were examined for proliferation by PCNA staining (a, b, e, and f) or for apoptosis by TUNEL staining (c and d). (C) Growth of MEFs from wild-type (▴) and Imp1−/− (□) E13.5 embryos. Cells were plated in 30-mm plates and counted for 4 consecutive days. Each value is an average of three independent experiments with duplicate plates. Growth of Imp1−/− MEF cells was significantly (P < 0.05) reduced after 24, 48, and 72 h. Scale bars: 200 μm (A and B), 300 μm (Card D), and 60 μm (E and F).
TABLE 1.
Reduction of villus, crypt and mucosal size in IMP1-deficient micea
| Organ and parameter | Value
|
P | % Reduction | |
|---|---|---|---|---|
| Imp1+/− mice | Imp1−/− mice | |||
| Small instestine | ||||
| No. of villi/mm | 11 (1.3) | 13 (2.0) | 0.07 | |
| Avg ht of villi (mm) | 0.50 (0.13) | 0.19 (0.11) | <0.01 | 62 |
| Avg width of villi (mm) | 0.062 (0.016) | 0.032 (0.001) | <0.01 | 49 |
| Mucosal thickness (mm) | 0.043 (0.001) | 0.020 (0.006) | <0.01 | 54 |
| Colon | ||||
| Avg height of crypts (mm) | 0.25 (0.01) | 0.12 (0.059) | <0.01 | 52 |
| Mucosal thickness (mm) | 0.18 (0.006) | 0.06 (0.0013) | <0.01 | 67 |
Tissues from two 1-month-old wild-type and Imp1−/− mice were dissected, fixed, and hematoxylin and eosin stained. The number of villi and the average height and width of the villi, crypts and mucosa were measured in three visual fields representing three different parts of the small intestine and colon, respectively. The results represent means (standard error of the mean).
FIG. 5.
Histological appearance of gut and kidney. Hematoxylin- and eosin-stained small intestine (compare A with C and D), colon (compare B with E and F), and kidney (compare G and H with I and J) sections from 1-month-old Imp1+/− (A, B, G, and H) and Imp1−/− (C to F, I, and J) littermate mice. Bars: A to F, 100 μm; G and I, 120 μm; H and J, 60 μm.
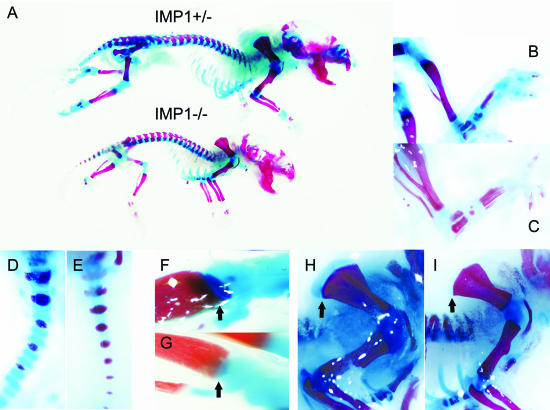
In the kidney, the abnormalities were less explicit, but the architecture of 3- to 15-day-old mouse kidneys exhibited features of the immature kidney. In homozygous mice, the glomeruli were small and hypoplastic, and the collecting tubules appeared irregular, with reduced lumen (compare Fig. 5G and H with I and J). In the liver, the changes were subtle and mainly affected the lobular structure. In the spleen, pancreas, skeletal muscle, lung, testis, and ovaries, we did not observe any histological abnormalities. Staining of the skeleton with alizarin red S, which stains mineralized bone, and alcian blue, which stains cartilage, demonstrated a distinct loss of cartilage in the lower extremities, the tail, the growth plate in the tibia, and on top of the blade of the scapula as well as in the mandible and the nasal bone (Fig. 6A to I). Otherwise, the sizes of the long bones were reduced in proportion to the whole mouse (data not shown).
FIG. 6.
Decreased cartilage staining in Imp1−/− mice. (A) Skeletons from newborn Imp1+/− and Imp1−/− mice were examined with alcian blue and alizarin red S. Mineralized bones and cartilages are stained red and blue, respectively. The hindlimbs (B and C), the tail (D and E), the growth plate in the tibia (arrows) (F and G), and the blade of scapula (arrow) (H and I) are shown at higher magnification. (B, D, F, and H) Imp1+/−; (C, E, G, and I) Imp1−/−.
Reduced cell proliferation in Imp1−/− mice.
To establish whether the reduced organ size resulted from hypoplasia and/or smaller cell size, we determined the amount of DNA and the wet and dry weights of kidneys from 3- to 4-month-old Imp1+/− (n = 3) and Imp1−/− (n = 3) mice. The amount of total DNA, the wet weight, and the dry weight were proportionally reduced (Fig. 7A), indicating that the smaller organs resulted mainly from hypoplasia. This was supported by direct measurement of cell proliferation and apoptosis in the liver and kidney from wild-type and Imp1−/− E17.5 embryos. Tissues were stained with PCNA-specific antibodies to identify cells in the S, G2, and M phases, and a TUNEL assay was used to detect apoptosis. In the liver and kidney, the relative percentage of PCNA-positive cells was reduced from 33% ± 4.9% to 23% ± 4.8% (P < 0.05) (Fig. 7B, a and b) and from 27% ± 5.7% to 16% ± 7.4% (P < 0.05), respectively. It was not possible to make a direct comparison of the frequency of PCNA-positive cells in the intestine due to the altered morphology (se above).
In the intestine of Imp1−/− mice, PCNA-positive cells were abundant all over the imperfectly developed villi, whereas in the wild-type intestine PCNA-positive cells were mainly located at the root of the villi, as normally seen at this stage of development (Fig. 7B, e and f). The number of apoptotic cells was not significantly changed in the liver (Fig. 7B, c and d) or in any of the other organs. Finally, we prepared cultures of MEFs from wild-type, Imp1+/−, and Imp1−/− E13.5 embryos and determined the proliferation rate of the cells by counting. Under the standard culture conditions used, the number of MEFs from Imp1−/− mice was 16% lower after 24 h (P < 0.05) and 24% lower after 72 h (P < 0.05) compared to wild-type MEFs (Fig. 7C). We infer that the growth reduction observed in homozygous mice results from hypoplasia.
