Activating mutations in phosphatidylinositol 3-kinase (PI3K)-AKT pathway components cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome (MPPH)1-3. We report that MPPH patients lacking upstream PI3K-AKT pathway mutations carry de novo mutations in cyclin D2 (CCND2), clustered around a residue that can be phosphorylated by glycogen synthase kinase 3β (GSK-3β)4. Mutant CCND2 was resistant to proteasomal degradation compared to wild type (WT) protein in vitro. The PI3K-AKT pathway modulates GSK-3β activity4 and accordingly, cells from patients with PIK3CA, PIK3R2 or AKT3 mutations similarly accumulated CCND2. CCND2 was expressed at higher levels in the brains of mouse embryos expressing activated Akt3. In utero electroporation of mutant CCND2 into the embryonic mouse brain produced more proliferating transfected progenitors and a smaller fraction exiting the cell cycle compared to cells electroporated with the WT sequence. These observations suggest that cyclin D2 stabilization, due to CCND2 mutation or PI3K-AKT activation, provides a unifying mechanism in PI3K-AKT related megalencephaly syndromes.
Germline mutations of PIK3R2 and AKT3 result in MPPH3, while postzygotic mutations of PIK3CA cause megalencephaly-capillary malformation syndrome (MCAP)3, and postzygotic mutations of PIK3CA and AKT3 cause isolated hemimegalencephaly1,5. We previously failed to detect mutations in PIK3R2 or AKT3 in 35% of probands with MPPH3. We hypothesized that mutation-negative patients have de novo mutations in one or more novel genes, and performed whole exome sequencing in two child parent trios (JT-144, LR11-424) and one child-parent pair (LR02-064).
We identified germline mutations in CCND2 in all three patients (p.Thr280Ala in one and p.Lys270* in 2 unrelated subjects), confirmed to be de novo by Sanger sequencing. None of the identified variants were present in dbSNP build 132, the 1000 Genomes Project, the NHLBI exome variant server of 6500 exomes, or in-house exome databases (Online Methods). We identified de novo heterozygous CCND2 mutations in nine further MPPH patients by conventional Sanger sequencing. The clinical and molecular findings of this cohort are shown in Figure 1, and summarized in Table 1, and Supplementary Table 1.
Figure 1.
Images of children with MPPH and CCND2 mutations. Photos and brain MRI of 4 affected individuals are shown: (a-d) LR07-041, (e-h) LR02-064, (i-l) JT-144, and (m-p) JT-210. (a, e, i, m, n) Photographs demonstrate macrocephaly with prominent forehead, and (b, f, j) postaxial polydactyly of the hands. (c, g, k, o) T1- or T2-weighted midsagittal images demonstrate large brain size in relation to facial structures, (d, h, l, p) while axial images show polymicrogyria that appears most severe in the perisylvian regions, but also involves other regions. All patients gave informed consent for publication.
Table 1.
Summary of the phenotypic and molecular data of 12 MPPH individuals with CCND2 mutations
| Subject ID | Sex | Age last assessed |
OFC | HYD or VMEG |
Shunt, age at shunting |
PMG** | Polydactyly§ | ID | cDNA change |
Amino-acid change |
Inheritance | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First OFC SD (age) |
Last OFC SD (age) |
|||||||||||
| LR02-064* | F | 6y | +3 (5days) | +3 (6y) | HYD | +, 6y | BPP grade 1 | ++/++ | Severe, no language |
c.808A>T | p.Lys270X | de novo |
| LR11-424* | F | 18m | +4 (birth) | +4 (18m) | VMEG | – | BPP grade 2 | +−/++ | Severe | c.808A>T | p.Lys270X | de novo |
| JT-144a* | F | 3y | +3 (birth) | +2.5 (3y) | VMEG | – | BPP grade 2 | ++/++ | Severe, no language |
c.838A>G | p.Thr280Ala | de novo |
| VI13216 | M | 7y | +4 (birth) | +7.5 (7y) | VMEG | – | BPP grade 1 | ++/−− | Severe | .838A>G | p.Thr280Ala | de novo |
| LR07-041b | F | 8m | +4 (6w) | +2 (8m) | VMEG (mild) |
– | BPP grade 1 | ++/−− | Severe, no language |
c.839C>A | p.Thr280Asn | de novo |
| LR11-352c | F | 13y | +4.5 (2w) | +5 (13y) | VMEG | – | BPP grade 2 | ++/++ | Mild LD, ambulatory |
c. 839C>A | p.Thr280Asn | de novo |
| JT-232 | M | 5y | +3.5 (2days) |
+6 (5y) | VMEG | – | BPP grade 2 | −−/++ | Severe, no language |
c.839C>A | p.Thr280Asn | Parents NA |
| LR11-346d | F | 28wk gestation |
MEGb | NA | VMEG (mild) |
– | Frontoparieta l PMG |
++/−− | NA | c.841C>T | p.Pro281Ser | de novo |
| LP95-025 | F | 20y | + 4 (3y) | +2.5 (8y) | HYD | +, 1m | BPP grade 2 | −−/−− | Severe, no language |
c.842C>G | p.Pro281Arg | de novo |
| JT-210e | F | 2y | +2 (birth) | +2 (2y) | VMEG (mild) |
– | BPP grade 2 | ++/−+ | Severe | c.842C>G | p.Pro281Arg |
Mother negative,
father NA |
| LR03-260 | M | 9y | +2 (birth) | +5 (9y) | VMEG | – | BPP grade 1 | ++/++ | Severe | c.842C>T | p.Pro281Leu |
Supplementary
Note |
| JT-238 | F | 11y | +2.5 (birth) | +3.5 (11y) | VMEG (L>R) |
– | BPP grade 1 | ++/++ | Severe | c.851T>G | p.Val284Gly |
Mother negative,
father NA |
Abbreviations: BPP, bilateral perisylvian polymicrogyria; F, female; HYD, hydrocephalus; ID, intellectual disability; LD, learning disability, m, months; M, male; MEG, megalencephaly; NA, not available/not applicable; OFC, occipitofrontal circumference; PMG, polymicrogyria; SD, standard deviations; VMEG, ventriculomegaly; w, weeks; y, years.
Subjects analyzed by exome sequencing. Mutation locations are based on reference gene accession NM_001759 (CCND2). All mutations were absent from the Exome Variant Server, which consists of whole-exome sequencing data from 6500 subjects (accessed January 2012).
