Abstract
The transcription factor T-bet controls the Th1 genetic program in T cells for effective antitumor responses. Anti-CTLA-4 immunotherapy elicits dramatic antitumor responses in mice and in human patients; however, factors that regulate T-bet expression during an antitumor response mediated by anti-CTLA-4 remain to be elucidated. We were the first to report that treatment with anti-CTLA-4 led to an increase in the frequency of T cells expressing inducible costimulator (ICOS). In both treated patients and mice our data revealed that CD4+ICOShi T cells can act as effector T cells, which produce the Th1 cytokine interferon gamma (IFNγ). We also showed in a small restrospective analysis that an increased frequency of CD4+ICOShi T cells correlated with better clinical outcome and the absence of ICOS or its ligand (ICOSL) in mouse models led to impaired tumor rejection. Here we show that CD4+ICOShi T cells from anti-CTLA-4-treated patients had an increase in signaling via the phospoinositide-3-kinase (PI3K) pathway and an increase in expression of T-bet. An ICOS-specific siRNA transfected into human T cells led to diminished PI3K-signaling and T-bet expression. Therefore, we hypothesized that ICOS, and specifically ICOS-mediated PI3K-signaling, was required for T-bet expression. We conducted studies in ICOS-deficient and ICOS-YF mice, which have a single amino acid change that abrogates PI3K-signaling by ICOS. We found that ICOS-mediated PI3K-signaling is required for T-bet expression during an antitumor response elicited by anti-CTLA-4 therapy. Our data provide new insight into the regulation of T-bet expression and suggest that ICOS can be targeted to improve Th1 antitumor responses.
Introduction
Anti-CTLA-4 blocks the cytotoxic lymphocyte antigen-4 (CTLA-4) inhibitory pathway on T cells thereby leading to enhanced T cell responses (1–3). We previously reported that patients treated with anti-CTLA-4 had an increase in the frequency of CD4 T cells expressing the inducible costimulator (ICOS), which correlated with clinical benefit (4–6). ICOS is expressed solely by T cells and the expression of ICOS increases upon T cell activation (7). ICOS interacts with ICOS-ligand, which is expressed by antigen-presenting cells (APC) (7). Although ICOS can be expressed on multiple T cell subsets, including Th2 cells, regulatory T cells and follicular helper T cells (8), we have previously shown that ICOS is expressed predominantly on effector T cells that produce the Th1 cytokine IFNγ after patients and mice received anti-CTLA-4 treatment (4, 9).
The transcription factor T-box expressed in T cells (T-bet) controls the Th1 genetic program in CD4 T cells for effective antitumor responses (10–13). Although T cell receptor signaling and CD28 co-stimulation regulate T-bet expression during the early stages of a T cell response (14–17, 7), other factors that regulate T-bet expression, especially during prolonged antitumor T cell responses in vivo, remain to be elucidated.
Here we show that increased ICOS expression correlated with increased phosphoinositide-3-kinase (PI3K)-signaling and increased T-bet expression in CD4 T cells from patients treated with anti-CTLA4. Therefore, we hypothesized that ICOS, which is a member of the CD28 family of proteins that can mediate signaling via the PI3K pathway, was an important factor in controlling T-bet expression. To test our hypothesis, we studied CD4 T cells ex vivo from ICOS−/− tumor-bearing mice treated with anti-CTLA-4. We found that CD4 T cells from ICOS−/− mice, which had intact CD28 expression, had diminished PI3K-signaling and T-bet expression, as compared to CD4 T cells from wild-type (WT) mice. To determine whether ICOS-mediated PI3K-signaling controlled T-bet expression, we conducted studies in ICOS-YF mice, which have a single amino acid change that abrogates PI3K-signaling by ICOS (18). These studies revealed that ICOS-mediated PI3K-signaling was critical for T-bet expression in the setting of anti-CTLA-4 therapy.
Our data provide new insight into the regulation of T-bet expression, which can be exploited for novel immunotherapy strategies to treat cancer and possibly infectious agents, by targeting ICOS to enhance PI3K-signaling and T-bet expression in Th1 effector CD4 T cells.
Materials and Methods
Mice, Cell lines and Reagents
C57BL/6 WT and ICOS−/− mice were obtained from the Jackson Laboratory (Bar Harbor, Maine). ICOS-YF mice were generated as previously described (18) and have been backcrossed more than ten generations into C57BL/6 background. B16/BL6 murine melanoma cell line was maintained, used and grown as previously published (9). No additional authentication was performed on the murine tumor cell lines used. Anti-CTLA-4 mAb (clone 9H10) was obtained from Bio X cell. Animals were cared for in accordance with National Institutes of Health and American Association for the Accreditation of Laboratory Animal Care International regulations. Experiments were performed according to an IACUC-approved protocol.
Tumor Growth and Treatment
B16/BL6 tumor cells were injected into mice and treatment with anti-CTLA plus Gvax was administered as previously described (9). Briefly, the anesthetized mice were injected with 1× 104 B16/BL6 melanoma cells at day 0, and then untreated or treated with anti-CTLA4 mAb plus Gvax on day 3, 6 and 9. The spleen, tumor-draining lymph nodes (DLN) and T cells from tumor-infiltrating lymphocytes (TILs) were obtained as previously published (9). Total CD4 T cells were isolated using microbeads (Miltenyi Biotec). CD4+ICOShi and CD4+ICOSlow cells were further sorted by using FACS Aria II sorter. Protein lysates from these cells were obtained for Western blot analyses.
Patients and Blood Processing
Patients with diagnoses of localized urothelial carcinoma received 2 doses of anti-CTLA-4 antibody ipilimumab at 3mg/kg or 10mg/kg, with a 3-week interval between doses on an IRB-approved clinical trial (2006-0080) as previously published (4–6). Ipilimumab is a fully human monoclonal immunoglobulin (IgG1) specific for human CTLA-4 (CD152). Blood was collected before the first dose; 3 weeks after dose #1; and 3–4 weeks after dose #2.