Target genes and global expression profiling of embryos and postnatal organs.
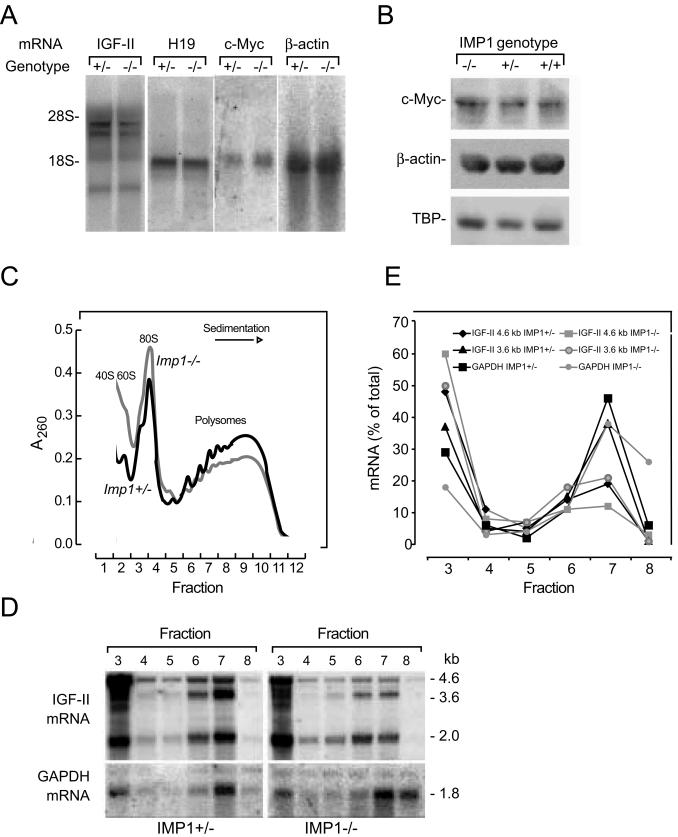
The known IMP1 targets include Igf2 leader 3, c-myc and β-actin mRNAs and H19 RNA, so we characterized the expression of these transcripts and proteins. Since Imp1 mRNA expression peaks at E12.5, the level of target mRNA and protein was examined at this point. Igf2 mRNA was detected with a coding region probe which binds to all Igf2 mRNAs. There was no significant change in the level of Igf2 leader 2, 3, or 4 mRNA (Fig. 8A) in Imp1−/− embryos compared to heterozygous embryos. H19 RNA levels were also unchanged, and the same was the case for β-actin and c-myc mRNA and protein levels (Fig. 8A and 8B) and for IMP2 and IMP3 protein levels (data not shown).
FIG. 8.
Expression of IMP1 target genes in E12.5 embryos. (A) Northern blot analysis of Igf2, H19, c-myc, and β-actin mRNAs in Imp1+/− and Imp1−/− E12.5 embryos. Total RNA (10 μg) was hybridized to cDNAs encoding Igf2, H19, c-myc, and β-actin. (B) Western blot analysis of c-Myc and β-actin in wild-type, Imp1+/−, and Imp1−/− whole E12.5 embryo lysates. Extracts were separated in SDS-10% polyacrylamide gels, transferred to Hybond-P membranes, and probed with antibodies specific for c-Myc and β-actin. As a loading control, the membranes were probed with an anti-TBP antibody. (C and D) Isolation of polysomes from E12.5 embryos. Cytoplasmic lysates from Imp1+/− and Imp1−/− embryos were separated in a 20 to 47% sucrose gradient. Panel C shows the A260 sedimentation profile of ribosomal subunits and polyribosomes, and panel D shows the Northern blot analysis of Igf2 mRNA in the fractions. The position of the major Igf2 leader 3 (4.6-kb) and leader 4 (3.6-kb) mRNA species derived from mouse promoters P2 and P3, respectively, are indicated. (E) Relative levels of Igf2 4.6-kb and 3.6-kb mRNAs in polysomes and monosomes, respectively. Data are represented as a percentage of total photon-stimulated luminescence for each transcript.
Because previous cell culture experiments indicated that IMP1 may be implicated in translational control of Igf2 (39), we examined the level of polysomal Igf2 mRNA in E12.5 embryos. Embryos were solubilized, and polysomes were isolated by sucrose gradient fractionation. Figure 8C shows the polysome profiles. A small redistribution of polysomes to monosomes was apparent in Imp1−/− embryos, but the relative amount of Gapdh mRNA in 20S-40S RNP fractions and polysomes was the same in Imp+/− and Imp1−/− embryos. Both the 3.6-kb and 4.6-kb Igf2 mRNAs were partially redistributed from polysomes to the top of the gradient in Imp1−/− embryos (Fig. 8D and E). Taken together, the results indicate that loss of IMP1 is associated with reduced translation of Igf2 mRNA. Since IMP1-deficient mice exhibited a postnatal growth deficiency and permanently remained smaller than their normal counterparts, we also measured the expression of Igf1 mRNA in the liver and of growth hormone mRNA in the brain by quantitative RT-PCR analysis. The analysis did not reveal any difference in the expression of these two factors (data not shown).
To obtain additional information about the cellular pathways involved in the phenotype, we performed global expression profiling of sex- and litter-matched E12.5 embryos and intestine, liver, and kidney from postnatal mice (Table 2). cRNA generated from total RNA was hybridized to Affymetrix U74Av2 arrays, which contain 12,480 different probe sets.
TABLE 2.
