Abstract
The carboxyl-terminal repeat domain (CTD) of RNA polymerase II is thought to help coordinate events during RNA metabolism. The mammalian CTD consists of 52 imperfectly repeated heptads followed by 10 additional residues at the C terminus. The CTD is required for cleavage and polyadenylation in vitro. We studied poly(A)-dependent termination in vivo using CTD truncation mutants. Poly(A)-dependent termination occurs in two steps, pause and release. We found that the CTD is required for release, the first 25 heptads being sufficient. Neither the final 10 amino acids nor the variant heptads of the second half of the CTD were required. No part of the CTD was required for poly(A)-dependent pausing—the poly(A) signal could communicate directly with the body of the polymerase. By removing the CTD, pausing could be observed without being obscured by release. Poly(A)-dependent pausing appeared to operate by slowing down the polymerase, such as by down-regulation of a positive elongation factor. Although the first 25 heptads supported undiminished poly(A)-dependent termination, they did not efficiently support events near the promoter involved in abortive elongation. However, the second half of the CTD, including the final 10 amino acids, was sufficient for these functions.
Termination of transcription at the ends of genes by RNA polymerase II depends on the same signals that are responsible for 3′-end processing of the RNA itself (53). Thus, in the case of polyadenylated mRNAs, the very same poly(A) signal that drives cleavage and polyadenylation also directs the polymerase to terminate (53). For many years it was thought that a poly(A) signal would require an assisting pause site in the downstream DNA in order to slow down the polymerase to allow time for the poly(A) signal to act. However, it is now clear that after it has been transcribed, the poly(A) signal alone can apprehend the polymerase as it moves down the template, without the assistance of any additional element in the DNA (49). Of course, various auxiliary elements are used to modify and regulate the core mechanism in specific circumstances (53).
Intriguingly, although no pause site in the DNA is required to assist the poly(A) signal, the first effect of the poly(A) signal on the polymerase is, nevertheless, to cause it to pause (49). Subsequently, as the polymerase continues on down the template, communication between the poly(A) signal and the polymerase is sustained until the final trigger to terminate is delivered (33). Thus, the poly(A) signal drives termination in two steps—poly(A)-dependent pausing followed by poly(A)-dependent release.
The design of the poly(A) signal in mammals is fairly straightforward (60). The core sequence consists of a highly conserved upstream element (AAUAAA), recognized by cleavage-polyadenylation specificity factor (CPSF), and a poorly defined downstream region (rich in Us or Gs and Us), bound by cleavage stimulation factor (CstF). The poly(A) cleavage site lies in between these two elements. However, the process of cleavage and polyadenylation has turned out to be far more complex than might be inferred from the simplicity of this basic arrangement (5). Apparently this reflects an important coordinating function of 3′-end processing which serves to integrate transcription with both splicing and transport (5, 24, 31). Thus, not only do splicing factors (43, 53), export factors (24, 31), and transcription initiation factors (5) interact functionally with the cleavage and polyadenylation apparatus, but even RNA polymerase II itself is a required participant in the cleavage and polyadenylation reaction (28).
The role of the polymerase in cleavage and polyadenylation has been studied both in vitro and in vivo. In vitro, the polymerase is a required cleavage and polyadenylation factor, and it is the carboxyl-terminal repeat domain (CTD) of its largest subunit (Rpb1) that provides the processing activity (29). This domain, in mammals, consists of a seven-amino-acid sequence repeated, with some deviations, 52 times followed by a unique 10-amino-acid motif at the C terminus. Ryan et al. (55) showed that at least half the length of the CTD is necessary for function in the 3′-end cleavage reaction in vitro, but the 10-amino-acid C-terminal motif is not required. Consistent with an important role for the CTD, several yeast and mammalian cleavage and polyadenylation factors (Pcf11, Yhh1, Ydh1, and CstF50) (52) have been shown to be CTD-binding proteins (3, 15, 19, 36).
However, the role of the CTD in vivo remains somewhat uncertain. Even in vitro its contribution to 3′-end processing depends on assay conditions (29), suggesting that in vivo its role in processing may depend on the local environment. Accordingly, the CTD is in fact not essential for cleavage and polyadenylation in vivo in yeast (16, 37), and CstF50, the only metazoan 3′-end processing factor known to bind the CTD (19), is not an essential protein in worms (40, 43), as defined by RNA interference (RNAi). Thus, the CTD participates in (16, 37) but may not be essential for 3′-end processing in vivo. Indeed, the burden may be shared by other parts of the polymerase. For example, Ssu72, an essential component of cleavage and polyadenylation factor in yeast (26), binds to Rpb2 (14), the second largest subunit of RNA polymerase II. Similarly, mammalian CPSF appears to bind to the polymerase (13), but evidently not via the CTD (19).
Fong et al. (20) studied the role of the CTD in vivo by transfecting cells with expression vectors coding for α-amanitin-resistant Rpb1 subunits having wild-type or mutant CTDs. By growing the cells in α-amanitin for 2 days, they inactivated the resident polymerases containing the endogenous Rpb1, thereby forcing the cells to survive on the ectopically provided α-amanitin resistant Rpb1 subunits. They then interrogated the cells for their content of cleaved and polyadenylated RNA. Consistent with the in vitro results of Ryan et al. (55), Fong et al. (20) found that cells surviving on less than half of a CTD contained little properly cleaved and polyadenylated RNA. However, in contrast to the in vitro results, Fong et al. (20) also found that even a full-length CTD was unable to support the production of polyadenylated RNA in vivo in the absence of the final C-terminal motif. Since the final 10 amino acids are required for efficient transcription per se (20), and the CTD itself is required for transcription in a manner that is promoter dependent (22), it is possible that pleiotropic effects are responsible for the failure of cells to produce polyadenylated RNA when they are forced to survive on these subfunctional polymerases. Thus, the nature of any direct involvement of the CTD in the 3′-end processing reaction in vivo remains unclear.
The CTD is thought also to be involved in transcription termination (42). Although this has not yet been tested directly, the fact that at least two of the CTD-binding cleavage and polyadenylation factors in yeast (Pcf11 and Yhh1) are also termination factors (3, 15) supports this view. However, though not obvious for processing, in the case of termination it is clear a priori that more than the CTD of the polymerase must be involved because any transcriptional transition, whether pausing or release, must obviously involve the catalytic core of the enzyme. This is consistent with the known Rpb2-binding activity of Ssu72 (14), which, in addition to being required for processing (26), appears also to be both a pausing and a termination factor in yeast (14, 56).
Here we report that the poly(A) signal communicates with the polymerase by both CTD-dependent and CTD-independent mechanisms. The Rpb1 CTD is required for poly(A)-dependent termination, but the body of the polymerase, with the CTD deleted, is sufficient for poly(A)-dependent pausing. We also show, as was found for cleavage and polyadenylation in vitro (29), that the 10-amino-acid C-terminal motif is not required for poly(A)-dependent termination.
MATERIALS AND METHODS
The plasmids used in this work have been described previously (19, 49). Transfection and G-less run-on transcription were carried out using COS cells as described by Kim and Martinson (33) except that the transfected DNA was a mixture of 0.9 μg of G-less cassette reporter plasmid plus 0.1 μg of α-amanitin-resistant Rpb1 expression plasmid, and α-amanitin was added to the nuclei prior to run-on to give a final concentration of 0.3 μg/ml. TRIzol was used to isolate the RNA, which was then digested with 1,000 (rather than 500) U of T1 RNase.