Peripheral blood mononuclear cells (PBMCs) were isolated from patients’ whole blood by density gradient centrifugation using lymphocyte separation Medium (Mediatech, Inc., Herndon, VA) and Leucosep tubes (Greiner Bio-one, Germany). Cells recovered from the gradient interface were washed twice in RPMI 1640 medium.
Reverse Phase Protein Array (RPPA) assays and analysis
CD4 T cells were obtained pre- and post-therapy from patients who were treated with anti-CTLA-4 mAb ipilimumab. CD4 T cells from the patient’s PBMCs were obtained by negative immunoselection using CD4+ T cell Isolation Kit II (Miltenyi Biotec Inc. Auburn, CA). Cells were lysed in RPPA lysis buffer [1% Triton X-100, 50 mM Hepes (pH 7.4), 150 mMNaCl, 1.5 mM MgCl2, 1 mmol/L EGTA, 100 mM NaF, 10 nmol/L NaPPi, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, whole proteinase and phosphatase inhibitor tablets (Boehrsinger/Roche, Mannheim, Germany)], then add 4× SDS/2-ME sample buffer (35% glycerol, 8% SDS, 0.25M Tris-HCl, pH 6.8; with 10% β-mercaptoethanol added before use) for 20 to 30 minutes with frequent vortexing on ice, centrifuged for 15 minutes at 14,000 rpm, and the supernatant collected. Lysates were transferred into a PCR 96-well plate. The plates were covered and incubated for 5 minutes at 95°C and then centrifuged for 1 minute at 2,000 rpm.
Protein extracts were serially diluted and probed for the expression of validated antibodies by RPPA (20, 21). Specifically, five serial 2-fold dilutions of the protein extracts were performed using RPPA lysis buffer containing 1% SDS. The diluted lysates were spotted on nitrocellulose-coated FAST slides (Whatman, Schleicher & Schuell BioScience, Inc., Keene, NH) by an Aushon 2470 arrayer (Aushon Biosystems, Burlington, MA) per manufacturer’s protocol. Each spotted slide was incubated with a primary antibody in the appropriate dilution. The specific protein-antibody interaction was recognized by biotin-conjugated secondary antibody and amplified by tyramide deposition. The analyte was detected by avidin-conjugated peroxidase reactive to its substrate chromogen diaminobenzidine (DAKO catalyzed signal amplification (CSA) system, DAKO, Carpinteria, CA). The stained RPPA slides were scanned by the Hewlett-Packard (HP) scanner and its companying scanning software, and the slide images were quantified for raw signal intensities by the software MicroVigene (VigeneTech Inc., Carlisle, MA) per manufacturer’s protocol. The raw signal intensities were then processed by the R package SuperCurve (20) developed by the Department of Bioinformatics and Computational Biology at M. D. Anderson Cancer Center (http://bioinformatics.mdanderson.org/Software/OOMPA/). The log2-scaled protein concentrations were normalized by global sample median normalization (GSMN). The median of all protein marker intensities for a single sample is subtracted from each of the data points (32, 33). To generate heat maps, Treeview and Cluster software were used (34). Student’s t-test was used to evaluate statistical significance of differences in protein expression levels between different samples groups. Box plots analysis of RPPA assay was generated by “mixed-effects model”. Mixed-effects models were fitted to incorporate multiple observations from an individual patient. All p-values were two-sided, at a significance level of 0.05.
Flow Cytometry Analyses
PBMCs from patients were assessed using anti-CD4 PerCP-Cy5.5 (BD Biosciences) and anti-ICOS PE-Cy7 (eBioscience). Flow Cytometry was carried out on a BD FACS Canto II. Data were analyzed by FACS Diva software. Gates were set according to appropriate isotype controls. In addition, CD4+ICOShi and CD4+ICOSlow cells were isolated by FACS Aria II cell sorter (BD Bioscience). For murine studies, antibodies used were CD4 (GK1.5), CD28 (37.51), ICOS (7E.17G9) and T-bet (eBio4B10) were all from eBioscience. Data were analyzed by FACS Diva and Flowjo software (TreeStar).
Western Blot Analysis and Antibodies
Human CD4 T cells were harvested and lysed in RPPA lysis buffer. The concentration of each sample was determined by a standard Bradford assay. Equal amounts of protein (20μg) from cells were subjected to Western blot analysis. Anti-phospho-Akt (Ser473), anti-Bcl-XL, anti-phospho-S6 ribosomal protein (Ser235/236), and β-actin were probed using appropriate murine or human antibodies available form Cell Signaling Technology (CST, Danvers, MA). Anti-Cyclin B1 (Epitopmics, Burlingame, CA), anti-TBX21/T-bet (CST), and anti-GATA-3 (CST) were obtained for assessment of human samples. Anti-cyclin B1 (CST); anti-TBX21/T-bet and anti-GATA3 were obtained for assessment of murine samples (Proteintech group, Chicago, IL). Semiquantification of protein was performed by densitometric digital analysis of protein bands (TIFF image) using Adobe Photoshop CS3 software. Band values (mean intensities) were measurement on each area of interest, subtracting the background value from the result and multiplying the remainder with the size of the area of interest. The relative amount of protein (normalized intensity) was calculated using the expression ratio of a target protein versus β-actin as a reference protein.