Differentially expressed transcripts in postnatal organs of Imp1−/− mice
| Organ | Family and genes | GenBank no. | Function(s)a | Change (fold) |
|---|---|---|---|---|
| Intestine | DNA and RNA binding | |||
| High-mobility-group nucleosomal binding domain 2 | X12944 | Modulator of chromatin structure | −2 | |
| Cell cycle control | ||||
| Cell division cycle 25 homolog A | U27323 | Cell cycle progression, mitosis | −3 | |
| Cytoskeleton and extracellular matrix | ||||
| Actin, alpha 1, skeletal muscle | M12347 | Microfilaments, motility | −3.5 | |
| Galectin-1 | X15986 | Extracell. matrix, myoblast diff. | −2.4 | |
| Lumican | AF013262 | Extracell. matrix comp. | −2.4 | |
| Procollagen type I α1 | U03419 | Extracell. matrix comp., cell adh. | −2.5 | |
| Procollagen type I α2 | X58251 | Extracell. matrix comp., cell adh. | −3 | |
| Procollagen type V α1 | AB009993 | Extracell. matrix comp., cell adh. | −2.3 | |
| Procollagen type VI α2 | Z18272 | Extracell. matrix comp., cell adh. | −2.2 | |
| Procollagen type VI α3 | AF064749 | Extracell. matrix comp., cell adh. | −2.9 | |
| Procollagen type XIV α1 | AJ131395 | Extracell. matrix comp., cell adh. | −2 | |
| Tenascin-C | X56304 | Extracell. matrix comp. | −2.3 | |
| Enzymes and metabolism | ||||
| Similar to angiotensin I converting enzyme 2 | AA690434 | Angiotensin-converting enzyme | 2.4 | |
| Carbonic anhydrase III | AJ006474 | Catalyze hydration of CO2 | −7 | |
| Cubilin | AJ010338 | Cobalamin uptake | 2 | |
| Apolipoprotein B-100 | AI787317 | Lipid transport | 2 | |
| Aminopeptidase A | M29961 | Angiotensin-converting enzyme | 2 | |
| Heat shock protein 47 | X60676 | Collagen biosynthesis | −2 | |
| Solute carrier family 5 | AA591002 | Glucose uptake | 2.7 | |
| Stearoyl-coenzyme A desaturase 1 | M21285 | Fatty acid synthesis | −4.3 | |
| Other | ||||
| Trefoil factor 2 | U78770 | Epithelial restitution | 2.7 | |
| Similar to regenerating islet-derived 1 | AV053434 | β-Cell regeration | 3.3 | |
| Similar to transmembrane 4 superfamily member 5 | AA718076 | Cell growth, adhesion, motility | 2.5 | |
| RIKEN cDNA gene | AV366654 | Unknown | −2.2 | |
| Liver | Hormones, receptors and signaling | |||
| Aquaporin-8 | AF018952 | Water transport | −2.6 | |
| Endothelial differentiation sphingolipid G-protein-coupled receptor 1 | AV347228 | Cell signaling, cell migration | −3.0 | |
| G protein-coupled receptor 49 | AF110818 | Cell signaling | −3.4 | |
| Inhibin beta-B | X69620 | Growth factor or hormone | −2.9 | |
| Acid-labile subunit | U66900 | Carrier protein | −3.4 | |
| Interleukin-11 receptor, alpha chain 2 | U69491 | Cell signaling | −2.2 | |
| Cytoskeleton and extracellular matrix | ||||
| Chemokine (C-X-C motif) ligand 12 | L12029 | Chemotaxis | −2.4 | |
| Claudin-2 | AF072128 | Cell-cell adhesion | −5.8 | |
| Midline-2 | Y18881 | Microtubule associated | −8.0 | |
| Procollagen type IIIα1 | AA655199 | Extracell. matrix comp., cell adh. | −2.6 | |
| Procollagen type XIV α1 | AJ131395 | Extracell. matrix comp., cell adh. | −2.5 | |
| Reelin | U24703 | Extracell. matrix comp., cell adh. | −2.2 | |
| Similar to rhomboid, veinlet-like 4 | AI847581 | Intramembrane protease, cell signaling | 2 | |
| Enzymes and metabolism | ||||
| Carbonic anhydrase III | AJ006474 | Catalyze hydration of CO2 | −4.0 | |
| Cytochrome P450IIJ5 | AI114881 | Electron transport, xenobiotic metab. | −2.1 | |
| Hydroxyacid oxidase 3 | AI648067 | Oxidative metabolism | −3.4 | |
| Solute carrier family 22a1 | U38652 | Organic cation transporter | −3.0 | |
| Other | ||||
| P311 | D45203 | Cellular growth | −3.3 | |
| Transforming growth factor beta 1-induced transcript 4 | X62940 | Transcription factor | −2.4 | |
| Expressed sequence tag | AA958903 | Unknown | −2.8 | |
| Kidney | Adenylyl cyclase-associated CAP protein homolog 1 | L12367 | Cell signaling, microfilaments, motility | −2.7 |
| ADP-ribosyltransferase 2b | X87612 | ATP biogenesis | 2.0 | |
| 25-Hydroxyvitamin D 1α-hydroxylase | AB006034 | Vitamin D synthesis | 2.2 | |
| Fc receptor, IgE, high-affinity I | J05020 | Cell signaling | −11.0 | |
| Insulin-like growth factor 2 mRNA-binding protein 1 | AF061569 | RNA localization | −2.4b | |
| Similar to outer dense fiber of sperm tails 2 | AV312779 | Centrosome matrix | 2.5 | |
| Similar to promyelocyte leukemia Zn finger protein | AI553024 | Transcription factor | 2.7 | |
| UDP-Gal:βGlcNAcβ1,4-galactosyltransferase | AA111141 | Glycosylation | 2.5 |
Extracell., extracellular; diff., differentiation; comp., component; adh, adherence.
Imp1 mRNA went from a present call to an absent call.