Transfections for RNase protection assays were in 100-mm plates (instead of the 35-mm plates used for run-on) and utilized a mixture of 6.2 μg of reporter DNA, 0.8 μg of Rpb1 DNA, and 0.5 μg of an RNA polymerase III transfection control plasmid. The control plasmid consisted of pCR2.1 (Invitrogen) into which a 111-nucleotide (nt) transcription unit under the control of the U6 promoter had been inserted. Nuclear RNA for the RNase protection was isolated using RNeasy columns (Qiagen). Cells were collected by scraping and lysed with buffer RLN (Qiagen). After removal of the cytoplasmic RNA fraction, the nuclear pellet was washed twice more in buffer RLN, homogenized on QIAshredder columns (Qiagen), and then processed following the manufacturer's instructions for obtaining animal cell total RNA. The isolated RNA was treated with DNase I (Roche) prior to the assay.
For immunoblotting, nuclei were prepared as previously described (49) except that after removal of the lysis buffer, nuclei were resuspended in sodium dodecyl sulfate (SDS) loading buffer and heated at 95° for 5 min. Immunoblotting was carried out according to the manufacturer's instructions (Roche) using polyvinylidene difluoride membrane for protein transfer. Anti-B10, N-20, and antisymplekin antibodies were purchased from Chemicon, Santa Cruz Biotechnology, and BD signal transduction, respectively. Proteins were separated using an SDS-5% polyacrylamide gel (Bio-Rad).
RESULTS
G-less run-on transcription assay for α-amanitin-resistant polymerases.
To study the role of the CTD in poly(A)-dependent termination, we transfected various Rpb1 expression plasmids (19) into COS cells. These plasmids encode either full-length Rpb1 subunits or mutant forms of the subunits bearing various deletions in the CTD. We then used run-on transcription of cotransfected G-less cassette reporter plasmids to measure the ability of polymerases containing either full-length or mutant CTDs to terminate (49, 58). The transiently expressed Rpb1 subunits contained, in addition to any deletions in the CTD, a mutation that confers resistance to α-amanitin (19). Adding α-amanitin to the nuclei during the run-on transcription procedure allowed us to restrict transcription in the assay to only those polymerases containing the ectopically expressed Rpb1 subunit (20).
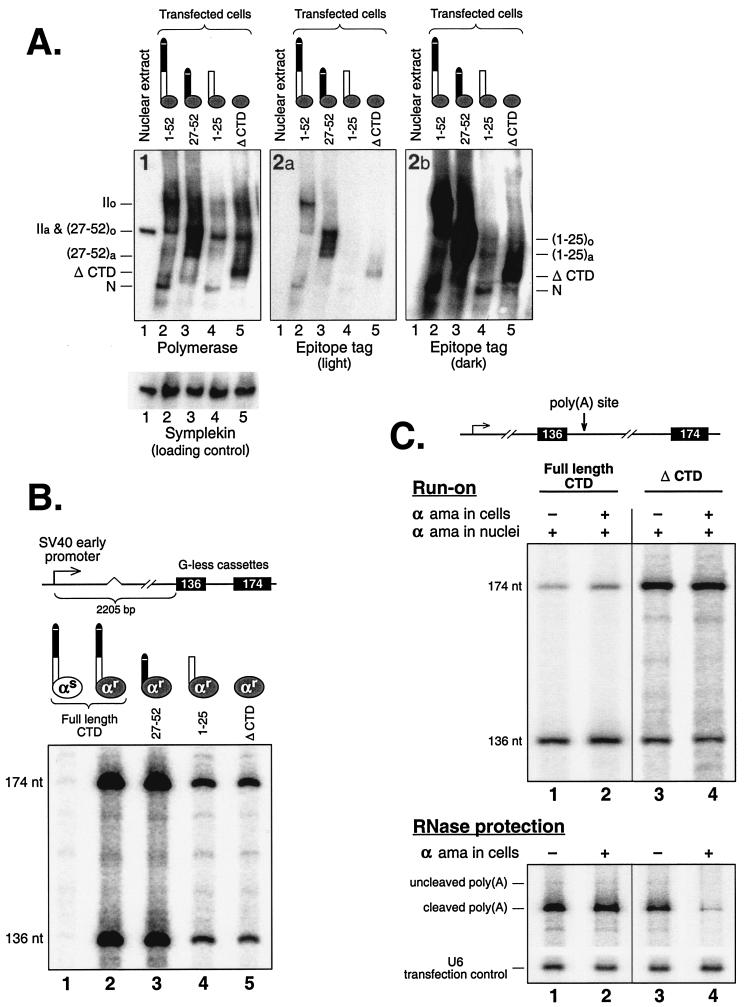
First we characterized the properties of the polymerases in our system. Figure 1A shows an immunoblot of the proteins from transfected nuclei (COS cells) and from a nuclear extract (nontransfected HeLa cells), probed and reprobed with various antibodies. For panel 1 in Fig. 1A we probed with N-20, an antibody directed against an endogenous epitope near the amino terminus of Rpb1. This epitope was present in all of the polymerase II enzymes in these samples, endogenous and recombinant. Nuclear extract from nontransfected cells (panel 1, lane 1) gave rise to only one prominent band, presumably hypophosphorylated RNA polymerase IIa. Nuclei, in contrast, gave substantial amounts of the highly phosphorylated RNA polymerase IIo as well (panel 1, lanes 2 to 5).
FIG.1.
G-less run-on assay for α-amanitin-resistant polymerases. (A) Expression levels of polymerases with different CTD lengths. Nuclear proteins prepared from cells transfected with the indicated epitope-tagged (B10) Rpb1 constructs were separated by SDS-5% polyacrylamide gel electrophoresis for immunoblotting. The membrane was probed with anti-B10, stripped, probed with antisymplekin, stripped, and probed with N-20. Panels 2a and 2b are from same anti-B10 probing but with different lengths of exposure. (B) Transcription activity in cells transfected with different Rpb1 expression plasmids. A scale drawing of the reporter construct, A3〈EmC0〉, is shown. This is the mutant version of construct number 1 in Fig. 5A of Orozco et al. (49). It contains 136- and 174-bp G-less cassettes located more than 2.2 kb downstream of the transcriptional start site. The 2.2-kb spacer includes the SV40 small intron as shown and is a patchwork of DNA sequences taken from the chicken βH-globin gene and pRSVcat (49, 58). A mixture of 0.9 μg of reporter construct and 0.1 μg of expression construct was transfected into COS cells for the various α-amanitin-sensitive (αs) and -resistant (αr) polymerases shown. After 2 days, nuclei were harvested and G-less run-on transcription was carried out in the presence of 0.3 μg of α-amanitin/ml. The reporter does not have a functional poly(A) signal and therefore yields a straightforward estimate of the transcriptional efficiencies of the different polymerases. (C) The CTD is required for poly(A)-dependent termination but not for poly(A)-dependent pausing. G-less run-on transcription was as for part B. When the cells were treated with α-amanitin (lanes 2 and 4), this was added to a concentration of 20 μg/ml approximately 9.5 h before harvest. These conditions were chosen, based on data in the work of Nguyen et al. (48), so as to minimize the time spent by the cells in the toxin. RNase protection was carried out on nuclear RNA. The positions of the protected RNAs corresponding to poly(A) cleaved (252 nt) and uncleaved (304 nt) RNA were estimated based on size markers run in the same gel.