RNA interference
On-TARGET plus SMARTpool targeting human ICOS (siRNA-ICOS) and non-targeting (Negative control, siRNA-NC) unrelated siRNAs were purchased from Dharmacon RNAi Technologies. Approximately 1 million post-therapy (week 7) PBMCs from patients were transfected with either human ICOS siRNAs (50nM) or control siRNAs (50nM) or mock transfected. Electroporation transfection method was performed according to manufacturer’s instructions of 4D Nucleofector® Device and Amaxa P3 primary cell 4D-Nucleofector X kit from Lonza (Basel, Switzerland). Some PBMCs were also transfected with pmax-GFPvector (1μg/sample, Lonza) to interrogate the transfection efficiency. Transfection efficiency was monitored by fluorescence microscopy and measured by flow cytometry at various time points. The post-transfected PBMCs were incubated in 12-well culture plates and CD4 T cells were sorted using a FACS Aria II sorter. Sorted CD4 T cells were stained with propidium iodide (BD) and assessed for viability by flow cytometry prior to performing Western blot analyses and real-time PCR.
Quantitative RT-PCR
Total RNA was isolated from CD4 T cells, as well as CD4+ICOShi and CD4+ICOSlow T cells using RNeasy kit (Qiagen) according to manufacturer’s instructions. cDNAs were generated using the SuperScript III Reverse Transcriptase kit (Invitrogen). Real-time quantitative PCR was performed by a 7500 Fast Real-Time PCR system (Applied Biosystems) according to the manufacturer’s instructions. Samples were used as templates in reactions to obtain the threshold cycle (Ct) that were normalized with the Ct of CD3-ε from the same sample (ΔCt) in T cells. To compare the relative levels of gene expression in different T cell types, ΔΔCt values were calculated with the ΔCt values associated with the lowest expression levels as 1. Fold-induction was calculated using 2ΔΔCt. Taqman Gene Expression Assays of ICOS, CD3-ε, IFNγ, IL-2, IL-4, IL-10, IL12A, IL17A, IL-21, TNF-α, T-bet, and GATA3 were purchased from Applied Biosystems. To determine the relative levels of gene expression in ICOS knockdown studies, ΔΔCt values were calculated with the ΔCt values associated with the control expression levels (transfected with siRNA-NC) as 1.
Statistical Analysis
All statistical analyses except for RPPA, which were described above, were conducted by using Prism 5.0 (GraphPad software, Inc). Results were represented as means±standard error mean (s.e.m) with a 2-sided student’s t test. Significant values were those with p≤0.05.
Results
ICOS expression correlates with increased PI3K-signaling, T-bet expression and Th1 cytokines
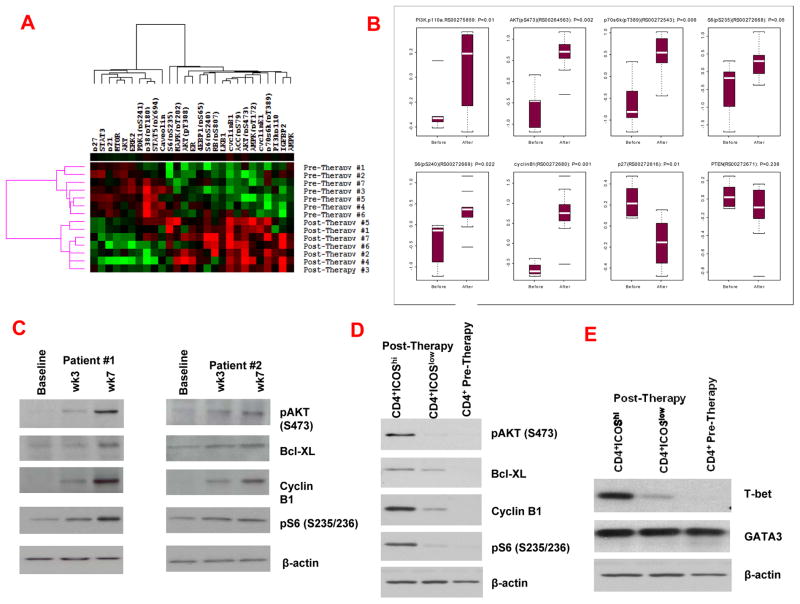
Ipilimumab, a fully human monoclonal immunoglobulin IgG1 that blocks a T cell inhibitory pathway mediated by CTLA-4 (1, 2) was FDA-approved for treatment of melanoma patients based on data from a Phase III clinical trial demonstrating survival benefits in treated patients (3). In some patients, anti-CTLA-4 therapy may take several months before tumor regression can be detected (19), suggesting the need for a prolonged T cell response that eventually facilitates tumor cell death. We evaluated the anti-CTLA-4 immune responses against bladder cancer in a clinical trial (Supplementary Fig. 1A) in which 12 patients received 2 doses of anti-CTLA-4, and peripheral blood was collected pre-therapy at week 0, post-therapy at week 3, and post-therapy at week 7 (4–6). We obtained matched pre- and post-therapy (week 7) purified CD4 T cells from 7 patients and conducted ex vivo studies to evaluate changes in different signaling pathways. Protein lysates from these cells were analyzed by reverse phase protein array (RPPA), which includes 84 validated antibodies representing multiple signaling pathways (20, 21) (Supplementary Fig. 1B). RPPA detected 27 proteins that were significantly changed (p<0.05) in post-therapy CD4 T cells as compared to pre-therapy CD4 T cells. Unsupervised hierarchical clustering of data from pre- and post-therapy samples is shown (Fig. 1A). There was a significant increase in proteins indicative of the PI3K-signaling (16) in post-therapy CD4 T cells as compared to pre-therapy cells, with marked increase in the expression of PI3Kp110, phospho (p)-AKT (S473), p70S6K (Tp389), pS6 (S235/236), pS6 (S240/244) and cyclin B1 (Fig. 1B). We also observed a decrease in the expression of p27 but there was no significant difference in the expression of PTEN (Fig. 1B). The RPPA data did not indicate a significant change in other signaling pathways such as apoptosis, angiogenesis, EGFR and Notch (Supplementary Figure 1B). The RPPA data related to PI3K-signaling was confirmed by Western blot analyses (Fig. 1C). Compared to baseline (pre-therapy), post-therapy CD4 T cells demonstrated greater expression of proteins indicative of an increase in PI3K-signaling at weeks 3 and 7 (Fig. 1C).