Bilateral perisylvian polymicrogyria (BPP) grades: grade 1, PMG extending beyond the perisylvian regions to the frontal or occipital poles or both; grade 2, PMG extending beyond the perisylvian regions but not to either pole
Postaxial polydactyly: both hands / both feet
Additional phenotype information:
JT-144: this child’s non-identical twin sister is healthy.
LR07-041: this child had progressively worsening irritability, severe episodes of opisthotonus, Cheyne-Stokes respirations, poor eye contact, and died at 8 months of age.
LR11-352: this child walked unassisted at 5 years. Her speech is limited to 2-word phrases.
LR11-346: this pregnancy was terminated at 28 weeks of gestation. On autopsy examination, the brain weighed 3x the normal weight.
JT-210: this child has cortical visual blindness and a ventricular septal defect that closed spontaneously
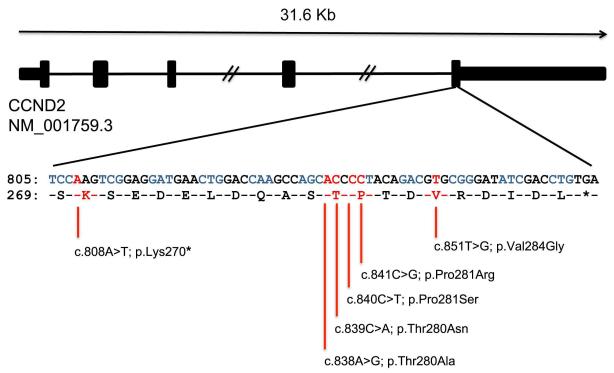
All identified mutations affect highly conserved amino acid residues (Fig. 2 and Supplementary Fig.1). Notably, 9 of 12 involved p.Thr280 or p.Pro281 (Table 1). The p.Thr280Asn mutation occurred in 3 subjects while p.Lys270*, p.Thr280Ala, and p.Pro281Arg were found in 2 subjects each. Murine cyclin D2 can be phosphorylated on p.Thr280 in vitro by GSK-3β, rendering Ccnd2 susceptible to ubiquitin/proteasome-dependent degradation4. The p.Thr280Ala mutant cyclin D2 cannot be phosphorylated, does not become polyubiquitinated, and thus is resistant to degradation4. Interestingly, all mutations in our MPPH cohort either alter highly conserved residues surrounding p.Thr280, or truncate the protein prior to this phosphorylation site (p.Lys270X). As the latter mutation lies in the final exon, it is predicted not to induce nonsense-mediated mRNA decay.
Figure 2.
Cyclin D2 (CCND2) mutations and protein conservation. Schematic showing the intron/exon structure of human CCND2. The cDNA sequence from c.805 and corresponding peptide sequence from p.S269 are shown with positions mutated in MPPH subjects marked in red. See also Supplementary Figure 1.
To test whether mutations at particular residues in MPPH patients result in accumulation of degradation-resistant cyclin D2, we transfected HEK293 cells with either WT or mutant (p.Thr280Ala, p.Pro281Arg, p.Val284Gly, and p.Lys270*) human CCND2 myc-tagged constructs. Cells were treated with cycloheximide at 0.05 mg/ml 24 hours post-transfection to block protein translation, and protein extracts were analyzed by Western blotting (Fig. 3a,b). A rapid reduction in cyclin D2 was seen in cells transfected with WT constructs following cycloheximide treatment. This was not observed in cells transfected with mutant constructs. Thus, MPPH-associated mutations in CCND2 result in accumulation of unphosphorylated degradation-resistant cyclin D2.
Figure 3.
Cyclin D2 (CCND2) protein stabilization by MPPH mutations. (a) Representative blots from CCND2 transfection experiments. Protein extracts from HEK293 cells transfected with WT or mutant CCND2 treated with cycloheximide (CHX) at time point 0 were analyzed by western blotting with an antibody against CCND2. WT CCND2 constructs demonstrate a drop in protein levels following inhibition of protein translation by CHX, while mutant constructs show apparently stable levels of the protein. (b) Quantification of CCND2 protein levels in transfected HEK293 cells as a proportion of β-actin levels averaged over three experiments. Relative densities of bands were determined relative to 1 at time point 0 for each experiment and normalized using the β-actin loading control. (c) Representative blots from patient lymphoblastoid experiments. Protein extracts from lymphoblastoid cells treated with CHX were subjected to Western blotting using an anti-CCND2 antibody. Control cells show a decline of endogenous CCND2 following CHX treatment while cells from patients with mutations in PI3K-AKT pathway genes previously implicated in MCAP/MPPH appear to maintain relatively stable levels of the protein. (d) Quantification of CCND2 protein levels in lymphoblastoid cell lines as a proportion of β-actin levels averaged over three experiments. Relative densities of bands were determined relative to 1 at time point 0 for each experiment and normalized using a β-actin loading control. Mean values and standard error bars are shown * p< 0.05, ** p<0.01, *** p< 0.0001 (e) Quantitative western blot of E14.5 brain from embryos homozygous for constitutively active Akt3-D219V shows equal loading of total GSK-3β, but elevated pS9GSK-3β and CCND2 levels. Mean values and standard error bars are shown, *p<0.05.
Activation of the PI3K-AKT pathway inhibits GSK-3β-mediated phosphorylation4. We hypothesized that the activating mutations previously observed in PIK3CA, PIK3R2, and AKT3 would also result in accumulation of degradation-resistant cyclin D2 through downstream inhibition of GSK-3β. Endogenous CCND2 levels were examined in lymphoblastoid cell lines derived from patients with mutations in PIK3CA (p.Glu453del), PIK3R2 (p.Gly373Arg), or AKT3 (p.Arg465Trp). Following cycloheximide treatment, we observed a decrease in CCND2 levels in control cells. In contrast, CCND2 in patient cells was resistant to degradation (Fig. 3c,d). These results suggest that megalencephaly-associated mutations in components of the PI3K-AKT pathway share the same functional endpoint as the mutations observed in CCND2: namely inhibition of proteasomal degradation of cyclin D2.