To look specifically at the recombinant Rpb1 subunits in the transfected cells, we used an antibody against the N-terminal B10 epitope tag present in all of these constructs (19). This antibody does not cross-react with any protein in the nontransfected extract (lanes 1 of panels 2a and 2b). Cartoons of the different recombinant polymerases used are shown at the top of each panel. These polymerases contain Rpb1 subunits bearing either an intact CTD (lane 2) or CTDs containing various deletions (lanes 3 to 5). As previously described by Fong and Bentley (19, 20), the intact Rpb1 subunit carries all 52 heptad repeats followed by the final 10-residue C-terminal motif (lanes 2), whereas the deletions carry only heptads 27 to 52 plus the C-terminal motif (lanes 3), only heptads 1 to 25 (lanes 4), or no CTD at all (lanes 5). The anti-epitope tag blot shows that most of the recombinant full-length Rpb1 is in the IIo form in the isolated nuclei (panel 2a, lane 2). Most of the 27-52 Rpb1 also is highly phosphorylated, with the predominant form migrating at the same position as the hypophosphorylated form of full-length Rpb1 (Fig. 1A, compare lane 3 with lane 2 in panel 2a).
The 1-25 Rpb1 and ΔCTD constructs are expressed less well than the others, as previously reported (19), perhaps because they lack the natural C terminus (7, 19). A significant fraction of 1-25 Rpb1 appears to be an N-terminal degradation product (panel 2b, lane 4) with a higher mobility on the gel than even the ΔCTD Rpb1 (Fig. 1A, compare lanes 4 and 5 in panel 2b). This species has been seen before (7, 30) and is also produced in cells expressing the full-length Rpb1 (see lanes 2 in all the panels). However, the 1-25 Rpb1 construct also yields a species, (1-25)a, having a mobility similar to that of the presumptive hypophosphorylated form of 27-52 Rpb1, (27-52)a, as would be expected from the similarity of their molecular weights (compare lane 4 of panel 2b with lane 3 of panel 2a). The third, most slowly migrating form of 1-25 Rpb1 is presumably highly phosphorylated (panel 2b, lane 4).
To characterize the transcription properties conferred on RNA polymerase II by the various ectopically expressed Rpb1 subunits, we carried out run-on transcription in the presence of α-amanitin for nuclei isolated from cells transfected with the various α-amanitin-resistant Rpb1 constructs (Fig. 1B). For this we used the G-less cassette reporter plasmid illustrated in Fig. 1B. Polymerase densities are monitored within the cassette regions by run-on transcription in the absence of GTP, followed by detection and quantitation of the cassette RNAs by RNase T1 digestion and gel electrophoresis (49, 58). For this initial characterization of the transcription properties of polymerases containing the various ectopically expressed Rpb1 subunits, we used a reporter that did not exhibit termination. Thus, the polymerase densities are expected to be the same on the upstream and downstream cassettes so that the run-on signal for each cassette should be essentially proportional to cassette length.
Figure 1B shows that only cells transfected with α-amanitin-resistant Rpb1 subunits (lanes 2 to 5) give significant run-on transcription under our assay conditions (which include 0.3 μg of α-amanitin/ml). Transfection with α-amanitin-sensitive Rpb1, as expected, yields nuclei that give a negligible run-on signal (lane 1). The near absence of signal in lane 1 also confirms that the endogenous RNA polymerase II is essentially fully inhibited under these conditions. Therefore, we assume that the bands in lanes 2 to 5 are almost exclusively the product of transcription by polymerases containing the ectopically expressed α-amanitin-resistant Rpb1 subunits. Consistent with the protein levels evident in panels 2a and 2b of the immunoblot in Fig. 1A, cells transfected with constructs containing the C-terminal half of the CTD, including the 10-amino-acid C-terminal motif (lanes 2 and 3), show more transcriptional activity than cells transfected with the 1-25 Rpb1 and ΔCTD Rpb1 constructs (lanes 4 and 5).
The CTD is not required for poly(A)-dependent pausing but is required for termination.
We showed previously that the poly(A) signal exerts two separate effects on transcription, pausing and release (49). Pausing takes hold almost as soon as the poly(A) signal is crossed, and then release ensues, continuing stochastically far down the template (49). The poly(A) signal is both necessary and sufficient to generate the signal that causes pausing (49), and sustained communication of the poly(A) signal with the polymerase is required in order to trigger release (33). The CTD is thought to be intimately involved in mediating communication between the polymerase and the poly(A) signal (52), but little is known about its role in poly(A)-dependent termination.
To address the role of the CTD in poly(A)-dependent termination, we compared the abilities of polymerases with and without a CTD to transcribe beyond a poly(A) signal. For this we used the construct shown in Fig. 1C in which the two G-less cassettes were placed far apart and the simian virus 40 (SV40) early poly(A) signal was inserted immediately downstream of the first cassette. Cells were transfected with either full-length or ΔCTD Rpb1 constructs, and then the nuclei were isolated and run-on transcription was carried out in the presence of α-amanitin. The cells also were grown either in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of α-amanitin prior to isolation of the nuclei. Growth in the presence of α-amanitin causes α-amanitin-sensitive polymerases to be destroyed (48). We wanted to know whether the presence of stalled, α-amanitin-sensitive polymerases on the template (for cells not treated with α-amanitin) may be a hindrance to the progress of the α-amanitin-resistant polymerases during the run-on assay.
The results show (Fig. 1C, top panel) that α-amanitin-resistant polymerases having a full-length CTD (lanes 1 and 2) respond normally to a poly(A) signal. The polymerase density in the upstream 136-bp cassette is substantially higher than it is in the downstream cassette on the other side of the poly(A) signal. In contrast, polymerases with no CTD cannot terminate (top panel, lanes 3 and 4). Indeed, the polymerase density increases rather than decreases following the poly(A) site, suggesting the presence of paused polymerases across the downstream template.
Prior growth of the cells in α-amanitin did not affect the results (Fig. 1C, top panel, compare lanes 1 and 3 with lanes 2 and 4). This is consistent with the fact that polymerases are spaced a minimum of 750 bp apart on our templates (58), while the greatest distance any polymerase would have to travel in the assay is the length of the longest cassette (174 bp). Thus, the probability of interference by sensitive polymerases stalled in the α-amanitin is remote. To confirm that our conditions of α-amanitin treatment were comparable to those of previous studies (19, 42) in their effects on the recovery of processed RNA from cells, we transfected and prepared nuclei as for the experiment described above but then isolated RNA from the nuclei for RNase protection assays instead of carrying out run-on transcription. The bottom panel of Fig. 1C confirms first that cells not grown in α-amanitin efficiently process the RNA transcribed from our reporter (lanes 1 and 3) and second that cells with an ectopic α-amanitin-resistant full-length Rpb1, even when grown in α-amanitin, also produce 3′-end-processed RNA (lane 2). Finally, the RNase protection results show that cells forced to depend on a ΔCTD polymerase by growth in α-amanitin can produce hardly any processed RNA (lane 4) despite the fact that transcription persists unabated (see lane 4 in the top panel of Fig. 1C). Note that the failure to produce processed RNA is not balanced by the recovery of an equivalent amount of unprocessed RNA (Fig. 1C, bottom panel, lanes 3 and 4). Similar results were observed previously (19, 42). Therefore, it is not known whether the defect is in processing or some other step of mRNA biogenesis. Because of these uncertainties with cells grown in α-amanitin, we chose to do all subsequent experiments with nuclei isolated from healthy (i.e., non-α-amanitin-treated) cells.