Figure 1. ICOS expression correlates with increased PI3K-signaling, T-bet expression and Th1 cytokines.
(A) RPPA data with unsupervised hierarchical clustering of 27 proteins that were significantly changed (p<0.05) in post-therapy CD4 T cells as compared to matched pre-therapy CD4 T cells. (B) Box plots to indicate changes in the expression levels of 7 proteins selected to represent PI3K-signaling in post-therapy (after) CD4 T cells as compared to pre-therapy (before) CD4 T cells. (C) Greater expression of proteins indicative of PI3K-signaling was detected in post-therapy CD4 T cells at weeks 3 and 7 as compared to pre-therapy CD4 T cells. (D) Greater expression of proteins indicative of PI3K-signaling was detected in post-therapy CD4+ICOShi T cells as compared to post-therapy CD4+ICOSlow T cells and pre-therapy CD4 T cells. (E) Greater expression of T-bet protein was detected in post-therapy CD4+ICOShi T cells as compared to post-therapy CD4+ICOSlow T cells and pre-therapy CD4 T cells. (F) Higher mRNA levels were observed for IFNγ and TNFα in post-therapy CD4+ICOShi T cells as compared to post-therapy CD4+ICOSlow T cells and pre-therapy CD4 T cells. *indicates p<0.05
Since the cytoplasmic tail of ICOS has an YMPM PI3K-binding motif (7,8), we investigated whether ICOS expression correlated with the increase in PI3K-signaling. We studied CD4 T cells from pre- and post-therapy (week 7) samples. CD4 T cells were sorted according to ICOS expression (hi vs. low) by flow cytometry using gating strategies based on isotype controls (Supplementary Fig. 1C) and as described (4–6). As previously published, ICOS expression can be denoted as “positive” vs. “negative” expression (9, 22, 23) or “hi” vs. “low” expression (4, 8, 24). Here we refer to the different populations of cells as ICOShi vs. ICOSlow (Supplementary Figure 1C). Both ICOShi and ICOSlow CD4 T cells consisted of activated T cells as measured by the expression of the IL2-receptor alpha subunit (CD25) (25) (Supplementary Fig. 1D). We sorted CD4+ICOShi and CD4+ICOSlow T cells from pre- and post-therapy (week 7) samples from 3 patients and pooled one million cells from each subset to obtain sufficient protein lysates for Western blot studies. We found that post-therapy CD4+ICOShi T cells had significantly greater expression of proteins indicative of PI3K-signaling compared to post-therapy CD4+ICOSlow T cells or pre-therapy CD4 T cells, with greater expression of pAKT (S473), cyclin B1, pS6 (S235/236) and Bcl-XL detected in CD4+ICOShi T cells (Fig. 1D). To determine whether the observed differences in PI3K-signaling were due to differences in CD28 expression, we analyzed CD28 expression on T cells from pre- and post-therapy samples as well as from CD4+ICOShi and CD4+ICOSlow T cells from the post-therapy samples. We did not observe a significant difference in the frequency of CD28 expression between pre- and post-therapy CD4 T cells or between CD4+ICOShi and CD4+ICOSlow T cells from post-therapy samples (Supplementary Fig. 1E).
To evaluate whether ICOS expression correlated with an increase in the expression of the transcription factor T-bet, which controls Th1 cytokine production by CD4 T cells, we studied ex vivo CD4 T cells from pre- and post-therapy patients’ samples. Post-therapy (week 7) CD4+ICOShi T cells had significantly greater T-bet expression compared to post-therapy CD4+ICOSlow T cells or pre-therapy CD4 T cells (Fig. 1E). There was no significant difference in the expression of the transcription factor GATA-3, which controls Th2 cytokine production by CD4 T cells (26). Additional studies revealed that post-therapy CD4+ICOShi T cells had higher expression of mRNA for the Th1 cytokines IFNγ and TNFα as compared to post-therapy CD4+ICOSlow and pre-therapy CD4 T cells (Fig. 1F). There were no differences in the expression of Th2 cytokines. These data indicate that ICOS expression correlated with the increase in PI3K-signaling, in T-bet expression and in Th1 cytokine production in CD4 T cells from anti-CTLA-4-treated patients.