The PI3K-AKT pathway inactivates GSK-3β by phosphorylation of the S9 residue6, which should interfere with GSK-3β dependent phosphorylation, ubiquitination, and proteasomal degradation of cyclin D2. Activating mutations previously described in PIK3R2 and PIK3CA result in activation of AKT33. To further test the relationship between cyclin D2 and this pathway in vivo, we examined the levels of CCND2 in E14.5 embryonic cortex of mice homozygous for a gain-of-function Akt3D219V mutant allele7. Quantitative western blot analysis revealed a 30% increase in the pS9, inactive GSK-3β phosphoprotein, associated with a 30-40% increase in cyclin D2, equivalent to the increase in cyclin D1 (Fig. 3e).
Neocortical neurons and glia are generated in the embryonic ventricular and subventricular zones (VZ and SVZ)8. Cyclin D2 promotes neuronal progenitor commitment to undergo a round of division and progress through the cell cycle9-11. In embryonic mouse brain, Ccnd2 is expressed in some VZ radial glial cells (RGCs) that co-label for Pax6, but primarily – though certainly not exclusively – Ccnd2- positive cells co-localize with Tbr2-positive intermediate progenitor cells (IPCs) in the SVZ12,13. IPCs are important drivers of brain size, since they allow for geometric expansion of cellular output from the SVZ10. Ccnd2 null mice display microcephaly and cerebellar hypoplasia14,15. In the human neocortex, CCND2 is expressed more robustly than cyclin D1 by 19 gestational weeks15. Thus, elevated CCND2 levels may enlarge brain size by maintaining cells in cycle and increasing progenitor numbers, while diminished CCND2 expression reduces brain volume by allowing cell cycle exit, resulting in progenitor pool depletion14.
We investigated the importance of CCND2 p.Thr280 and the neighboring p.Pro281 residue for VZ/SVZ divisions by testing mutations, including those found in MPPH patients, using in utero electroporation (IUEP) in mouse embryos (Online Methods). Western blot of HEK293 cells transfected with these constructs demonstrated that CCND2 sequences placed upstream of the IRES-eGFP cassette were actively translated into protein (Supplementary Fig. 2). Immunostaining of embryonic cortex following IUEP confirmed that expression of nuclear CCND2 in GFP-positive cells was elevated after transfection with mutant forms of CCND2 compared to cells transfected with GFP only or with untransfected cells (i.e. GFP-) (Supplementary Fig. 2).
IUEP was performed at embryonic day 13.5 (E13.5) and embryos were collected 48 hours later. Immunohistochemistry using anti-GFP and anti-Ki67 antibodies (Fig. 4) showed that 85% of control cells expressing GFP alone did not co-label for Ki67, and therefore, had exited the cell cycle; a large proportion had migrated to the cortical plate (Fig. 4 a,f,k). Significantly more GFP-positive cells expressing WT cyclin D2 also co-labeled for Ki67 and were actively dividing (27.6 ± 3.0% of GFP+ WT cyclin D2 vs. 14.6 ± 2.6% of GFP+ only controls) (Fig. 4 b,g,k). Electroporation of a mutant construct mimicking phosphorylation at Thr280 (p.Thr280Asp) (Fig. 4 d,i,k) yielded results statistically indistinguishable from the WT CCND2, suggesting that the transfected WT protein was near-maximally phosphorylated at residue 280. However, compared to the exogenous WT protein, cells expressing mutant CCND2 (p.Thr280Ala) had a significantly higher percentage of GFP+/Ki67+ co-labeled cells (54.9 ± 4.9% of GFP+ p.Thr280Ala vs. 27.6 ± 3.0% of GFP+ WT cyclin D2 and 35.5 ± 5.3% GFP+ p.Thr280Asp) (Fig. 4 c,h,k). Another MPPH-mutation (p.Pro281Arg) also produced a higher percentage of proliferating GFP+ cells than the WT sequence (60.5 ± 4.2% GFP+ p.Pro281Arg vs. 27.6 ± 3.0% of GFP+ WT cyclin D2, 35.5 ± 5.3% GFP+ p.Thr280Asp) (Fig 4 e,j,k). The significance of these comparisons is summarized in Supplementary Table 2. These data support a mechanism in which aberrantly stabilized CCND2 mediates abnormal proliferation leading to increased neural progenitor numbers.
Figure 4.
Comparative ability of mutant and WT cyclin D2 (CCND2) to enhance neural progenitor proliferation in embryonic mouse brain. Two patient mutations (p.Thr280Ala and p.Pro281Arg) are compared to WT and phosphomimetic (p.Thr280Asp) CCND2. WT and mutant cDNAs were cloned into an IRES eGFP vector affording independent translation of both CCND2 and, from an internal ribosome entry site, eGFP. Electroporated (GFP+) cells expressed either GFP alone or together with human WT or mutant CCND2. Constructs (1 μg each) were delivered to cortex by IUEP at E13.5 and embryos were harvested at E15.5. (a-e) Cortical sections were immunostained with an anti-GFP antibody. (f-j) The same sections were double immunolabeled (yellow) with anti-GFP (green) and proliferation marker, anti-Ki67 (red), antibodies. (a,f) Cells receiving vector without CCND2 are mostly no longer dividing and many have migrated up to the cortical plate. (b,g) More cells continue to divide (yellow cells, mostly in the SVZ/IZ) when overexpressing WT CCND2. (c,h,e,j) Progenitors expressing the phosphodeficient p.Thr280Ala or putative phosphodeficient p.Pro281Arg CCND2 found in MPPH patients show strikingly more proliferation and fewer GFP+ cells residing in the cortical plate. (d,i) A phosphomimetic (p.Thr280Asp) form of CCND2 is significantly less effective in promoting proliferation than the patient mutations. (k) Quantification of the proportions of transfected (GFP+) cells that are proliferating (GFP+Ki67+/GFP+ total cells) *p<0.05, **p<0.001 with respect to GFP-only (control); §p<0.0001 with respect to WT-CCND2 or p.Thr280Asp as indicated. Full statistical summary is shown in Supplementary Table 2. n=7 embryos (a), n=5 embryos each (b-e), n=embryos, each value average of 5 sections/embryo. MZ=marginal zone, CP=cortical plate, IZ=intermediate zone, SVZ=subventricular zone, VZ=ventricular zone. Scale bar = 20 μm.