In E12.5 embryos, there were essentially no differences in gene expression. Only three probe sets corresponding to the same number of transcripts were changed more than twofold. As expected, the Imp1 transcript was completely absent (138-fold downregulation). Otherwise, there was only downregulation of a rearranged immunoglobulin G transcript (GenBank M30208) and upregulation of an expressed sequence tag (GenBank C77112) of unknown function.
In the intestine, a total of 26 probe sets corresponding to 24 different transcripts were changed more than twofold. Among the transcripts involved in gene expression or cell cycle regulation, only the high-mobility group nucleosomal binding domain 2 and cdc25 mRNAs were changed. Among enzymes and metabolic factors, the most striking changes involved the carbonic anhydrase III (CaIII) and stearoyl-coenzyme A desaturase 1 (Scd1) transcripts, which were downregulated seven- and fourfold, respectively. Moreover, changes were observed in a whole series of transcripts encoding proteins associated with the extracellular matrix. Galectin-1, lumican, and tenascin-C as well as several procollagen transcripts, including type I α1 and α2, type V α1, type VI α2 and α3, and type XIV α1, and heat shock protein 47 (Hsp47), which is involved in collagen biosynthesis (35), were downregulated. Finally, we note that the trefoil factor 2 (Tff2) transcript was upregulated.
In the liver, 20 different transcripts were changed more than twofold. As in the intestine, transcripts encoding extracellular matrix-associated factors, including procollagen type III α1, and type XIV α1, were downregulated. Moreover, transcripts encoding midline-2 and claudin-2, which are microtubule and tight-junction associated, respectively (5, 14), were decreased, and CaIII mRNA was also downregulated. Among hormones and receptors, the acid-labile subunit mRNA was decreased. In the kidney, there were only minor changes. Corresponding to the expression of IMP1 protein in the postnatal kidney, the level of the Imp1 transcript was reduced to background levels (absent call) in the Imp1−/− mice.
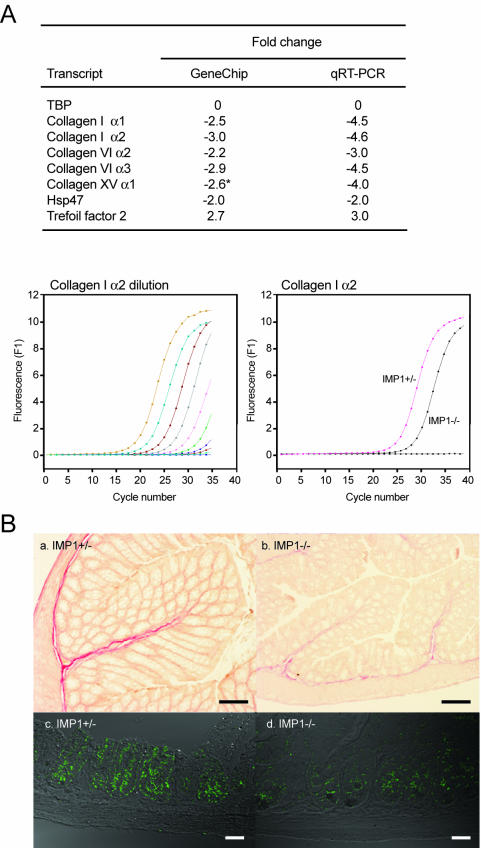
To verify the microarray findings, we determined the expression of collagen type I α1 and α2, collagen type VI α2 and α3, Hsp47, and Tff2 transcripts by quantitative RT-PCR. Collagen type XV α1, which was downregulated 2.6-fold but not included in Table 2 because the P value was 0.07, was also included in the RT-PCR analysis (Fig. 9A). In all cases, the results were roughly in agreement with the microarray data. The reduced mRNA levels of collagens in the gut were further corroborated by collagen protein staining of the colon (Fig. 9B, a and b). Moreover, immunofluorescence staining showed that collagen type XV α1 immunoreactivity in small intestine was also reduced (Fig. 9B, c and d).
FIG. 9.
Expression of collagens in the gut of Imp1−/− mice. (A) Quantitative RT-PCR was used to verify the changes in collagen, Hsp47, and trefoil factor 2 mRNA levels observed in the microarray analysis. The asterisk indicates that collagen type XV α1 exhibited a P value of only 0.07 in the microarray analysis. The lower panel shows the threefold dilution curve (left) and the relative expression of collagen type I α2 in Imp1+/− and Imp1−/− mice (right). (B) Colon sections from 1-month-old Imp1+/− and Imp1−/− mice were analyzed for collagen by staining with saturated picric acid solution containing acid fuchsin (a and b). Collagen fibers are shown in red. Immunohistochemistry was applied to detect collagen type XV α1 in small intestine sections from 1-month-old Imp1+/− and Imp1−/− mice (c and d). Bars: a and b, 100 μm; c and d, 35 μm.
Taken together, the results demonstrate that fetal (E12.5) loss of IMP1 does not change mRNA levels, whereas IMP1 deficiency in the postnatal gut is associated with decreased synthesis of extracellular matrix and microfilament network components.
DISCUSSION
To characterize the physiological role of IMP1, we generated IMP1-deficient mice exhibiting a gene trap in intron 2 of the Imp1 gene. The gene trap leads to deletion of the second RRM and the four KH modules, which have been demonstrated to be essential for RNA binding, cytoplasmic granule formation, and subcytoplasmic localization (37).
Imp1 promoter activity was detected as early as E10.5 and peaked at E12.5 in the fore- and hindbrain as well as in the limb buds. Moreover, Imp1 mRNA expression was detected in both mesenchymal and parenchymal cells of the internal organs, in particular in the heart, lung, and liver. At E17.5, Imp1 mRNA expression decreased in most places, but expression remained high in the epithelial lining of the intestine and in the kidney. In fact, Imp1 mRNA could be detected in significant amounts until postnatal day 12, and low levels were found even in the adult organs. As described previously, the fetal expression pattern overlaps the spatial distribution of Imp3/KOC mRNA (31, 34), and this may have implications for the phenotype of the IMP1-deficient mouse, since the IMPs exhibit a similar RNA-binding specificity and may bind RNA as heterodimers (J. Christiansen, unpublished data). Although variations in the genetic background of the mutant may play a role, redundancy of IMP2 and IMP3 may also contribute to the variation in the severity of the Imp1−/− phenotype that we observed. Moreover, it may also explain why IMP1-deficient mice do not exhibit more generalized defects corresponding to the widespread fetal expression of IMPs.