ICOS-specific siRNA diminishes PI3K-signaling, T-bet expression and Th1 cytokines
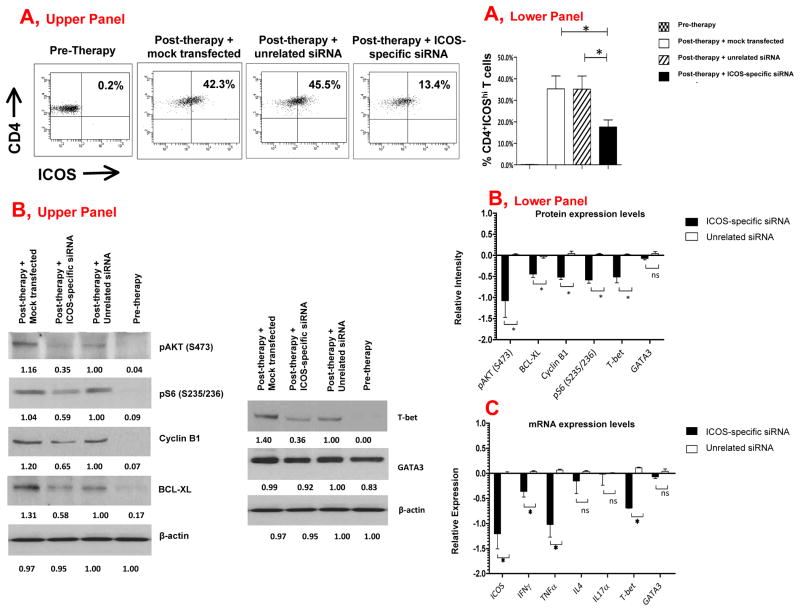
To determine whether ICOS was directly driving PI3K-signaling and expression of T-bet and Th1 cytokines, we utilized an ICOS-specific siRNA to decrease ICOS expression in CD4 T cells from three patients treated with anti-CTLA-4 mAb (N=3). Transfection of ICOS-specific siRNA decreased ICOS expression (Fig. 2A, representative patient data, Upper Panel and summary data, Lower Panel) and diminished the expression of proteins indicative of PI3K-signaling, with decreased expression of pAKT, pS6, cyclin B1 and Bcl-XL protein (Fig. 2B, representative Western Blot, Upper Panel and summary data, Lower Panel). The ICOS-specific siRNA also led to diminished protein expression for T-bet (Fig. 2B). In addition, ICOS-specific siRNA significantly decreased the expression of mRNA for T-bet and for the Th1 cytokines IFNγ and TNFα (Fig. 2C). The ICOS-specific siRNA did not have any detectable impact on the expression of mRNA for IL-4, a Th2 cytokine, or for IL-17α (Fig. 2C). These data indicate that ICOS expression directly impacted PI3K-signaling, T-bet expression and Th1 cytokine production in activated human T cells.
Figure 2. ICOS-specific siRNA diminishes PI3K-signaling, T-bet expression and Th1 cytokines.
(A) An ICOS-specific siRNA led to the decreased frequency of CD4+ICOShi T cells in post-therapy samples (Upper Panel, representative patient and Lower Panel, summary data from 3 different patients). (B) An ICOS-specific siRNA led to the decreased protein expression of T-bet and proteins indicative of PI3K-signaling (Upper Panel, representative experiment, and Lower Panel, summary data). Numerical values listed under each band represent calculated intensity for protein expression. (C) An ICOS-specific siRNA led to the decreased mRNA levels for ICOS, IFNγ, TNFα and T-bet (Lower panel). *indicates p<0.05; ns indicates no statistical significance.
ICOS expression is required for increased PI3K-signaling and T-bet expression during an in vivo antitumor immune response
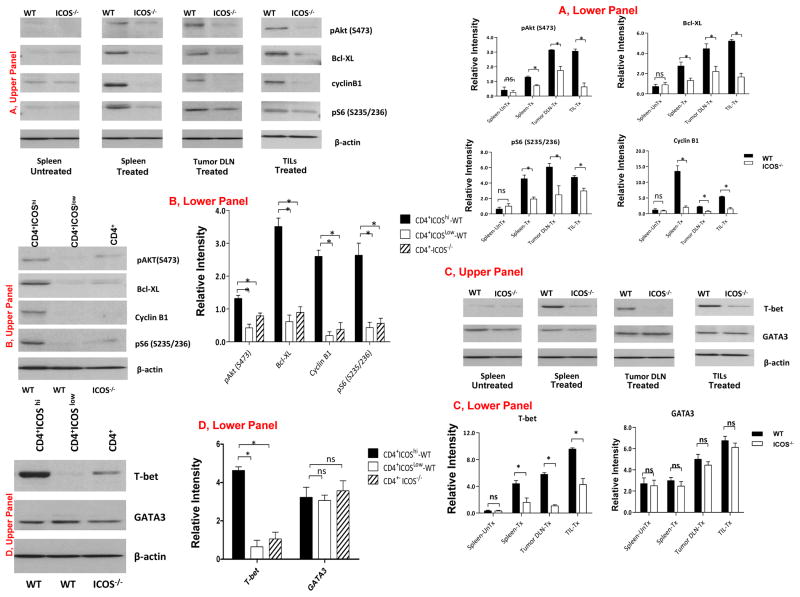
To investigate PI3K-signaling and T-bet expression during prolonged antitumor T cell responses, we performed ex vivo analyses of CD4 T cells harvested 7 days after the mice have completed treatment with anti-CTLA-4. We found that CD4 T cells from the spleen, tumor-draining lymph nodes (DLN) and tumor-infiltrating lymphocytes (TILs) of the treated WT mice had greater expression of proteins indicative of PI3K-signaling as compared to CD4 T cells from treated ICOS−/− mice (Fig. 3A, representative experiment, Upper Panel, and summary data, Lower Panel). Proteins indicative of PI3K-signaling were expressed to a much greater extent in CD4+ICOShi cells as compared to CD4+ICOSlow T cells from WT mice or CD4 T cells from ICOS−/− mice (Fig. 3B, representative experiment, Upper Panel, and summary data, Lower Panel). We also found that T-bet expression was markedly higher in T cells from the spleen, DLN and TILs of WT mice as compared to those from ICOS−/− mice (Fig. 3C, representative experiment, Upper Panel, and summary data, Lower Panel). CD4+ICOShi T cells from WT mice also had greater T-bet expression as compared to CD4+ICOSlow T cells from WT mice or CD4 T cells from ICOS−/− mice (Fig. 3D, representative experiment, Upper Panel, and summary data, Lower Panel). There was no difference in the expression of GATA-3. These data demonstrated significantly lower expression of proteins indicative of PI3K-signaling and substantially less T-bet expression in CD4 T cells from ICOS−/− tumor-bearing mice treated with anti-CTLA-4 therapy, as compared to CD4 T cells from anti-CTLA-4-treated WT mice.
Figure 3. ICOS expression is required for increased PI3K-signaling and T-bet expression during an in vivo antitumor immune response.