MPPH-associated CCND2 mutation effects on cell cycle function were investigated by determining the proportion of electroporated (GFP-positive) cells undergoing mitosis. We used immunodetection of phosphohistone H3 (PH3), a marker of cells in M-phase, following transfection with WT, phosphodeficient (p.Thr280Ala), and phosphomimetic (p.Thr280Asp) forms of CCND2. Electroporation of the WT CCND2 sequence resulted in 2.83 ± 0.37% of GFP+ cells also being labeled for PH3, most of which were located within the VZ and SVZ (Fig. 5 a,d). The phosphodeficient CCND2 construct significantly increased cell numbers labeled for GFP and PH3 (Fig. 5 b,d) compared to the WT sequence (6.41 ± 0.22% p.Thr280Ala vs. 2.83 ± 0.37% WT CCND2, p=0.000054). Lastly, expression of the phosphomimetic sequence yielded a 3.39 ± 0.49% overlap between GFP and PH3-immunoreactivity (Fig. 5 c,d), which was significantly different from the phosphodeficient, but not the WT CCND2, construct (Fig. 5d). Thus, the phosphodeficient form of CCND2 is more effective in promoting mitosis compared to transfection with either the WT or phosphomimetic CCND2 sequences.
Figure 5.
Phosphodeficient CCND2 more effectively promotes proliferation and prevents neural progenitor cells from exiting the cell cycle. (a-d) Tissue sections immunostained with anti-GFP (green) and anti-PH3 (purple) antibodies following electroporation with WT, phosphodeficient (p.Thr280Ala), and phosphomimetic (p.Thr280Asp) CCND2. (a,b) Compared to WT CCND2 introduction of the phosphodeficient CCND2 resulted in a significant increase in the number of GFP+ and PH3+ double-labeled cells in the VZ/SVZ, (c) while the phosphomimetic CCND2 was no different from WT. (d) Quantification shows significantly more GFP+ PH3+ cells after introduction of phosphodeficient CCND2 compared to the WT (p=0.000054) or phosphomimetic sequences (p=0.00049). There was no difference between the phosphomimetic and WT CCND2 forms (p=0.41). (e-l) Comparison of actively proliferating (P-Fraction) and non-proliferating (Exit Fraction) cortical progenitors transfected with WT or mutant CCND2 constructs. (e-g) Cells were co-labeled with anti-GFP (green) and anti-BrdU (red) antibodies following BrdU injection at E14.5 (24 hours post-IUEP) and harvest at E15.5. (h-j) The same samples were also co-labeled with anti-Ki67 (blue) antibodies. (e,h,k) Expression of either the WT or (g,j,k) phosphomimetic (p.Thr280Asp) forms of CCND2 resulted in a similar increase in the percentage of progenitors in the exit fraction over the P-fraction (see Methods for calculations), thereby indicating more cells had exited the cell cycle 24 hours post-BrdU injection. (f,i,k) Introduction of the phosphodeficient (p.Thr280Ala) form of CCND2 significantly increased the number of progenitors in the P-fraction (still proliferating 24 hours post-BrdU pulse) and correspondingly decreased the exit fraction, compared to both the WT and phosphomimetic forms of CCND2. Thus, phosphodeficient CCND2 was more effective in promoting mitotic events and keeping cells in cycle. Full statistical analyses are in Supplementary Table 3. GFP/PH3 experiment, n=4 WT CCND2, n=5 p.Thr280Ala, and n=5 p.Thr280Asp. GFP/BrdU/Ki67 experiment, n=5 WT CCND2, n=5 p.Thr280Ala, and n=5 p.Thr280Asp. n=embryos, each value average of 5 sections/embryo. Data are presented as mean ± standard error measurement. MZ=marginal zone, CP=cortical plate, IZ=intermediate zone, SVZ=subventricular zone, VZ=ventricular zone. Scale bar in (c) and (j) = 20 μm. *p<0.02 compared to WT CCND2, and §p<0.01 compared to p.Thr280Ala.
To examine the effect of WT and mutant CCND2 expression on cell cycle exit, bromodeoxyuridine (BrdU) was injected intraperitoneally into pregnant dams 24 hours post-IUEP. After 24 hours tissues were collected and sections stained with anti-GFP, anti-BrdU, and anti-Ki67 antibodies to determine the proliferative fraction (P-fraction, ie: cells that remain in the cell cycle, GFP+/BrdU+/Ki67+) and exit fraction (ie: those cells that have exited the cell cycle 24 hours post-BrdU injection, GFP+/BrdU+/Ki67-). Following electroporation of the WT CCND2, 40.8 ± 4.4% of GFP+ cells were in the P-fraction, while 59.2 ± 4.4% of cells accumulated in the exit fraction (Fig. 5e,h,k, Supplementary Table 3). After transfection with the phosphodeficient CCND2 mutation, progenitors displayed a higher P-fraction (59.3 ± 3.6%) and corresponding decrease in the exit fraction (40.7 ± 3.6%) relative to the WT CCND2 sequence (Fig. 5e,f,h,i,k, Supplementary Table 3). The phosphomimetic CCND2 construct yielded P-fractions (45.4 ± 1.4%) and exit fractions (54.6 ± 1.4%) statistically identical to the WT CCND2 sequence (Fig. 5e,g,h,j,k, Supplementary Table 3). However, expression of the phosphomimetic CCND2 construct resulted in a lower P-fraction and higher exit fraction compared to the phosphodeficient CCND2 construct (Fig. 5f,g,i,j,k, Supplementary Table 3). Thus, expression of phosphodeficient forms of CCND2 increases progenitor proliferation and the proportion of cells undergoing mitosis, with a concomitant decrease in the exit fraction.
Lastly, we investigated how transfection of cortical progenitors with WT and mutant CCND2 forms would impact the RGC and IPC populations. Previous work has detailed the transition of progenitors from Pax6-expressing RGCs to Tbr2-positive IPCs during development within the cortex16,17. Therefore, we examined the expression of Pax6 and Tbr2 in electroporated progenitors. First, we examined the proportion of electroporated progenitors that co-express Pax6. Introduction of the GFP only control generated 21.5 ± 1.5% of GFP+ cells that were positive for Pax6 as well (Supplementary Fig.3a). Electroporation with either the WT CCND2 or the phosphomimetic CCND2 construct did not result in an increase in the proportion of progenitors that were immunopositive for Pax6 (20.1 ± 1.1%, and 23.0 ± 0.6%, respectively). In contrast, electroporation of the phosphodeficient CCND2 construct caused a significant increase in the proportion of GFP+ progenitors that co-express Pax6, over all other groups (27.7 ± 1.4%), indicating that phosphodeficient CCND2 leads to expansion of RGCs in the developing cortex. We then found that the GFP only control resulted in 9.4 ± 0.7% of GFP+ cells also expressing Tbr2 (Supplementary Fig.3b). In contrast, the WT CCND2 sequence and expression of the phosphomimetic sequence generated significant increases in the proportion of electroporated cells (17.6 ± 2.1%, 15.3 ± 1.7%, respectively) that were also immunopositive for Tbr2, signifying an expansion of the IPC population. The phosphodeficient CCND2 mutant produced an even greater, 27.4 ± 2.9%, expansion of progenitors expressing both GFP and Tbr2, which was significantly different from the GFP control, WT CCND2, and phosphomimetic groups. Thus, the phosphodeficient, degradation resistant form of CCND2 causes an expansion of both the RGC and IPC population. We suggest it is this expansion that underlies the megalencephaly in MPPH patients. The WT CCND2 sequence resulted in an increase in the proportion of transfected progenitors that co-express Tbr2, but not Pax6, suggesting that WT CCND2 might preferentially promote expansion of the IPC population.