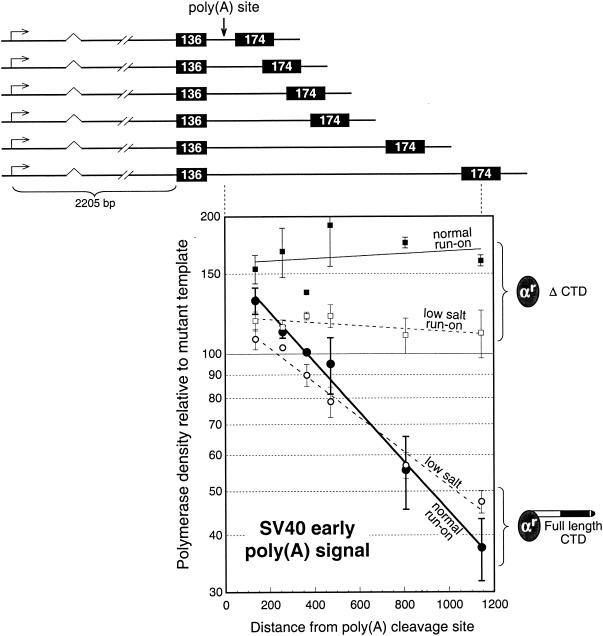
To address the role of the CTD in poly(A)-dependent pausing and release quantitatively, we repeated the experiments of Orozco et al. (49), using the transfected α-amanitin-resistant Rpb1 subunits with or without a CTD (Fig. 2). To measure pausing and release as a function of distance, we cotransfected the series of G-less cassette reporter constructs shown in Fig. 2 (which are identical to the constructs described in detail in Fig. 5 of reference 49). In these constructs a 136-bp G-less cassette was followed by an SV40 early poly(A) signal as for Fig. 1C, but the position of the downstream 174 bp G-less cassette was systematically varied as shown in Fig. 2. We previously showed that endogenous monkey (COS cell) RNA polymerase II terminated downstream of the poly(A) site both gradually and stochastically, resulting in a progressive decrease in polymerase density in the 174-bp cassette as it was placed at increasing distances downstream (49). As shown in Fig. 2, ectopically expressed human α-amanitin-resistant Rpb1 with a full-length CTD behaves similarly.
FIG. 2.
The CTD is required for termination but not for poly(A)-dependent pausing. The reporter constructs shown, containing the SV40 early poly(A) signal (the same as those in Fig. 5A of reference 49), were transfected into COS cells along with expression plasmids for full-length Rpb1 or Rpb1 with its CTD deleted. Each reporter was transfected in parallel with its poly(A) signal mutant (the two SV40 early AAUAAA hexamers, each inactivated by two nucleotide changes; see Fig. 3 of reference 49). After run-on transcription, the 174-nt cassette signal from the wild-type reporter was expressed as a percentage of that for the mutant (following normalization of all 174-nt signals to their respective 136-nt signals; see reference 49). The lines are exponential fits to the data. In the case of polymerases with the full-length CTD, these fits illustrate the first-order nature of poly(A)-dependent termination (49). In the case of the ΔCTD polymerases, the lines have no theoretical significance. Normal and low-salt conditions refer to run-on in the presence of 280 and 28 mM ammonium sulfate, respectively. Points without error bars represent single determinations. All other data, except for ΔCTD under normal run-on conditions, show the average and range for two separate transfections. For ΔCTD under normal run-on conditions, the mean ± standard deviation for three separate transfections is shown.
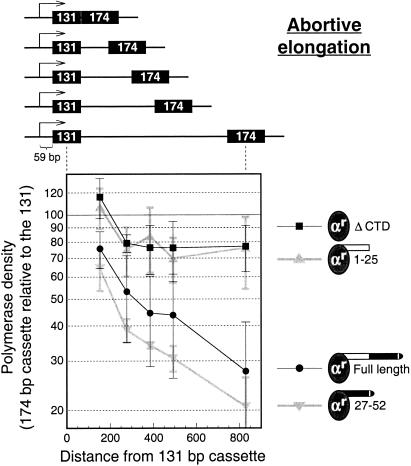
FIG. 5.
Promoter-proximal activity of the 1-25 CTD construct resembles that of the ΔCTD construct. Abortive elongation was analyzed in the same manner and using the same reporter constructs as for Fig. 4 of Orozco et al. (49). The same normalization procedure using the same fully processive reporter construct was also carried out. The data points represent the mean ± standard deviation for at least three separate transfections.
The ectopically expressed full-length Rpb1 also exhibits pausing like its endogenous counterpart. This is indicated in Fig. 2 by polymerase densities greater than 100% at short distances downstream of the poly(A) site. These percentages refer to the polymerase densities downstream of functional poly(A) signals after normalization to the polymerase densities obtained from identical plasmids whose poly(A) signals had been inactivated by mutation (49). Therefore, the increase in polymerase density (pausing) is directly attributable to the functionality of the poly(A) signal. Thus, the ectopically expressed full-length Rpb1 subunits support poly(A)-dependent pausing followed by release (Fig. 2) in the same manner as do the endogenous polymerases (49).
In contrast to full-length Rpb1, truncated Rpb1 lacking the entire CTD (ΔCTD) does not support termination (Fig. 1C). The data shown in Fig. 2 confirm this and show that there is no decrease in polymerase density across the entire 1,000 bp of assay space. Interestingly, in spite of its complete inability to support termination, the ΔCTD Rpb1 continues to exhibit robust poly(A)-dependent pausing. Indeed, the increase in polymerase density immediately downstream of the poly(A) signal is greater for the ΔCTD construct than for the full-length CTD (compare data for the two normal run-on reactions in Fig. 2). Moreover, this high polymerase density for the ΔCTD construct persists for more than 1 kb farther downstream, suggesting a process of iterative pausing (i.e., slower elongation), as would be expected, for example, from modification of an elongation factor. The apparent increase in the level of downstream pausing by the ΔCTD Rpb1 compared to that by the full-length Rpb1 may indicate an increase in the propensity of the ΔCTD Rpb1 to pause or it may reflect the absence of polymerase attrition through termination.
We wanted an independent indication that the high polymerase density downstream of a functional poly(A) signal reflects pausing or slowing down of transcription. Run-on transcription, as typically carried out, uses high salt, Sarkosyl, or heparin to relieve impediments to elongation so that all elongationally competent polymerases can be counted (21, 27). However, by using milder run-on conditions it is possible to obtain a better approximation of the level of transcriptional elongation activity actually occurring within the cell (17, 18, 21, 35, 54). If the high density of ΔCTD polymerases observed downstream of the wild-type poly(A) signal under normal run-on conditions (Fig. 2) reflects pausing or slowing down relative to the mutant template, then run-on transcription under conditions of low salt that better retain this functional difference should diminish this increase in signal. For example, if polymerases slow down by a factor of 2, the polymerase density will increase by a factor of 2, but the polymerase flux through the region would not change (e.g., twice the density of polymerases, each transcribing at half the rate). Therefore, if the increased run-on signal observed downstream of the wild-type poly(A) site under normal assay conditions reflects an increase in the number of polymerases from pausing or slowing down, then carrying out run-on transcription under low-salt conditions to try to retain the difference in elongation rates of the polymerases on the wild-type and mutant templates should reduce or eliminate the wild-type-specific increase in signal (49).