(A) Greater expression of proteins indicative of PI3K-signaling in CD4 T cells from the spleen, draining lymph nodes (DLN) and tumor-infiltrating lymphocytes (TILs) of treated WT mice as compared to those in CD4 T cells from treated ICOS−/− mice (Upper Panel, representative experiment, and Lower Panel, summary data from 3 independent experiments with mean and sem). (B) Greater expression of proteins indicative of PI3K-signaling in CD4+ICOShi cells from treated WT mice as compared to CD4+ICOSlow cells from treated WT mice and CD4 T cells from treated ICOS−/− mice (Upper Panel, representative experiment, and Lower Panel, summary data from 3 independent experiments with mean and sem). (C) Greater expression of T-bet protein in CD4 T cells from the spleen, draining lymph nodes (DLN) and tumor-infiltrating lymphocytes (TILs) of treated WT mice as compared to those of treated ICOS−/− mice (Upper Panel, representative experiment, and Lower Panel, summary data from 3 independent experiments with mean and sem). (D) Greater expression of T-bet protein in CD4+ICOShi cells from treated WT mice as compared to T-bet protein in CD4+ICOSlow cells from treated WT mice and in CD4 cells from treated ICOS−/− mice (Upper Panel, representative experiment, and Lower Panel, summary data from 3 independent experiments with mean and sem).
Of note, both WT and ICOS−/− mice had similar frequencies of CD28+ CD4 T cells (Supplementary Fig. 2B). Therefore, the differences in CD28 expression could not be the reason for the observed differences in PI3K-signaling and T-bet expression. In addition, short-term in vitro stimulation with anti-CD3 and anti-CD28 elicited similar levels of PI3K-signaling and expression of T-bet in CD4 T cells from both ICOS−/− and WT mice (Supplementary Fig. 2C). This in vitro data indicated that CD4 T cells from both ICOS−/− and WT mice had intact functional pathways via T cell receptor- and CD28-signaling to mediate PI3K-signaling and T-bet expression. However, despite having intact functional T cell receptor- and CD28-signaling pathways, the CD4 T cells from ICOS−/− mice had significantly diminished PI3K-signling and T-bet expression during a prolonged in vivo antitumor response. Taken together, these data suggest an important role for ICOS-mediated PI3K-signaling in regulating T-bet expression.
T-bet expression requires ICOS-mediated PI3K-signaling during an in vivo antitumor immune response
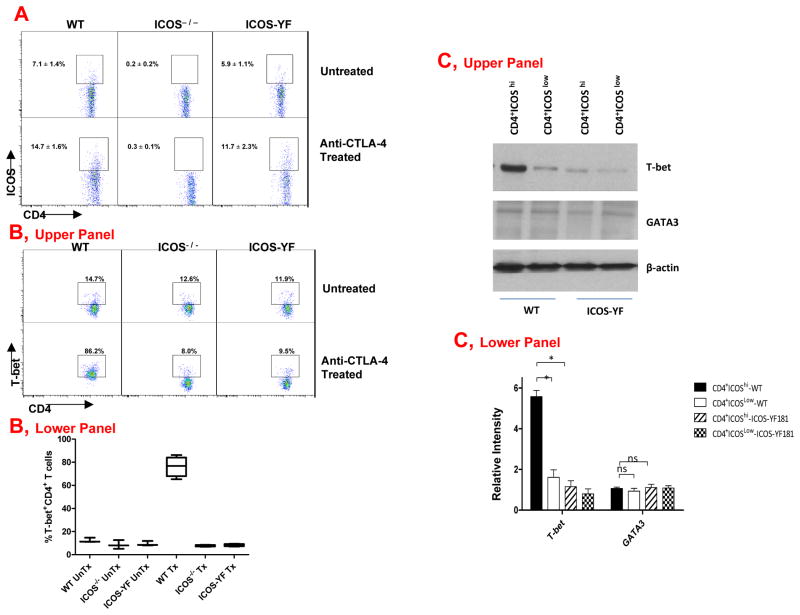
To determine whether ICOS controlled T-bet expression by signaling through the PI3K pathway, we studied T cell responses in ICOS-YF mice, which have a single amino acid change that abrogates the PI3K binding site within the ICOS cytoplasmic tail thereby significantly decreasing the expression of proteins indicative of PI3K-signaling in T cells from these mice (18) (Supplementary Fig. 3). Similar to T cells from WT mice, T cells from ICOS-YF mice demonstrated an increase in the surface expression of ICOS after tumor-bearing mice were treated with anti-CTLA-4 (Fig. 4A). However, despite this increase in the surface expression of ICOS, T cells from ICOS-YF mice did not have an increase in the frequency of T-bet+ T cells after treatment with anti-CTLA-4 (Fig. 4B). In contrast, CD4 T cells from treated WT mice had a 9-fold increase in the frequency of T-bet+ T cells (Fig. 4B, Upper Panel, representative experiment and Lower Panel, summary data). We sorted CD4+ICOShi and CD4+ICOSlow T cells from WT and ICOS-YF mice and confirmed by Western blot that T-bet expression was highest in CD4+ICOShi T cells from WT mice and significantly lower in T cells from ICOS-YF mice (Fig. 4C, representative experiment, Upper Panel, and summary data, Lower Panel). There was no difference in the expression of GATA-3. Collectively, our data indicate that anti-CTLA-4 therapy increases ICOS expression, which mediates an increase in T-bet expression through PI3K-signaling.
Figure 4. T-bet expression requires ICOS-mediated PI3K-signaling during an in vivo antitumor immune response.