Polymicrogyria in MPPH patients may be related to an altered relationship between basal progenitors and radial glia. A homozygous loss of function mutation in EOMES (TBR2), expressed in IPCs, was previously associated with a microcephaly-polymicrogyria phenotype18. In fetal brain, CCND2 is expressed mostly in IPCs and altered cyclin D2 levels are expected to disrupt the proportion of IPC- vs. RGC-derived neurons and/or the differentiation program of IPCs14. We note that CCND2-mutated MPPH patients have polydactyly more frequently than those with PI3K-AKT pathway mutations (Supplementary Table 1), but the mechanism underlying this phenotypic difference remains to be explored.
Thus, we have identified a highly specific pattern of de novo mutations in CCND2 in 12 subjects with MPPH. We, and others previously reported activating mutations in PI3K-AKT pathway elements upstream of CCND2 in other patients with MPPH and overlapping megalencephaly syndromes1-3. Evidence presented here supports that these two types of mutational events result in a common functional endpoint, namely increased cyclin D2 activity in neural precursors due to its intracellular accumulation. The data suggest that pathological stabilization of cyclin D2 in the developing cerebral cortex may represent a unifying mechanism for the development of megalencephaly and polymicrogyria in MPPH and other PI3K-AKT associated megalencephaly syndromes (Supplementary Figure 4).
ONLINE METHODS
Whole exome sequencing
Alignment and variant calling (JT144)
Target capture was performed using the Agilent SureSelect All Exon v4 exome enrichment kit, according to standard manufacturer protocols. Sequencing of 150-bp paired end reads was performed using an Illumina MiSeq. Reads were aligned to GRCh37 using Novoalign (Novocraft Technologies, Selangor, Malaysia) and processed using the Genome Analysis Toolkit (GATK) and Picard (see URLs) to perform realignment of short insertions and deletions (indels) and remove duplicate reads. Depth of coverage of the consensus coding sequence (CCDS) was assessed using the GATK, which showed that >94 % of CCDS bases were covered by at least five good quality reads (minimum Phred-like base quality 17 and minimum mapping quality 20). Single nucleotide variants (SNVs) and indel variants were called using the UnifiedGenotyper feature of the GATK19,20.
Filtering (JT144)
Custom Perl scripts were used to remove variants present in dbSNP132 or with a minor allele frequency ≥ 0.1 % and to annotate functional consequences. We identified variants present in the child, but not their parents with a minimum Phred-like genotype quality of 30 and selected for indels within coding regions, non-synonymous SNVs and splice-site variants. Variants present the 1000 Genomes dataset (Nov. 2011), the NHLBI GO exomes and another 22 ethnically matched in-house exomes were also removed. This left four de novo variants (CCND2 [NM_001759] c.838A>G, p.Thr280Ala; HN1L [NM_144570] c.58C>G, p.Pro20Ala; GLG1 [NM_012201] c.1229A>G, p.His410Arg; BTG3 [NM_001130914] c.387del, p.Ala131Argfs*16) which were confirmed present in the child and absent in the parents by Sanger sequencing.
Alignment and variant calling (LR11-424 and LR02-064)
Target capture was performed using the Agilent SureSelect All Exon 50 MB exome enrichment kit according to standard manufacturer protocols. Sequencing of 100-bp paired end reads was performed using an Illumina HiSeq 2000, which generated over 13 Gb of sequence for each sample. We removed adaptor sequences and quality trimmed reads using the Fastx toolkit (see URLs) and then used a custom script to ensure that only read pairs with both mates present were subsequently used. Reads were aligned to hg19 with BWA 0.5.921, and indel realignment was done using GATK19,20. Duplicate reads were marked using Picard (see URLs) and excluded from downstream analyses. We assessed coverage of CCDS bases using the GATK, which showed that all samples had >=92.7% of CCDS bases covered by at least 10 reads, and >=88.4% of CCDS bases covered by at least 20 reads. SNVs and indels were called using samtools mpileup with the extended base alignment quality (BAQ) adjustment (-E), and were then quality filtered to require at least 20% of reads supporting the variant call22. Variants were annotated using both Annovar23, and custom scripts to annotate functional consequences, and whether they had previously been seen in dbSNP132, the 1000 Genomes dataset (Nov. 2011), the NHLBI GO exomes or in approximately 570 exomes previously sequenced at our centres.
Filtering (LR11-424 and LR02-064)
We removed all variants seen in the parents of the affected subjects, or in any of our 570 controls samples, the 6500 NHLBI GO exomes or the 1000 Genomes project. We focused on nonsynonymous SNVs, indels and splice site variants. This analysis revealed a single de novo variant, p.Lys270X in CCND2, in patient LR11-424. Surprisingly, LR02-064 had the same CCND2 variant, p.Lys270X, which was not present in the single parent that was exome sequenced.
Constructs for site-directed mutagenesis and transfection
A C-terminal myc-tagged cDNA clone of CCND2 transcript NM_001759 was purchased from Origene (Origene, Rockville, MD, USA, cat. RC210316). The constructs with p.Thr280Ala, p.Pro281Arg or p.Val284Gly mutations were created using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) as per manufacturer’s instructions with the primer pairs as in Supplementary Table 4.