When we repeated the run-on transcription assay for ΔCTD Rpb1-transfected cells using low-salt conditions, the excess run-on signal downstream of the wild-type poly(A) site was substantially reduced (Fig. 2). This supports the interpretation that the excess polymerase density downstream of the wild-type poly(A) signal in ΔCTD Rpb1-transfected cells reflects an increased density of polymerases that suffer from impaired elongation. We also carried out a low-salt run-on assay for full-length-CTD-transfected cells (Fig. 2). Although there are minor differences compared to experiments with endogenous polymerases (49), the results generally confirm the observation of Orozco et al. (49) that the excess in polymerase density immediately downstream of the poly(A) signal detected by the normal run-on assay also corresponds to pausing or slowing down.
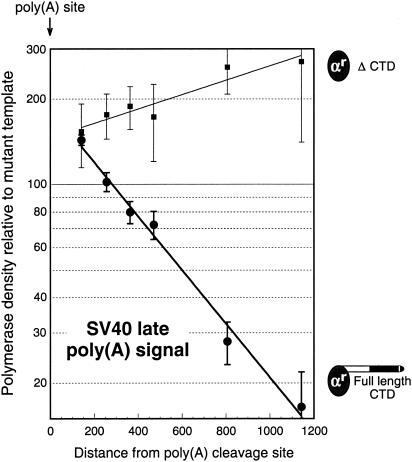
The results shown in Fig. 2 were obtained with the SV40 early poly(A) signal. This is a fairly typical poly(A) signal, its principal unusual feature being that it overlaps with another typical, but weaker, poly(A) signal that starts about 30 nt farther upstream (11). We wanted to see whether truncation of the CTD would abrogate termination by a stronger poly(A) signal. The SV40 late poly(A) signal is reported to be about fivefold stronger than the SV40 early signal (6) and is characterized by the possession of both upstream and downstream enhancers (2, 39). We compared the ability of full-length and ΔCTD versions of Rpb1 to pause and terminate downstream of the SV40 late poly(A) site (Fig. 3). In agreement with the results reported previously (49) for endogenous COS cell polymerases, polymerases containing the full-length ectopically expressed α-amanitin-resistant Rpb1 pause and terminate efficiently downstream of the SV40 late poly(A) site (Fig. 3). In contrast, when the Rpb1 subunit lacks its CTD, termination is completely abolished but robust downstream pausing remains (Fig. 3). Therefore, the requirement for a CTD in poly(A)-dependent termination applies to two very different poly(A) signals. Likewise, the CTD-independent nature of poly(A)-dependent pausing may also be general.
FIG. 3.
The SV40 late poly(A) signal also requires the CTD for termination but not for pausing. The data were obtained and analyzed as for Fig. 2. The reporter constructs resemble those shown in Fig. 2 and are the same as those used for Fig. 7 of Orozco et al. (49). For the full-length CTD, the average and range of values for two separate transfections is shown. For ΔCTD, the mean ± standard deviation for two or three separate transfections is shown.
Either half of the CTD suffices for termination, and the 10-residue C-terminal motif is not required.
Fong and Bentley (19) have reported that Rpb1 carrying only the first half of the CTD (1-25 Rpb1) cannot support efficient cleavage and polyadenylation in vivo, whereas Rpb1 carrying the second half of the CTD (27-52 Rpb1) can. More recently, Fong et al. (20) have attributed this effect primarily to the fact that the 27-52 Rpb1 construct also contains the 10-residue C-terminal motif.
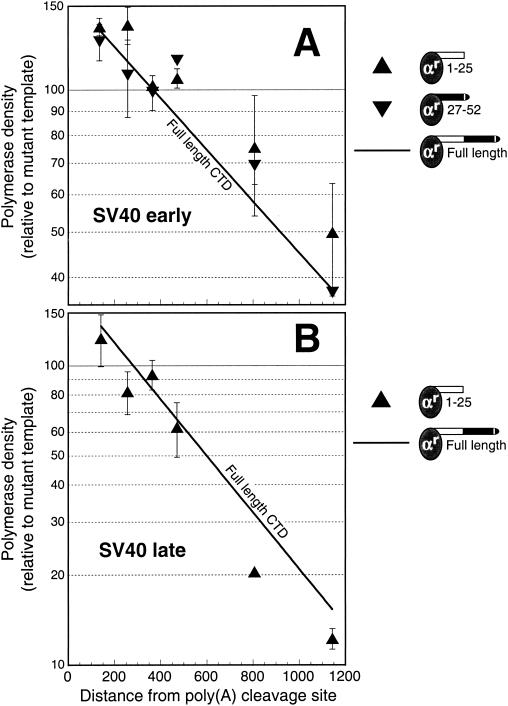
We wondered whether these two partial deletions of the CTD would exhibit similar contrasting effects on poly(A)-dependent termination. Surprisingly, Fig. 4A shows that 1-25 Rpb1 and 27-52 Rpb1 behave similarly in a poly(A)-dependent termination assay. The line in Fig. 4A, taken directly from Fig. 2, shows the results for the full-length CTD. The data points for the two CTD partial deletions fall reasonably close to the line, indicating that there is little deficit in their ability to support poly(A)-dependent termination.
FIG. 4.
Both halves of the CTD are equivalent to full-length CTD in the ability to support poly(A)-dependent termination. (A) SV40 early poly(A) signal. The data were obtained and analyzed as for Fig. 2. The 1-25 data points represent the average and range for two separate transfections, except for the value at 365 bp, which is a single data point. The 27-52 values show the mean ± standard deviation for two or three separate transfections. For comparison, the line for full-length CTD from Fig. 2 (normal run-on conditions) is shown. (B) SV40 late poly(A) signal. The data were obtained and analyzed as for Fig. 3. The data points represent the average and range for two separate transfections, except for the value at 807 bp, which is a single data point. For comparison, the line for full-length CTD from Fig. 3 is shown.
Given its reported inability to support efficient cleavage and polyadenylation, we were surprised that 1-25 Rpb1 resembled full-length CTD in its ability to respond to a poly(A) signal by terminating. We therefore repeated the termination assay for 1-25 Rpb1 using a different poly(A) signal. The results of Fig. 4A were for the SV40 early poly(A) signal. Figure 4B shows that the 1-25 Rpb1 construct supports efficient termination by the SV40 late poly(A) signal as well. The line in Fig. 4B is taken directly from Fig. 3 and shows the termination profile for the SV40 late poly(A) signal as delivered by Rpb1 carrying a full-length CTD. A comparison of the line with the data points shows that poly(A)-dependent termination by the 1-25 Rpb1 is as effective as that for the full-length CTD.
ΔCTD and 1-25 CTD constructs are deficient in promoter-proximal abortive elongation.