The most striking features of the Imp1−/− phenotype are the small size of the mice and the significant perinatal lethality. Growth reduction was detectable from E17.5, and at birth Imp1−/− pups were on average 20 to 25% smaller than their wild-type and heterozygous littermates. Until weaning, the relative size of Imp1−/− mice was further reduced, so adult animals were on average 40% smaller than their normal littermates. With exception of a tendency to a shorter snout and a relatively small trunk, Imp1−/− mice were proportionally reduced in size. Similar to several other dwarfing mutations. such as inactivation of Igf1 and Igf2 (7, 28), growth reduction in Imp1−/− mice resulted from hypoplasia rather than reduced cell size. Since Imp1 is mainly expressed during embryogenesis, the mechanisms underlying fetal growth retardation may be different from the postnatal growth reduction and involve reduced Igf2 translation as well as the smaller placenta (29). Additional targets encoding intracellular factors are also likely to play a role, since MEFs failed to exhibit normal growth in vitro. Moreover, we noticed a minor reduction of polysomal mRNAs in Imp1−/− embryos, indicating that synthesis of other proteins may be affected by the loss of IMP1. The postnatal growth retardation may result primarily from the impaired function of the small intestine (see below).
About half of the Imp1−/− pups died within the first 3 days of life. Survivors were in general viable and seemed to have normal life expectancy compared to wild-type and heterozygous littermates. There were no indications of late embryonal death, suggesting that death resulted from an inability to make the transition from intrauterine to extrauterine life. Although we cannot exclude competing causes of death, the macroscopic and histological appearance of the gut, the anemic appearance, the hepatic steatosis, and the general failure of the animals to thrive all indicate that intestinal dysfunction may play a significant role. Moreover, the gut was characterized by a prolonged Imp1 expression that may reflect the fact that morphogenesis of the mouse intestine is not complete at birth but continues into the third postnatal week (44). Therefore, the gut may be particularly susceptible to IMP1 deficiency.
In the dying and dehydrated pups, the small intestine was hypoplastic, with a strongly reduced villus size, and in the colon the crypts were short and irregular. Both Paneth and goblet cells were present in the intestine, suggesting that IMP1 does not have major effects on lineage allocation. The mucosa strongly resembled that of Nkx2-3−/− and Fkh6−/− mice, both of which exhibit similar postnatal growth reduction and increased perinatal lethality (22, 44). Based on the measurements of average villus height and width, it can be estimated that the mucosal (villus) surface in the small intestine of IMP1-deficient mice may be only 10 to 20% of that in wild-type mice. In this way, IMP1-deficient mice resemble patients with coeliac disease, who exhibit a major loss of villi and develop short stature and failure to attain puberty due to severe malabsorption. Secondary bacterial invasion may accelerate intestinal damage and lead to postinfective malabsorption, similar to the situation in patients with tropical sprue, but the fact that the morphological changes are already visible at E17.5 indicates that loss of mucosal lining is a direct result of IMP1 deficiency.
As mentioned above, the histological changes among Imp1−/− mice varied in severity, ranging from essentially complete loss of villi to an almost normal appearance, and in the surviving adult animals, the histological features were almost normal, similar to the Nkx2-3−/− and Fkh6−/− mice described above. The only other organs in which we observed significant histological abnormalities were the kidneys, which also exhibited prolonged IMP1 expression. Here, animals from P3 to P30 exhibited small and hypomorphic glomeruli and immature collecting tubules, resembling the changes observed in Notch2 knockout mice (30). As in the gut, the histological changes were gradually normalized, indicating that IMP1-deficient mice exhibit delayed maturation of the organs.
The molecular pathways leading to dwarfism and intestinal hypoplasia are likely to involve several mechanisms. Loss of IGF-II (7) or c-Myc (51) is associated with dwarfism, whereas lack of H19 RNA is associated with increased growth of the embryo (26). At E12.5, none of the target mRNAs were globally up- or downregulated, and neither were the β-actin and c-Myc protein levels. IMP1 was originally isolated as an RNA-binding protein associating with the 5′ UTR of Igf2 leader 3 mRNA (mouse 4.6-kb mRNA), but all Igf2 transcripts possess an additional binding site in the common 3′ UTR (J. Christiansen, unpublished data). In agreement with previous data, we found that Igf2 leader 3 mRNA is present mainly in 20 to 40S RNP at E12.5, whereas the 3.6-kb leader 4 transcript is largely polysomal. We noted a relative redistribution of both transcripts from polysomes to RNP in IMP1-deficient mice, indicating that Igf2 translation is decreased in the knockout mice. This result contrasts with our previous finding that overexpression of IMP1 selectively mediated translational repression of an Igf2 leader 3-luciferase reporter mRNA in cultured cells (39) and may indicate that the 3′ UTR element is essential for proper regulation of the transcripts by IMP1 in the endogenous situation. In rodents, Igf2 expression ceases shortly after birth (4), and reduced levels of IGF-II may therefore be primarily relevant for the embryonal and placental growth reduction of IMP1-deficient animals.