Transfection and protein extractions
HEK293 cells were transfected with WT or mutant CCND2 constructs using Lipofectamine™ 2000 Transfection Reagent (Life Technologies, Paisley, UK) as per the manufacturer’s instructions. Media was changed 24 hours post-transfection for media containing 50 μg/ml cycloheximide (Sigma-Aldrich, Gillingham, UK) to block protein translation. Protein was extracted at 0, 60, 120 and 240 minute time points, using NP-40 lysis buffer (150 mM sodium chloride, 1.0% NP-40, 50 mM Tris, pH 8.0) containing protease and phosphatase inhibitors (Thermo Scientific, Loughborough, UK).
Lymphoblastoid cell line treatment and protein extraction
Lymphoblastoid cell lines derived from patients with mutations in PIK3R2 (p.Gly373Arg), PIK3CA (p.Glu453del) or AKT3 (p.Arg465Trp) were treated with 25 μg/ml cycloheximide (Sigma-Aldrich) for 0, 30, 60 or 90 minutes and protein extracted as above.
Western blotting of CCND2-myc construct transfected cell line and patient lymphoblastoid protein extracts
Equal amounts of protein were heated to 70°C for 10 mins with NuPAGE® LDS Sample Buffer (4X) (Life Technologies) and run on NuPAGE® Novex® 4-12% Bis-Tris gels (Life Technologies) and transferred to PVDF membranes (Life Technologies). Membranes were blocked for one hour at room temperature (RT) with 5% dried skimmed milk (Marvel) in PBS with 0.1% Tween-20. Membranes were then incubated with primary antibody at a dilution of 1:200 for CCND2 (ab3085, Abcam, Cambridge, UK) and 1:10,000 for β-actin (ab8226, Abcam) overnight in blocking solution at 4°C with slight agitation. The membranes were washed with 0.1% Tween-20 in PBS and then incubated with secondary antibody for 1 hour at RT. The secondary antibody (polyclonal goat anti-mouse immunoglobulins/HRP, P0447, Dako, Ely UK) was diluted 1:10,000 in blocking solution. Membranes were then washed in PBS with 0.1% Tween-20, developed with SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) and visualized using a Geldoc XR system (Biorad, Hemel Hempstead, UK). Experiments were performed in triplicate and .tiff images were analyzed using ImageJ24. Bands were quantified and intensities determined relative to the corresponding ‘0’ time point. CCND2 intensity was normalized with β-actin.
Generation of constructs for in utero electroporation
The open reading frame of human CCND2 was cloned into the pCAGIG vector (IRES-eGFP, Addgene, Cambridge, MA, USA) at the multiple cloning site following digestion with EcoRI and XhoI. Phosphodeficient (p.Thr280Ala), phosphomimetic (p.Thr280Asp) and patient (p.Pro281Arg) mutations were made using the QuikChange Site-Directed Mutagenesis Kit (Supplementary Table 4; Agilent Technologies, Santa Clara, CA, USA) as per manufacturer’s instructions. The p.280 phosphodeficient mutation involved the following change: ACC (Thr) to GCC (Ala), the phosphomimetic mutation entailed altering the first mutant: GCC (Ala) to GAC (Asp) and the p.281 mutation changed CCT (Pro) to CGT (Arg). Constructs were then sequenced to confirm the mutations. There were five experimental groups: GFP control, WT CCND2, p.Thr280Ala, p.Thr280Asp, and p.Pro281Arg CCND2 mutants.
Animals
Timed pregnant female wild-type CD-1 mice at least 10 weeks of age were obtained from Charles River Laboratories (Wilmington, MA, USA) and then maintained as part of our laboratory colony and were housed at 23°C with a 12h light/dark cycle in Thoren units. The day of plug discovery (at 12 noon) was designated as embryonic day 0.5 (E0.5). All experiments were performed in accordance with protocols approved by the Weill Cornell Medical College Institutional Animal Care and Use Committee.
In utero electroporation and BrdU injections
Electroporations were carried out at E13.5. The pregnant dam was anesthetized with Isoflurane (Butler Schein, Dublin, OH, USA) and a laparotomy was performed to expose the uterus. DNA was diluted 1:20 in Fast Green (Sigma-Aldrich) to achieve a final concentration of 1 μg/μl. Then, 1 μg DNA was injected into the lateral ventricle of each embryo. Tweezertrodes (7 mm platinum, BTX, Harvard Apparatus, Holliston, MA, USA) were placed over the injection site and then five 50-ms pulses at 42 V were delivered with 1-s intervals between each pulse using a BTX-ECM830 electroporator (BTX, Harvard Apparatus) to introduce DNA into the cortex. Embryos were returned to the abdominal cavity, the wound was sutured; dams were allowed to recover and were closely monitored. Embryos were harvested at 48 hours post-IUEP at E15.5. Embryo brains were collected in cold 1x phosphate buffered saline (PBS) and screened for GFP expression using a stereomicroscope (Leica M165 FC, Buffalo Grove, IL, USA). Brains were transferred to 4% paraformaldehyde in 1x PBS with 7% sucrose overnight at 4°C. The following day, brains were transferred to 15% sucrose and then 30% sucrose in 1x PBS, shaking at 4°C, until properly sunk. Brains were then embedded in optimal cutting temperature solution (Electron Microscopy Sciences, Hatfield, PA, USA) and stored at −20°C.
A subset of pregnant dams were injected with bromodeoxyuridine (BrdU; Sigma-Aldrich, St. Louis, MO, USA) at 50 mg/kg at E14.5 (24 hours post-IUEP). All tissue was then harvested at E15.5 (48 hours post-IUEP).
Immunohistochemistry
Brains were sectioned coronally at 12 μm on a cryostat (HM505E, Microm, Mars, PA, USA) and collected on glass slides. Sections to be stained were washed 3x with 1x PBS with 0.3% Triton X-100 (PBS-T) and then subjected to antigen retrieval with steam in 1 mM EDTA, 5 mM Tris pH 8 (GFP and Pax6 dual labeling) for 20 mins or 10 mM sodium citrate buffer, pH 6 for 15 mins (for all other staining). Slides were allowed to come to RT, rinsed in 1x PBS-T, and then blocked for 1 h at RT in 10% normal donkey serum (Jackson Immunoresearch Laboratories, Inc., West Grove, PA, USA) in 1x PBS-T. Antibodies were diluted in blocking medium and were used as follows: anti-GFP (600-101-215, goat IgG, Rockland Immunochemicals, Inc., Gilbertsville, PA, USA, 1:10,000), anti-Ki67 (RM-9106-S1, rabbit IgG, Thermo Scientific, Waltham, MA, USA, 1:1000), anti-phosphohistone H3 (PH3, 06-570, rabbit IgG, Millipore, Billerica, MA, USA, 1:2000), anti-BrdU, 555627, mouse IgG, BD Biosciences, San Jose, CA, USA, 1:500), anti-Pax6 (PRB-278P, rabbit IgG, Covance, Princeton, NJ, USA, 1:1000), and anti-Tbr2 (ab23345, rabbit IgG, Abcam, 1:2000). Primary antibodies were incubated overnight at 4°C. The following day, slides were washed 3x in 1x PBS-T and incubated with Alexa fluorescence-conjugated secondary antibodies (Molecular Probes, Life Technologies, Grand Island, NY, USA, 1:1000 in blocking solution) for 2 h at RT. Slides were then washed and coverslipped with ProLong Antifade Kit (Life Technologies).