(A) Tumor-bearing WT mice that received treatment with anti-CTLA-4 had an increased frequency of ICOS+ CD4 T cells, which was not found for CD4 T cells from ICOS−/− mice but was observed for CD4 T cells from ICOS-YF mice. (B) Treated WT mice had a significant increase in the frequency of T-bet+ CD4 T cells, which was not observed in the treated ICOS−/− and ICOS-YF mice (Upper Panel, representative experiment, and Lower Panel, summary data from 3 independent experiments with mean and sem). (C) Sorted CD4+ICOShi and CD4+ICOSlow T cells from WT and ICOS-YF mice revealed greater protein expression of T-bet in CD4+ICOShi cells from WT mice as compared to CD4+ICOSlow cells from WT mice or CD4+ICOShi and CD4+ICOSlow cells from ICOS-YF mice (Upper Panel, representative experiment and Lower Panel, Summary data from 3 independent experiments (mean and sem). UnTx indicates untreated; Tx indicates treated; *indicates p<0.05; ns indicates no statistical significance.
Discussion
We have analyzed samples from patients treated with anti-CTLA-4 (ipilimumab, BMS) to investigate and identify the immunologic biomarkers and/or mechanisms that play important roles in mediating antitumor immune responses. We were the first to detect an increase in CD4+ICOShi T cells in both patients (4, 6) and mice (9) after treatment with anti-CTLA-4. Other investigators also reported a similar increase in the frequency of CD4+ICOShi T cells after patients received treatment with a different anti-CTLA-4 antibody known as tremelimumab (27). We were also the first to report that the increased frequency of ICOS+ CD4 T cells may serve as a biomarker of clinical benefit for patients treated with ipilimumab (6) and similar findings were recently reported by another group (28). We were also the first to report that the increased frequency of ICOS+ CD4 T cells may serve as a pharmacodynamic biomarker for anti-CTLA-4 therapy (29). We also evaluated ICOS expression on T cells in the context of another immunotherapy strategy (gp100 vaccine); however, we did not observe an increase in the frequency of ICOS+ CD4 T cells after that vaccination (29), which suggests that the increased frequency of ICOS+ CD4 T cells may serve as a specific biomarker for anti-CTLA-4 therapy. In addition to serving as a biomarker of therapy, we reported that ICOS and its ligand (ICOSL) played important functional roles in mediating optimal antitumor immune responses, including the production of the Th1 cytokine IFNγ and tumor rejection in murine models (9).
It is well-established that early T cell immune responses rely on CD28-mediated PI3K-signaling (14, 15, 7); however, it has remained unclear whether CD28-mediated PI3K-signaling also plays a role in later T cell responses. We did not detect a change in the frequency of CD28-expressing T cells either after patients received anti-CTLA-4 treatment or after tumor-bearing mice received anti-CTLA-4 treatment. Our data demonstrated an important role for ICOS in antitumor T cell responses, and since the cytoplasmic tail of ICOS has an YMPM PI3K-binding motif (7, 16), we hypothesized that ICOS may enable PI3K-signaling to increase T-bet expression for later T cell responses in the setting of anti-CTLA-4 therapy. Our results demonstrated the expression of T-bet in ICOS-deficient T cells after 48 hours of in vitro activation (Supplementary Fig. 2C) but the expression of T-bet was impaired in ICOS-deficient T cells 7 days after in vivo anti-CTLA-4 therapy (Fig. 3C). These results suggest that T-bet expression is regulated temporally such that the initial T cell activation via the T cell receptor and CD28 costimulation can induce PI3K-signaling and T-bet expression but, this is a limited event, and sustained expression of T-bet, which may be required for Th1 antitumor responses and tumor rejection, requires ICOS-mediated PI3K-signaling. Additional studies will be needed to further test this concept.
Our studies indicated impaired PI3K-signaling and T-bet expression in CD4 T cells obtained ex vivo from ICOS−/− mice as compared to those from WT mice. To determine whether there was a link between ICOS-specific PI3K-signaling and T-bet expression, we used ICOS-YF knock-in mice, which have a single amino acid change (Y to F) thus preventing PI3K-signaling by ICOS. In studies with ICOS-YF mice, we found impaired T-bet expression, which was similar to the data observed in ICOS−/− mice. Collectively, our studies provide evidence for ICOS-mediated PI3K-signaling as an important regulator of T-bet expression for Th1 antitumor immune responses that occur after treatment with anti-CTLA-4.
Additional studies will be required to understand the mechanisms that occur during later T cell responses to facilitate ICOS-mediated PI3K-signaling as opposed to CD28-mediated PI3K-signaling. Previous studies highlighted the possibility that CD28-PI3K complexes may undergo preferential internalization thus limiting further CD28 signaling, or ICOS may have higher affinity for certain adaptor subunits of PI3K thus outcompeting CD28 for PI3K-signaling (7). Moreover, T-bet expression can be driven by IL-12/STAT4 signaling (12) and future studies will be required to determine whether the IL-12/STAT4 signaling pathway is distinct from the ICOS-mediated PI3K-signaling pathway. In addition, ICOS has been shown to play a role in Th2 immune responses (7) and it will be necessary to evaluate whether, under specific conditions, ICOS regulates the expression of GATA-3. Furthermore, PI3K-signaling has been established as a critical pathway for tumorigenesis and tumor progression (30). Therefore, PI3K-inhibitors are being tested in the clinic as therapeutic agents for cancer patients (31). However, the impact of these PI3K-inhibitors on human immune responses has not been well-studied and will need to be addressed.
In summary, we report here the regulation of T-bet expression through ICOS-mediated PI3K-signaling, which will impact design of future cancer immunotherapy strategies. We propose that CTLA-4 blockade enhances CD28-mediated signaling during the early phases of a T cell immune response thus facilitating increased expression of ICOS, which regulates PI3K-signaling and T-bet expression for the late-phases of a T cell response. Novel combination treatment with anti-CTLA-4 plus an agonistic agent to target ICOS should be developed and tested as a strategy to improve antitumor responses and tumor rejection. Importantly, our studies suggest that ICOS-mediated PI3K-signaling can regulate T-bet expression for the late phases of in vivo Th1 responses that may be necessary for other immune responses such as the eradication of prolonged bacterial and viral infections, which should be tested in future studies and considered for the development of novel therapeutic strategies to target ICOS.