Polymerases that fail to achieve high processivity following escape from the promoter undergo a process of premature termination called abortive elongation (51). The mechanism of premature termination immediately downstream of the promoter differs from that of poly(A)-dependent termination downstream of a poly(A) signal (49). We wondered whether the CTD requirement for premature termination (abortive elongation) also differs from that for poly(A)-dependent termination.
To measure abortive elongation, we used the set of reporter constructs shown in Fig. 5 (same as the constructs in Fig. 4 of reference 49). In these constructs, a 131-bp cassette is placed immediately downstream of the promoter and is followed by a 174-bp cassette placed at increasing distances downstream for each member of the series. Figure 5 shows that polymerases containing ectopically expressed full-length Rpb1 abortively elongate in a manner similar to that previously reported for the endogenous COS cell polymerases (49), such that most of the polymerases are lost from the template within several hundred base pairs. In contrast, the results for ΔCTD Rpb1 (Fig. 5) suggest a reduced amount of abortive elongation that occurs only very close to the promoter. A similar observation was made previously by McCracken et al. (44). Interestingly, 1-25 Rpb1 and 27-52 Rpb1, which both functionally resemble full-length Rpb1 in poly(A)-dependent termination (Fig. 4), behave quite unlike each other in abortive elongation. This further underscores the fact that poly(A)-dependent termination and abortive elongation are mechanistically distinct processes.
DISCUSSION
CTD required for poly(A)-dependent termination.
Poly(A)-dependent termination involves two activities, downstream pausing and release (49). Under normal circumstances there is a transient rise in polymerase density immediately downstream of a poly(A) signal, reflecting pausing. Then, the polymerase density begins a sustained decline down the template as termination ensues (49). Here we have shown that these two aspects of poly(A)-dependent termination can be uncoupled. Figures 2 and 3 illustrate that robust termination occurs when the polymerase possesses a CTD, but that no termination at all can be detected across the same region of template when the CTD is absent.
The absolute CTD-requirement for poly(A)-dependent termination suggests the involvement of a CTD-binding processing-termination factor. An obvious candidate is CstF, the only mammalian cleavage and polyadenylation factor so far confirmed as a CTD-binding protein (19). CstF has three subunits, p50, p64, and p77. The p64 subunit binds to the downstream U/GU-rich element of the poly(A) signal (60), which is known to be required for efficient termination (11). Consistent with a role for mammalian CstF in termination, a termination factor identified genetically in Schizosaccharomyces pombe has been found to be a homolog of CstF64 (1). A C-terminal truncation of this protein supported normal cleavage and polyadenylation but not termination.
CstF binds to the CTD through its p50 subunit (19). Surprisingly, however, CstF50 has been reported not to be an essential protein in Caenorhabditis elegans (40, 43). However, these conclusions were based on RNAi, so it is possible that CstF is abundant and stable and therefore a poor candidate for RNAi depletion. On the other hand, another protein with no RNAi phenotype, SRm160, was synthetically lethal with CstF50, indicating that there was indeed a functional decrease in CstF50 protein concentration (43). If CstF50 is indeed nonessential, it may be that other cleavage and polyadenylation factor(s) also contribute to binding the CTD so that CstF50 is dispensable. In addition, it is likely that there are redundant pathways to ensure termination even when poly(A)-dependent termination fails (see below).
The CTD requirement for poly(A)-dependent termination can be fulfilled even if half of the CTD has been deleted (Fig. 4). Importantly, the first 25 heptads alone of the CTD are sufficient. Thus, the 1-25 Rpb1 construct, lacking not only the 27 downstream heptads but also the final 10-residue C-terminal motif, was equal to the full-length CTD in the efficiency with which it supported poly(A)-dependent termination (Fig. 4). Interestingly, Fong et al. reported that constructs lacking the 10-residue C-terminal motif, including the 1-25 Rpb1 construct, cannot support the production of cleaved and polyadenylated transcripts in vivo (19, 20). This might mean that partial deletions of the CTD can uncouple processing from termination. Alternatively, it might reflect the different experimental methods used. In the work reported here, cells were cultured under normal conditions and then the nuclei were isolated and exposed to α-amanitin for the run-on transcription. In the work of Fong et al., the cells were cultured in the presence of α-amanitin for 2 days prior to isolation and analysis of the RNA (19, 20). Growth in the presence of α-amanitin forces the cells to survive on the truncated, but α-amanitin-resistant, Rpb1 that was provided by transfection. Since CTD truncations have promoter-specific effects on transcription (22), the failure to recover processed RNA could reflect both direct and indirect effects of the impaired Rpb1 on RNA metabolism.
In vitro, 25 heptads support cleavage and polyadenylation only weakly—the full 52-heptad length is required for maximal activity (55). The in vitro reaction is also modestly favored by the variant heptads in the distal half of the CTD (55). For poly(A)-dependent termination in vivo, we find neither this length preference nor any advantage conferred by the variant heptads in the second half of the CTD (Fig. 4). We do not know whether these different requirements reflect differences between the processing and the termination reactions or differences between in vitro and in vivo conditions.
CTD-less polymerase supports poly(A)-dependent pausing.
Instead of responding with a decrease in polymerase density, CTD-less polymerases respond to the poly(A) signal with an increase in polymerase density that persists far down the template (Fig. 2 and 3). The downstream pausing for CTD-less polymerases is much more dramatic than for normal polymerases. This may reflect the absence of any offsetting termination. These data, together with previous work (49), allow us to say the following about poly(A)-dependent pausing. First, pausing is unquestionably triggered by the poly(A) signal, or at least its AAUAAA hexamer, because all poly(A)-dependent effects are defined exclusively by wild type-mutant comparisons (where mutants differ from wild types only by two or three positions in their hexamers). Second, the poly(A) signal causes pausing to occur at positions downstream on the template. Third, as illustrated by the CTD-less polymerases, pausing does not occur at a discrete distance downstream of the poly(A) site but rather takes place across a large region (Fig. 2 and 3). Fourth, the lack of a requirement for the CTD indicates that the signal to begin pausing is delivered directly to the body of the polymerase. We now discuss these last two points at greater length.
By eliminating termination, the CTD-less polymerases reveal that poly(A)-dependent pausing is not a unique event that occurs at a particular distance downstream of the poly(A) site. This distinguishes poly(A)-dependent pausing clearly from the conventional idea of pause “sites.” Most likely, poly(A)-dependent pausing reflects a long-lasting change in the polymerase, causing it to pause iteratively as it proceeds down the template. In fact, polymerases normally have some probability of pausing at every step of nucleotide addition (10, 47), and average rates of transcription reflect largely the action of elongation factors that modify the length and probability of this pausing (10). Therefore, it is likely that the poly(A) signal down-regulates one or more positive elongation factors or up-regulates a negative one. Interestingly, recent chromatin immunoprecipitation experiments with yeast suggest that some elongation factors dissociate from the transcription complex as it crosses the poly(A) signal (32).