To obtain additional leads to the phenotype of the mice, we examined global gene expression profiles in the embryos and in the postnatal gut, kidney, and liver. In embryos, only two transcripts in addition to Imp1 mRNA were changed, emphasizing that IMP1 is mainly involved in posttranscriptional events. In the gut, however, we noted a striking downregulation of extracellular matrix components, such as the collagen scaffold, the adhesive glycoproteins tenascin-C and lumican, and galectin-1. Moreover, the mRNA expression of the procollagen chaperone Hsp47 was decreased (35). The association between IMPs and production of extracellular matrix factors is in agreement with recent data showing that overexpression of IMP3/KOC was associated with increased deposition of collagen type I and fibronectin in the pancreas of transgenic mice (53). Moreover, reduced expression of collagen in mesenchymal cells may explain the reduced cartilage formation in distinct parts of the skeleton.
During mouse development, deposition of extracellular matrix begins in early embryogenesis and lasts until a steady state is reached postnatally, at about 4 to 5 months of age. Several studies have indicated that formation of the villi and crypts during gut ontogeny requires reciprocal interactions between the epithelium and mesenchyme (reviewed in reference 3). This is illustrated by the similar phenotypes of platelet-derived growth factor A- and platelet-derived growth factor receptor alpha-deficient mice, in which the absence of the proper mesenchymal cells leads to disrupted villus formation (23). IMP1-deficient mice have the same number of villi as normal mice, indicating that IMP1 loss does not affect the initial budding of the villus. Moreover, the developing villi were rich in proliferative activity. We therefore propose that the disrupted villus formation is caused by the lack of extracellular matrix scaffold which, together with growth-promoting signals from platelet-derived growth factor and/or bone morphogenetic proteins, may be prerequisites for proper villus formation.
Among other transcripts that were downregulated, we noted carbonic anhydrase III (CaIII), which is expressed in both colon and liver (49). Carbonic anhydrase activity and CaII mRNA were recently found to be upregulated in the pancreas of an IMP3/KOC transgene mouse (53), indicating that IMPs and carbonic anhydrase expression may be closely associated. Stearoyl-coenzyme A desaturase 1 (Scd1) (41), which regulates body adiposity, increased insulin sensitivity, and resistance to diet-induced obesity (6, 42), was also downregulated. Decreased levels of Scd1 may contribute to the postnatal loss of weight and emaciation that take place during the first month of life. In the gut, trefoil factor 2 (Tff2), which is involved in mucosal healing and epithelial regeneration (13, 50), was also upregulated. In the liver, the acid-labile subunit (Als) mRNA, which sequesters IGF-I or IGF-II with IGF-binding protein 3 (IGFBP-3) or IGFBP-5 (2) and thereby extends their half-lives, was downregulated. Inactivation of the Als gene in mice results in mild postnatal growth retardation (52), and although we found normal Igf1 mRNA levels in the liver in Imp1−/− mice, reduction in the Als level may reduce the level of IGF-I protein. Finally, in relation to the role of IMP1 in localization of β-actin mRNA to the leading edge (43, 45), we note that cyclase-associated protein 1 (Cap1) was downregulated in the kidney. CAP1 is a bifunctional component of the actin-cofilin complex, which binds to Ras-responsive adenylate cyclases and inhibits actin polymerization (19). CAP1 effectively recycles actin and cofilin, allowing rapid turnover of actin filaments, which is an essential driving force behind cell motility (32).
Whereas the significance of RNA-binding proteins in spatial and temporal control of gene expression during development is evident in invertebrate organisms such as D. melanogaster and C. elegans, the function of RNA-binding proteins during mammalian development has been less clear. With the exception of FMRP, Dazla, and Nova-1, which are associated with mental retardation, azospermia, and neuronal viability, respectively (1, 21, 47), the majority of the mammalian RNA-binding proteins are constitutively expressed and exhibit a relatively broad RNA-binding specificity. The IMP proteins are therefore unusual in the sense that the proteins exhibit restricted spatiotemporal expression and probably recognize a relatively small subset of mRNA targets.
The manifest phenotype of the Imp1−/− mice stands in contrast to the subtle changes in the global expression profile at E12.5, when IMP expression peaks, and provides a striking example of posttranscriptional control of mammalian gene expression. IMP1, -2, and -3 have been implicated in a variety of posttranscriptional processes, such as mRNA localization, turnover, and translational control (reviewed in references 36 and 56), and recent data have shown that they control the migration of neural crest cells (55). Moreover, overexpression of IMP3/KOC leads to remodeling and hyperplasia of the exocrine pancreas and fibrosis (53). Our data substantiate that IMP1 is essential for normal embryonal and postnatal growth, in particular for the adaptation to extrauterine life via control of intestinal development.
Acknowledgments
We gratefully acknowledge Pernille Ekstrøm, Joan Christiansen, and Bo Lindberg for technical assistance.
This work was supported by the Danish Medical and Natural Science Research Councils, the Neye Foundation, the Novo Nordisk Foundation, and the Toyota Foundation.