An additional set of brains was processed for paraffin sectioning as previously described14. Sections were collected at 4 μm and first processed for anti-CCND2 immunohistochemistry (MS-221-P0, mouse IgG, Thermo Scientific, 1:100)14. After the primary antibody incubations, sections were incubated in biotinylated secondary antibody (1:400, 1 hr), and then peroxidase-conjugated tertiary antibody for 1 hr following manufacturer’s instructions (Covance). Tissue was then subjected to a 3,3-diaminobenzidine (DAB) reaction (Covance). Next, tissue was thoroughly washed and then blocked, and incubated with anti-GFP antibodies (as above, 1:500) overnight. Fluorescent secondary antibody incubations were carried out as described above.
Microscopy and Cell Quantifications
Sections were imaged with a 20x objective using an epifluorescent microscope (Leica DM550B with Leica Application Suite Advanced Fluorescence 3.0.0 build 8134 software, Leica Microsystems Inc., Buffalo Grove, IL, USA). Sections through the entire electroporated area (rostro-caudal) were analyzed. Embryos that expressed GFP through the cortex were analyzed. Those that did not fit these criteria were eliminated from analyses and all embryos that met these criteria were quantified. For all IUEP experiments, embryo brains were assigned a random number that the investigator performing the counts was not privy to, so as to reduce bias. After the counts were completed, embryos were matched to their respective numbers and assigned to appropriate experimental groups.
The number of total GFP+ cells and the number of GFP+/Ki67+ cells were counted and computed so that n=# of embryos, five sections/embryo (n=7 GFP, n=5 WT CCND2, n=5 p.Thr280Ala, n=5 p.Thr280Asp, and n=5 p.Pro281Arg. At least 7000 GFP+ cells were analyzed for each experimental group. For the GFP/PH3 co-labeling experiment, at least 5300 GFP+ cells/experimental group were analyzed (n=4 WT CCND2, n=5 p.Thr280Ala, and n=5 p.Thr280Asp, 5 sections/embryo). Lastly, for the GFP/BrdU/Ki67 experiment, at least 6800 GFP+ cells were analyzed/group (n=5 WT CCND2, n=5 p.Thr280Ala and n=5 p.Thr280Asp, 5 sections/embryo). To calculate the P-Fraction (proliferative fraction) and exit fractions we employed the following formulas:
P-Fraction
(total number of GFP+/BrdU+/Ki67+ cells)/(total number of GFP+/BrdU+ cells) × 100
Exit fraction
(total number of GFP+/BrdU+ cells) − (total number of GFP+/BrdU+/Ki67+ cells)/(total number of GFP+/BrdU+ cells) × 100
For the GFP/Pax6 analysis, at least 8500 GFP+ cells/experimental group were counted (n=6 GFP, n=8 WT CCND2, n=5 p.Thr280Ala, and n=5 p.Thr280Asp, 5 sections/embryo). Lastly, for the GFP/Tbr2 experiment, at least 6400 GFP+ cells/experimental group were counted (n=6 GFP, n=6 WT CCND2, n=5 p.Thr280Ala, and n=5 p.Thr280Asp, 5 sections/embryo).
Final counts are presented as percentages ± standard error measurement. Unless otherwise noted, a two-tailed unpaired Student’s t-test was performed with a p< 0.05 considered significant. For Figure 4 and the Pax6 and Tbr2 experiments (Supplementary Figure 3), a one-way ANOVA was used where a p <0.05 was deemed significant. Post hoc tests (LSD) were then performed to corroborate the independent t-tests. Pairwise comparisons were not adjusted for the multiple comparisons because all groups were defined a priori. In this case, the statistical values shown were from the post hoc tests following an ANOVA analysis. Statistics were calculated using Microsoft Excel and SPSS (Version 22, IBM). All analyzed data met the assumptions of the two-tailed unpaired Student’s t-test or the one-way ANOVA and the variance between experimental groups was similar. For all in utero experiments, we estimated we would need 4-7 embryos/group based on studies previously performed in our laboratory13. Graphs were made in Microsoft Excel and final figures were compiled in Adobe Photoshop 12.0 (Adobe Systems, San Jose, CA, USA).
Cell Culture
HEK293 cells (from ATCC) were transfected with the IUEP constructs using XFect (Clonetech Laboratories Inc., Mountain View, CA, USA) according to manufacturer’s instructions and then collected 48 hrs later. Cells were lysed using Mammalian Protein Extraction Reagent (Thermo Scientific) with protease and phosphatase inhibitors in addition to EDTA and then flash frozen until needed for western blotting.
Western blotting of GFP constructs
Samples were reduced and boiled at 70°C for 10 minutes. Equal amounts of protein were run on polyacrylamide gels as above (Invitrogen) and then transferred to nitrocellulose membranes (Whatman, Piscataway, NJ, USA). Membranes to be stained with anti-CCND2 and anti-GFP antibodies were blocked in 5% non-fat dry milk in 1x Tris-buffered saline with 0.05% Tween 20 (TBS-T). Membranes were then incubated in primary antibodies as follows: anti-CCND2 (sc-53637/sc-56305, mouse IgG, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA, 1:1000) and anti-GFP (see above, goat IgG, 1:4,000), diluted in block solution overnight at 4°C with slight agitation. The following day, membranes were washed and then incubated in secondary antibody for 1 hr at RT diluted in Odyssey LI-COR solution with 0.05% Tween 20 using LI-COR secondary antibodies (1:10,000, donkey anti-mouse IRDye 800SCW and donkey anti-goat IRDye 680LT). Blots were then visualized on the Odyssey FC imaging system (LI-COR Biosciences, Lincoln, Nebraska, USA). Quantitation was determined by serial dilution of lysates and fluorescent values within the linear range were used for determining relative levels, normalized to the loading control of total GSK-3β. Primary antibodies used in the quantitative westerns included anti-CCND2 (as above), anti-CCND1 (ab134175, rabbit IgG, Abcam, 1:4000), anti-GSK-3β (9832, mouse IgG, Cell Signaling, Danvers, MA, USA, 1:2000), and anti-pS9-GSK-3β (9323, rabbit IgG, Cell Signaling, 1:1000).