Since the poly(A) signal does not require the CTD in order to enhance pausing, it probably communicates with one or more factors that bind the body of the polymerase. One candidate is CPSF, which reportedly binds to the polymerase (13) yet does not interact well with the CTD (19). The common perception that CPSF does bind the CTD may reflect the fact that CPSF and CstF exist together as a complex (57) and so bind the CTD through CstF50 (19). CPSF recognizes the AAUAAA hexamer of the poly(A) signal (45). Perhaps CPSF rides the body of the polymerase (13), binds the poly(A) signal hexamer when it emerges (42), and then signals the polymerase to slow down. It is possible that CPSF itself possesses elongation factor activity; alternatively, CPSF may interact with an elongation factor or, finally, the poly(A) signal may interact with other elongation factors directly. Candidate elongation factors that are known or thought to bind the body of the polymerase include TFIIF (9, 59), Fcp1 (34, 41), DSIF (38), NELF (46), and Ssu72 (14). All of these are multifunctional factors (for example, Fcp1 is best known as a CTD phosphatase) whose repertoires include direct effects on elongation. Thus, TFIIF, Fcp1, and DSIF act to accelerate elongation under appropriate conditions in vitro, whereas NELF and Ssu72 intensify pausing.
Although iteratively pausing (slowly elongating) polymerases persist for at least 1 kb downstream of the poly(A) site in the ΔCTD experiments (Fig. 2 and 3), they do not persist for ever. The way in which our assay detects pausing is through a higher polymerase density in the downstream cassette than in the upstream cassette. If the polymerases remained slow indefinitely, they would encounter the G-less cassettes multiple additional times as they circled the plasmid, and the distinction in polymerase density between the upstream and downstream cassettes would vanish. Therefore, at some point farther downstream, either the slow polymerases terminate by a different mechanism or they resume transcription at normal rates and then respond to the poly(A) signal again the next time around. Termination by an alternative mechanism is consistent with the fact that the CTD-binding CstF50 appears to be dispensable in C. elegans (see above). Possibly the polymerases, having been modified by the poly(A) signal at the pause step, become substrates for backup, non-CTD-dependent mechanisms of termination, such as transcript release factor 2 (25) or other DNA binding proteins (4, 12).
The two-step, pause-release mechanism of poly(A)-dependent termination.
We have shown that poly(A)-dependent termination involves both enhanced pausing (slowing down), which depends only on the body of the polymerase, and release, a step that requires also the CTD. Data presented here and previously (33, 49) show that both steps require the poly(A) signal. Thus, we know that downstream pausing requires the poly(A) signal because, as pointed out above, pausing is observed for wild-type but not mutant poly(A) signals. And we know that release requires the poly(A) signal because cis antisense inactivation of the poly(A) signal following the onset of pausing prevents release (33). Thus, it appears that these two processes, release and downstream pausing, are mechanistically independent—they depend on different parts of the polymerase, and they require separate input from the poly(A) signal.
Is the poly(A) signal a composite element in which separate pausing and release functions are superimposed? If so, one might expect to find elements having only one but not both of these functions. Such elements may exist. Peterson et al. (50) described an element, unrelated to poly(A) signals, that leads to increased polymerase densities downstream but no processing or termination. Thus, the element appears to induce downstream pausing in the absence of release. If poly(A)-dependent pausing is a processing and termination checkpoint (49), candidates for elements that cause termination but not pausing may include the 3′-end processing signals of RNA polymerase II genes whose transcripts are not subject to complex processing alternatives for which a checkpoint would be required. The replication-dependent histone genes, whose transcripts are not spliced or polyadenylated but whose 3′-end processing signals are nevertheless required for termination (8, 23), may fall in this class.
The CTD and abortive elongation.
The premature termination that takes place during abortive elongation near the promoter differs mechanistically from poly(A)-dependent termination (49). Here we have shown that these two types of termination also have different CTD requirements. Poly(A)-dependent termination is supported equally by full-length Rpb1 or by Rpb1 constructs lacking either the first or the second half of the CTD (Fig. 4). All of these constructs differ dramatically from the ΔCTD construct in their ability to support poly(A)-dependent termination (Fig. 2 and 3). In contrast, for premature termination the 1-25 Rpb1 construct behaves identically to the ΔCTD construct and very differently from the 27-52 and full-length CTD constructs (Fig. 5).
It is possible that the abortive elongation shown in Fig. 5 does not reflect premature termination but, instead, promoter proximal pausing. Thus, copious pausing near the promoter would drive up the polymerase density in the 131-bp cassette, and the gradual resumption of normal elongation downstream would yield decreasing polymerase densities in the 174-bp cassette. According to this interpretation, the ΔCTD and 1-25 Rpb1 constructs would engage poorly in promoter-proximal pausing relative to the full-length and 27-52 Rpb1 constructs. Although we cannot presently distinguish between the premature termination and the promoter-proximal pausing possibilities, the general considerations discussed below are similar by either model. In what follows we will simply refer to abortive elongation.
The abortive elongation illustrated in Fig. 5 takes place across the first several hundred base pairs downstream of the promoter. In contrast, the SV40 early poly(A) site (Fig. 2 and 4A) lies 2.4 kb downstream of the transcriptional start site, and the SV40 late poly(A) site (Fig. 3 and 4B) lies 1.2 kb downstream. Thus, it is only polymerases that survive abortive elongation in our constructs that manage to get far enough downstream to engage in poly(A)-dependent termination (Fig. 2 to 4). This shows that whatever differences may exist in the efficiency with which full processivity is established near the promoter (Fig. 5), the full or partial CTD-containing polymerases that ultimately survive go on to respond to the poly(A) signal similarly (Fig. 4).
Importantly, although 1-25 Rpb1 polymerases are indistinguishable from ΔCTD polymerases during abortive elongation (Fig. 5), their response to the poly(A) signal downstream is dramatically different (termination for 1-25 construct versus pausing for ΔCTD construct). The absence of any discernible effect of heptads 1-25 at the beginning of transcription suggests that the termination ability conferred by these heptads reflects interactions of the CTD during elongation, such as with proteins recruited by the poly(A) signal (55), rather than indirect effects stemming from events at the promoter.
Acknowledgments
We thank David Bentley for the Rpb1 expression plasmids and for helpful discussions.
This work was supported by NIH grant GM50863.