REFERENCES
- 1.Anonymous. 1994. Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell 78:23-33. [PubMed] [Google Scholar]
- 2.Baxter, R. C., and J. L. Martin. 1989. Structure of the Mr 140,000 growth hormone-dependent insulin-like growth factor binding protein complex: determination by reconstitution and affinity-labeling. Proc. Natl. Acad. Sci. USA 86:6898-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birchmeier, C., and W. Birchmeier. 1993. Molecular aspects of mesenchymal-epithelial interactions. Annu. Rev. Cell Biol. 9:511-540. [DOI] [PubMed] [Google Scholar]
- 4.Brown, A. L., D. E. Graham, S. P. Nissley, D. J. Hill, A. J. Strain, and M. M. Rechler. 1986. Developmental regulation of insulin-like growth factor II mRNA in different rat tissues. J. Biol. Chem. 261:13144-13150. [PubMed] [Google Scholar]
- 5.Buchner, G., E. Montini, G. Andolfi, N. Quaderi, S. Cainarca, S. Messali, M. T. Bassi, A. Ballabio, G. Meroni, and B. Franco. 1999. MID2, a homologue of the Opitz syndrome gene MID1: similarities in subcellular localization and differences in expression during development. Hum. Mol. Genet. 8:1397-1407. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, P., M. Miyazaki, N. D. Socci, A. Hagge-Greenberg, W. Liedtke, A. A. Soukas, R. Sharma, L. C. Hudgins, J. M. Ntambi, and J. M. Friedman. 2002. Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science 297:240-243. [DOI] [PubMed] [Google Scholar]
- 7.DeChiara, T. M., A. Efstratiadis, and E. J. Robertson. 1990. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345:78-80. [DOI] [PubMed] [Google Scholar]
- 8.Deshler, J. O., M. I. Highett, T. Abramson, and B. J. Schnapp. 1998. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 8:489-496. [DOI] [PubMed] [Google Scholar]
- 9.Deshler, J. O., M. I. Highett, and B. J. Schnapp. 1997. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 276:1128-1131. [DOI] [PubMed] [Google Scholar]
- 10.Doyle, G. A., N. A. Betz, P. F. Leeds, A. J. Fleisig, R. D. Prokipcak, and J. Ross. 1998. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res. 26:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle, G. A., J. M. Bourdeau-Heller, S. Coulthard, L. F. Meisner, and J. Ross. 2000. Amplification in human breast cancer of a gene encoding a c-myc mRNA-binding protein. Cancer Res. 60:2756-2759. [PubMed] [Google Scholar]
- 12.Eom, T., L. N. Antar, R. H. Singer, and G. J. Bassell. 2003. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 23:10433-10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrell, J. J., D. Taupin, T. J. Koh, D. Chen, C. M. Zhao, D. K. Podolsky, and T. C. Wang. 2002. TFF2/SP-deficient mice show decreased gastric proliferation, increased acid secretion, and increased susceptibility to NSAID injury. J. Clin. Investig. 109:193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuse, M., K. Fujita, T. Hiiragi, K. Fujimoto, and S. Tsukita. 1998. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141:1539-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gress, T. M., F. Muller-Pillasch, M. Geng, F. Zimmerhackl, G. Zehetner, H. Friess, M. Buchler, G. Adler, and H. Lehrach. 1996. A pancreatic cancer-specific expression profile. Oncogene 13:1819-1830. [PubMed] [Google Scholar]
- 16.Havin, L., A. Git, Z. Elisha, F. Oberman, K. Yaniv, S. P. Schwartz, N. Standart, and J. K. Yisraeli. 1998. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 12:1593-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayano, T., M. Yanagida, Y. Yamauchi, T. Shinkawa, T. Isobe, and N. Takahashi. 2003. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J. Biol. Chem. 278:34309-34319. [DOI] [PubMed] [Google Scholar]
- 18.Herrick, D. J., and J. Ross. 1994. The half-life of c-myc mRNA in growing and serum-stimulated cells: influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol. Cell. Biol. 14:2119-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubberstey, A. V., and E. P. Mottillo. 2002. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 16:487-499. [DOI] [PubMed] [Google Scholar]
- 20.Ioannidis, P., T. Trangas, E. Dimitriadis, M. Samiotaki, I. Kyriazoglou, C. M. Tsiapalis, C. Kittas, N. Agnantis, F. C. Nielsen, J. Nielsen, J. Christiansen, and N. Pandis. 2001. C-MYC and IGF-II mRNA-binding protein (CRD-BP/IMP-1) in benign and malignant mesenchymal tumors. Int. J. Cancer. 94:480-484. [DOI] [PubMed] [Google Scholar]
- 21.Jensen, K. B., B. K. Dredge, G. Stefani, R. Zhong, R. J. Buckanovich, H. J. Okano, Y. Y. Yang, and R. B. Darnell. 2000. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25:359-371. [DOI] [PubMed] [Google Scholar]
- 22.Kaestner, K. H., D. G. Silberg, P. G. Traber, and G. Schutz. 1997. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 11:1583-1595. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson, L., P. Lindahl, J. K. Heath, and C. Betsholtz. 2000. Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development 127:3457-3466. [DOI] [PubMed] [Google Scholar]
- 24.Kim-Ha, J., K. Kerr, and P. M. Macdonald. 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81:403-412. [DOI] [PubMed] [Google Scholar]
- 25.Leeds, P., B. T. Kren, J. M. Boylan, N. A. Betz, C. J. Steer, P. A. Gruppuso, and J. Ross. 1997. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene 14:1279-1286. [DOI] [PubMed] [Google Scholar]
- 26.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34-39. [DOI] [PubMed] [Google Scholar]
- 27.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, J. P., J. Baker, A. S. Perkins, E. J. Robertson, and A. Efstratiadis. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75:59-72. [PubMed] [Google Scholar]
- 29.McCarthy, J. C. 1967. Effects of litter size and maternal weight on foetal and placental weight in mice. J. Reprod Fertil. 14:507-510. [DOI] [PubMed] [Google Scholar]
- 30.McCright, B., X. Gao, L. Shen, J. Lozier, Y. Lan, M. Maguire, D. Herzlinger, G. Weinmaster, R. Jiang, and T. Gridley. 2001. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128:491-502. [DOI] [PubMed] [Google Scholar]
- 31.Mori, H., S. Sakakibara, T. Imai, Y. Nakamura, T. Iijima, A. Suzuki, Y. Yuasa, M. Takeda, and H. Okano. 2001. Expression of mouse igf2 mRNA-binding protein 3 and its implications for the developing central nervous system. J. Neurosci Res. 64:132-143. [DOI] [PubMed] [Google Scholar]