Western blotting of AKT3-D219V embryo brain lysates
Embryo brains were harvested at E14.5. Lysates were prepared and equal amounts of protein were loaded onto polyacryl-amide gels for Western blot transfer and immunodetection as for cell cultures above. A total of three WT/Akt3-D219V/D219V littermate pairs were analyzed, providing three biological replicates. Each WT/Akt3-D219V/D219V pair was processed in parallel as follows: 21, 18, 15, 12, 9 or 6 μl of lysate was loaded on a gel. Out of 6 data points for each biological replicate at least three were in the linear range, giving three technical replicates for each sample, for a total n=9. WT and mutants were then compared using a sign test and for every WT-mutant pair from the same litter and of the same dilution the sign of the difference was the same (e.g. CCND2 or CCND1 was greater in the mutant than WT littermate), for a two-sided level of significance of 0.02.
URLs
FASTX-Toolkit, http://hannonlab.cshl.edu/fastx_toolkit/; GATK, http://www.broadinstitute.org/gsa/wiki/; Picard tools, http://picard.sourceforge.net/; Samtools, http://samtools.sourceforge.net/, Homologene, http://www.ncbi.nlm.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the study patients and their families, without whose participation this work would not be possible. We thank Prof Mark O’Driscoll (University of Sussex) for advice and help. This work was funded by the Government of Canada through Genome Canada, the Canadian Institutes of Health Research (CIHR), the Ontario Genomics Institute (OGI-049), Genome Quebec and Genome British Columbia (to K.M.B.). The work was selected for study by the FORGE Canada Steering Committee, consisting of K. Boycott (U. Ottawa), J. Friedman (U. British Columbia), J. Michaud (U. Montreal), F. Bernier (U. Calgary), M. Brudno (U. Toronto), B. Fernandez (Memorial U.), B. Knoppers (McGill U.), M. Samuels (U. de Montreal), and S. Scherer (U. Toronto). Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health under award numbers P01-NS048120 (to M.E.R.), NRSA F32 NS086173 (to K.A.G.), and R01NS058721 (to W.B.D.), and by The Baily Thomas Charitable Fund (to D.T.P.), and the Sir Jules Thorn Charitable Trust and Great Ormond Street Children’s Hospital Charity (to E.G.S.).
Footnotes
AUTHOR CONTRIBUTIONS: ES, WBD, DTP, MER, KMB, GMM, DAP, AEF and KAG designed the study and experiments;
GMM, AEF, CA, DTB, KPJB, LF, KWG, GMSM, KP, ES, HvE, NV, DW, DTP, WBD and ES identified, consented and recruited the study subjects and provided clinical information;
GMM, AEF, DTP and WBD evaluated the magnetic resonance imaging;
GMM, DAP, CJ, JS, MV, CL and NR developed the bioinformatics scripts, and performed genetic data analysis and confirmation studies;
RDH and RFH provided the Akt3 mouse mutant brain samples;
DAP and CVL performed the protein stability experiments;
KAG, SS, SSK and MER performed and analyzed the IUEP experiments; and quantitative western blot analyses of Akt3-D219V
GMM, DP, AEF, KAG, DTP, MER, WBD and ES wrote the manuscript.
REFERENCES
- 1.Lee JH, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nature genetics. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindhurst MJ, et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nature genetics. 2012;44:928–933. doi: 10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riviere JB, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nature genetics. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kida A, Kakihana K, Kotani S, Kurosu T, Miura O. Glycogen synthase kinase-3β and p38 phosphorylate cyclin D2 on Thr280 to trigger its ubiquitin/proteasome-dependent degradation in hematopoietic cells. Oncogene. 2007;26:6630–6640. doi: 10.1038/sj.onc.1210490. [DOI] [PubMed] [Google Scholar]
- 5.Poduri A, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 7.Tokuda S, et al. A novel Akt3 mutation associated with enhanced kinase activity and seizure susceptibility in mice. Human Molecular Genetics. 2011;20:988–999. doi: 10.1093/hmg/ddq544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Cerdeno V, Noctor SC, Kriegstein AR. The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cerebral Cortex. 2006;16(Suppl 1):i152–161. doi: 10.1093/cercor/bhk017. [DOI] [PubMed] [Google Scholar]
- 10.Nonaka-Kinoshita M, et al. Regulation of cerebral cortex size and folding by expansion of basal progenitors. The EMBO Journal. 2013 doi: 10.1038/emboj.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 12.Glickstein SB, Alexander S, Ross ME. Differences in Cyclin D2 and D1 Protein Expression Distinguish Forebrain Progenitor Subsets. Cerebral Cortex. 2007;17:632–642. doi: 10.1093/cercor/bhk008. [DOI] [PubMed] [Google Scholar]
- 13.Glickstein SB, et al. Selective cortical interneuron and GABA deficits in cyclin D2-null mice. Development. 2007;134:4083–4093. doi: 10.1242/dev.008524. doi:10.1242/dev.008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glickstein SB, Monaghan JA, Koeller HB, Jones TK, Ross ME. Cyclin D2 Is Critical for Intermediate Progenitor Cell Proliferation in the Embryonic Cortex. The Journal of Neuroscience. 2009;29:9614–9624. doi: 10.1523/JNEUROSCI.2284-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huard JM, Forster CC, Carter ML, Sicinski P, Ross ME. Cerebellar histogenesis is disturbed in mice lacking cyclin D2. Development. 1999;126:1927–1935. doi: 10.1242/dev.126.9.1927. [DOI] [PubMed] [Google Scholar]
- 16.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Baala L, et al. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nature genetics. 2007;39:454–456. doi: 10.1038/ng1993. doi:10.1038/ng1993. [DOI] [PubMed] [Google Scholar]
- 19.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. doi:10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. doi:10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. doi:10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.