REFERENCES
- 1.Aranda, A., and N. Proudfoot. 2001. Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell 7:1003-1011. [DOI] [PubMed] [Google Scholar]
- 2.Bagga, P. S., L. P. Ford, F. Chen, and J. Wilusz. 1995. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 23:1625-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barilla, D., B. A. Lee, and N. J. Proudfoot. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:445-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossone, S. A., C. Asselin, A. J. Patel, and K. B. Marcu. 1992. MAZ, a zinc finger protein, binds to c-MYC and C2 gene sequences regulating transcriptional initiation and termination. Proc. Natl. Acad. Sci. USA 89:7452-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvo, O., and J. L. Manley. 2003. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 17:1321-1327. [DOI] [PubMed] [Google Scholar]
- 6.Carswell, S., and J. C. Alwine. 1989. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol. Cell. Biol. 9:4248-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, R. D., B. Palancade, A. Lang, O. Bensaude, and D. Eick. 2004. The last CTD repeat of the mammalian RNA polymerase II large subunit is important for its stability. Nucleic Acids Res. 32:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chodchoy, N., N. B. Pandey, and W. F. Marzluff. 1991. An intact histone 3′-processing site is required for transcription termination in a mouse histone H2a gene. Mol. Cell. Biol. 11:497-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, W. H., J. L. Craighead, W. H. Chang, C. Ezeokonkwo, A. Bareket-Samish, R. D. Kornberg, and F. J. Asturias. 2003. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell 12:1003-1013. [DOI] [PubMed] [Google Scholar]
- 10.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 11.Connelly, S., and J. L. Manley. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 2:440-452. [DOI] [PubMed] [Google Scholar]
- 12.Connelly, S., and J. L. Manley. 1989. RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol. Cell. Biol. 9:5254-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dantonel, J. C., K. G. Murthy, J. L. Manley, and L. Tora. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389:399-402. [DOI] [PubMed] [Google Scholar]
- 14.Dichtl, B., D. Blank, M. Ohnacker, A. Friedlein, D. Roeder, H. Langen, and W. Keller. 2002. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 10:1139-1150. [DOI] [PubMed] [Google Scholar]
- 15.Dichtl, B., D. Blank, M. Sadowski, W. Hubner, S. Weiser, and W. Keller. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 21:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dower, K., and M. Rosbash. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA nuclear export. RNA 8:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyer, C., and P. Hausen. 1978. On the activity of RNA polymerase B in lysates from Ehrlich ascites cells. Eur. J. Biochem. 86:241-253. [DOI] [PubMed] [Google Scholar]
- 18.Eick, D., F. Kohlhuber, D. A. Wolf, and L. J. Strobl. 1994. Activation of pausing RNA polymerases by nuclear run-on experiments. Anal. Biochem. 218:347-351. [DOI] [PubMed] [Google Scholar]
- 19.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fong, N., G. Bird, M. Vigneron, and D. L. Bentley. 2003. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 22:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gariglio, P., M. Bellard, and P. Chambon. 1981. Clustering of RNA polymerase B molecules in the 5′ moiety of the adult beta-globin gene of hen erythrocytes. Nucleic Acids Res. 9:2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerber, H. P., M. Hagmann, K. Seipel, O. Georgiev, M. A. West, Y. Litingtung, W. Schaffner, and J. L. Corden. 1995. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 374:660-662. [DOI] [PubMed] [Google Scholar]
- 23.Gu, X., and W. F. Marzluff. 1996. 3′ processing and termination of mouse histone transcripts synthesized in vitro by RNA polymerase II. Nucleic Acids Res. 24:3797-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammell, C. M., S. Gross, D. Zenklusen, C. V. Heath, F. Stutz, C. Moore, and C. N. Cole. 2002. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22:6441-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara, R., C. P. Selby, M. Liu, D. H. Price, and A. Sancar. 1999. Human transcription release factor 2 dissociates RNA polymerases I and II stalled at a cyclobutane thymine dimer. J. Biol. Chem. 274:24779-24786. [DOI] [PubMed] [Google Scholar]
- 26.He, X., A. U. Khan, H. Cheng, D. L. Pappas, Jr., M. Hampsey, and C. L. Moore. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 17:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirayoshi, K., and J. T. Lis. 1999. Nuclear run-on assays: assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 304:351-362. [DOI] [PubMed] [Google Scholar]
- 28.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 29.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs, E. Y., I. Ogiwara, and A. M. Weiner. 2004. Role of the C-terminal domain of RNA polymerase II in U2 snRNA transcription and 3′ processing. Mol. Cell. Biol. 24:846-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen, T. H., K. Dower, D. Libri, and M. Rosbash. 2003. Early formation of mRNP. License for export or quality control? Mol. Cell 11:1129-1138. [DOI] [PubMed] [Google Scholar]
- 32.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, S. J., and H. G. Martinson. 2003. Poly(A)-dependent transcription termination: continued communication of the poly(A) signal with the polymerase is required long after extrusion in vivo. J. Biol. Chem. 278:41691-41701. [DOI] [PubMed] [Google Scholar]
- 34.Kimura, M., H. Suzuki, and A. Ishihama. 2002. Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell. Biol. 22:1577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krumm, A., T. Meulia, M. Brunvand, and M. Groudine. 1992. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 6:2201-2213. [DOI] [PubMed] [Google Scholar]
- 36.Kyburz, A., M. Sadowski, B. Dichtl, and W. Keller. 2003. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res. 31:3936-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licatalosi, D. D., G. Geiger, M. Minet, S. Schroeder, K. Cilli, J. B. McNeil, and D. L. Bentley. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101-1111. [DOI] [PubMed] [Google Scholar]
- 38.Lindstrom, D. L., and G. A. Hartzog. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159:487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lutz, C. S., and J. C. Alwine. 1994. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 8:576-586. [DOI] [PubMed] [Google Scholar]
- 40.MacMorris, M., C. Brocker, and T. Blumenthal. 2003. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9:847-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandal, S. S., H. Cho, S. Kim, K. Cabane, and D. Reinberg. 2002. FCP1, a phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol. Cell. Biol. 22:7543-7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 43.McCracken, S., D. Longman, I. L. Johnstone, J. F. Caceres, and B. J. Blencowe. 2003. An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J. Biol. Chem. 278:44153-44160. [DOI] [PubMed] [Google Scholar]
- 44.McCracken, S., E. Rosonina, N. Fong, M. Sikes, A. Beyer, K. O'Hare, S. Shuman, and D. Bentley. 1998. Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harb. Symp. Quant. Biol. 63:301-309. [DOI] [PubMed] [Google Scholar]
- 45.Murthy, K. G., and J. L. Manley. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9:2672-2683. [DOI] [PubMed] [Google Scholar]
- 46.Narita, T., Y. Yamaguchi, K. Yano, S. Sugimoto, S. Chanarat, T. Wada, D. K. Kim, J. Hasegawa, M. Omori, N. Inukai, M. Endoh, T. Yamada, and H. Handa. 2003. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 23:1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neuman, K. C., E. A. Abbondanzieri, R. Landick, J. Gelles, and S. M. Block. 2003. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell 115:437-447. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen, V. T., F. Giannoni, M. F. Dubois, S. J. Seo, M. Vigneron, C. Kedinger, and O. Bensaude. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 24:2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orozco, I. J., S. J. Kim, and H. G. Martinson. 2002. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem. 277:42899-42911. [DOI] [PubMed] [Google Scholar]
- 50.Peterson, M. L., S. Bertolino, and F. Davis. 2002. An RNA polymerase pause site is associated with the immunoglobulin μs poly(A) site. Mol. Cell. Biol. 22:5606-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Proudfoot, N., and J. O'Sullivan. 2002. Polyadenylation: a tail of two complexes. Curr. Biol. 12:R855-R857. [DOI] [PubMed] [Google Scholar]
- 53.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 54.Rougvie, A. E., and J. T. Lis. 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54:795-804. [DOI] [PubMed] [Google Scholar]
- 55.Ryan, K., K. G. Murthy, S. Kaneko, and J. L. Manley. 2002. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol. Cell. Biol. 22:1684-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinmetz, E. J., and D. A. Brow. 2003. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 23:6339-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takagaki, Y., and J. L. Manley. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20:1515-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeung, G., L. M. Choi, L. C. Chao, N. J. Park, D. Liu, A. Jamil, and H. G. Martinson. 1998. Poly(A)-driven and poly(A)-assisted termination: two different modes of poly(A)-dependent transcription termination. Mol. Cell. Biol. 18:276-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, C., H. Yan, and Z. F. Burton. 2003. Combinatorial control of human RNA polymerase II (RNAP II) pausing and transcript cleavage by transcription factor IIF, hepatitis delta antigen, and stimulatory factor II. J. Biol. Chem. 278:50101-50111. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]