- 32.Moriyama, K., and I. Yahara. 2002. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 115:1591-1601. [DOI] [PubMed] [Google Scholar]
- 33.Mueller-Pillasch, F., U. Lacher, C. Wallrapp, A. Micha, F. Zimmerhackl, H. Hameister, G. Varga, H. Friess, M. Buchler, H. G. Beger, M. R. Vila, G. Adler, and T. M. Gress. 1997. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene 14:2729-2733. [DOI] [PubMed] [Google Scholar]
- 34.Mueller-Pillasch, F., B. Pohl, M. Wilda, U. Lacher, M. Beil, C. Wallrapp, H. Hameister, W. Knochel, G. Adler, and T. M. Gress. 1999. Expression of the highly conserved RNA binding protein KOC in embryogenesis. Mech. Dev. 88:95-99. [DOI] [PubMed] [Google Scholar]
- 35.Nagai, N., M. Hosokawa, S. Itohara, E. Adachi, T. Matsushita, N. Hosokawa, and K. Nagata. 2000. Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. J. Cell Biol. 150:1499-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen, F. C., J. Nielsen, and J. Christiansen. 2001. A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand. J. Clin. Lab. Investig. Suppl. 234:93-99. [PubMed] [Google Scholar]
- 37.Nielsen, F. C., J. Nielsen, M. A. Kristensen, G. Koch, and J. Christiansen. 2002. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J. Cell Sci. 115:2087-2097. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen, J., S. K. Adolph, E. Rajpert-De Meyts, J. Lykke-Andersen, G. Koch, J. Christiansen, and F. C. Nielsen. 2003. Nuclear transit of human zipcode-binding protein IMP1. Biochem. J. 376:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen, J., J. Christiansen, J. Lykke-Andersen, A. H. Johnsen, U. M. Wewer, and F. C. Nielsen. 1999. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 19:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen, J., F. Cilius Nielsen, R. Kragh Jakobsen, and J. Christiansen. 2000. The biphasic expression of IMP/Vg1-RBP is conserved between vertebrates and Drosophila. Mech. Dev. 96:129-132. [DOI] [PubMed] [Google Scholar]
- 41.Ntambi, J. M. 1999. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. J. Lipid Res. 40:1549-1558. [PubMed] [Google Scholar]
- 42.Ntambi, J. M., M. Miyazaki, J. P. Stoehr, H. Lan, C. M. Kendziorski, B. S. Yandell, Y. Song, P. Cohen, J. M. Friedman, and A. D. Attie. 2002. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. USA 99:11482-11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oleynikov, Y., and R. H. Singer. 2003. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr. Biol. 13:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pabst, O., R. Zweigerdt, and H. H. Arnold. 1999. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development 126:2215-2225. [DOI] [PubMed] [Google Scholar]
- 45.Ross, A. F., Y. Oleynikov, E. H. Kislauskis, K. L. Taneja, and R. H. Singer. 1997. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17:2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross, J., I. Lemm, and B. Berberet. 2001. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene 20:6544-6550. [DOI] [PubMed] [Google Scholar]
- 47.Ruggiu, M., R. Speed, M. Taggart, S. J. McKay, F. Kilanowski, P. Saunders, J. Dorin, and H. J. Cooke. 1997. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 389:73-77. [DOI] [PubMed] [Google Scholar]
- 48.Runge, S., F. C. Nielsen, J. Nielsen, J. Lykke-Andersen, U. M. Wewer, and J. Christiansen. 2000. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J. Biol. Chem. 275:29562-29569. [DOI] [PubMed] [Google Scholar]
- 49.Spicer, S. S., Z. H. Ge, R. E. Tashian, D. J. Hazen-Martin, and B. A. Schulte. 1990. Comparative distribution of carbonic anhydrase isozymes III and II in rodent tissues. Am. J. Anat. 187:55-64. [DOI] [PubMed] [Google Scholar]
- 50.Taupin, D., and D. K. Podolsky. 2003. Trefoil factors: initiators of mucosal healing. Nat. Rev. Mol. Cell. Biol. 4:721-732. [DOI] [PubMed] [Google Scholar]
- 51.Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G. R. Martin, and J. M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414:768-773. [DOI] [PubMed] [Google Scholar]
- 52.Ueki, I., G. T. Ooi, M. L. Tremblay, K. R. Hurst, L. A. Bach, and Y. R. Boisclair. 2000. Inactivation of the acid labile subunit gene in mice results in mild retardation of postnatal growth despite profound disruptions in the circulating insulin-like growth factor system. Proc. Natl. Acad. Sci. USA 97:6868-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner, M., S. Kunsch, D. Duerschmied, M. Beil, G. Adler, F. Mueller, and T. M. Gress. 2003. Transgenic overexpression of the oncofetal RNA binding protein KOC leads to remodeling of the exocrine pancreas. Gastroenterology 124:1901-1914. [DOI] [PubMed] [Google Scholar]
- 54.Wisdom, R., and W. Lee. 1991. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 5:232-243. [DOI] [PubMed] [Google Scholar]
- 55.Yaniv, K., A. Fainsod, C. Kalcheim, and J. K. Yisraeli. 2003. The RNA-binding protein Vg1 RBP is required for cell migration during early neural development. Development 130:5649-5661. [DOI] [PubMed] [Google Scholar]
- 56.Yaniv, K., and J. K. Yisraeli. 2002. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene 287:49-54. [DOI] [PubMed] [Google Scholar]
- 57.Zaina, S., R. V. Newton, M. R. Paul, and C. F. Graham. 1998. Local reduction of organ size in transgenic mice expressing a soluble insulin-like growth factor II/mannose-6-phosphate receptor. Endocrinology 139:3886-3895. [DOI] [PubMed] [Google Scholar]
- 58.Zambrowicz, B. P., G. A. Friedrich, E. C. Buxton, S. L. Lilleberg, C. Person, and A. T. Sands. 1998. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature 392:608-611. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, J. Y., E. K. Chan, X. X. Peng, and E. M. Tan. 1999. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J. Exp. Med. 189:1101-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Q., K. Yaniv, F. Oberman, U. Wolke, A. Git, M. Fromer, W. L. Taylor, D. Meyer, N. Standart, E. Raz, and J. K. Yisraeli. 1999. Vg1 RBP intracellular distribution and evolutionarily conserved expression at multiple stages during development. Mech. Dev. 88:101-106. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]