Supplementary Material
Acknowledgments
This work was supported in part by a Department of Defense Idea Development Award, W81XWH-10-1-0073 01, and NIH/NCI R01 grant, CA1633793-01 (all to PS). We thank Willem Overwijk, Mike Davies and Jim Allison for critical reading of this manuscript.
Footnotes
The authors do not have any conflict of interest to declare related to this manuscript
References
- 1.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. Journal of Experimental Medicine. 1995;182:459–65. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–36. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 3.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. New England Journal of Medicine. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liakou CI, Kamat A, Tang DN, Chen H, Sun J, Troncoso P, et al. CTLA-4 blockade increases IFNγ-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proceedings of the National Academy of Sciences USA. 2008;105:14987–92. doi: 10.1073/pnas.0806075105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Liakou CI, Kamat A, Pettaway C, Ward JF, Tang DN, et al. Anti-CTLA-4 therapy results in higher CD4+ICOShi T cell frequency and IFNγ levels in both nonmalignant and malignant prostate tissues. Proceedings of the National Academy of Sciences USA. 2009;106:2729–34. doi: 10.1073/pnas.0813175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carthon BC, Wolchok JD, Yuan J, Kamat A, Ng Tang DS, et al. Preoperative CTLA-4 blockade: Tolerability and immune monitoring in the setting of a presurgical clinical trial. Clinical Cancer Res. 2012;16:2861–71. doi: 10.1158/1078-0432.CCR-10-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudd CE, Schneider H. Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat Rev Immunol. 2003;3:544–56. doi: 10.1038/nri1131. [DOI] [PubMed] [Google Scholar]
- 8.Simpson TR, Quezada SA, Allison JP. Regulation of CD4 T cell activation and effector function by inducible costimulator (ICOS) Curr Opin Immunol. 2010;22:326–32. doi: 10.1016/j.coi.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Fu T, He Q, Sharma P. The ICOS/ICOSL pathway is required for optimal antitumor responses mediated by anti-CTLA-4 therapy. Cancer Res. 2011;71:5445–54. doi: 10.1158/0008-5472.CAN-11-1138. [DOI] [PubMed] [Google Scholar]
- 10.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 11.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–42. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 12.Lazarevic V, Glimcher LH. T-bet in disease. Nature Immunol. 2011;12:597–606. doi: 10.1038/ni.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;11:836–48. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–89. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama T, Yamashita M. The TCR-mediated signaling pathways that control the direction of helper T cell differentiation. Seminars in Immunol. 2010;22:303–9. doi: 10.1016/j.smim.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 16.Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nat Rev Immunol. 2003;3:317–30. doi: 10.1038/nri1056. [DOI] [PubMed] [Google Scholar]
- 17.Harding FA, McArthur JG, Gross JA, Raulet DH, Allison JP. CD28-mediated signalling co-stimulates murine T cells and prevents induction of anergy in T cell clones. Nature. 1992;356:607–9. doi: 10.1038/356607a0. [DOI] [PubMed] [Google Scholar]
- 18.Gigoux M, Shang J, Pak Y, Xu M, Choe J, Mak TW, et al. Inducible costimulator promotes helper T cell differentiation through phosphoinositide 3-kinase. Proceedings of the National Academy of Sciences USA. 2009;106:20371–76. doi: 10.1073/pnas.0911573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saenger YM, Wolchok JD. The heterogeneity of the kinetics of response to ipilimumab in metastatic melanoma: patient cases. Cancer Immun. 2008;8:1. [PMC free article] [PubMed] [Google Scholar]
- 20.Hu J, He X, Baggerly KA, Coombes KR, Hennessy BT, Mills GB. Non-parametric quantification of protein lysate arrays. Bioinformatics. 2007;23:1986–94. doi: 10.1093/bioinformatics/btm283. [DOI] [PubMed] [Google Scholar]
- 21.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular Cancer Therapeutics. 2006;5:2512–21. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 22.Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlate with antibody responses to influenza vaccination. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onabajo OO, George J, Lewis MG, Mattapallil JJ. Rhesus macaque lymph node PD-1(hi)CD4+ T cells express high levels of CXCR5 and display a CCR7(lo)ICOS+Bcl6+ T-follicular helper (Tfh) cell phenotype. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–63. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 25.Kmieciak M, Gowda M, Graham L, Godder K, Bear HD, Marincola FM, et al. Human T cells express CD25 and FoxP3 upon activation and exhibit effector/memory phenotypes without any regulatory/suppressor function. J Transl Med. 2009;7:89–95. doi: 10.1186/1479-5876-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 27.Vonderheide RH, LoRusso PM, Khalil M, Gartner EM, Khaira D, Soulieres D, et al. Tremelimumab in combination with exemestane in patients with advanced breast cancer and treatment-associated modulation of inducible costimulator expression on patient T cells. Clin Cancer Res. 2010;16:3485–94. doi: 10.1158/1078-0432.CCR-10-0505. [DOI] [PubMed] [Google Scholar]
- 28.Di Giacomo AM, Calabrò L, Danielli R, Fonsatti E, Bertocci E, Pesce I, et al. Long-term survival and immunological parameters in metastatic melanoma patients who responded to ipilimumab 10 mg/kg within an expanded access programme. Cancer Immunol Immunother. 2013;62:1021–8. doi: 10.1007/s00262-013-1418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng Tang D, Shen Y, Sun J, Wen S, Wolchok JD, Yuan J, et al. Increased frequency of ICOS+ CD4 T cells as a pharmacodynamic biomarker for anti-CTLA-4 therapy. Cancer Immunol Res. 2013;1:1–6. doi: 10.1158/2326-6066.CIR-13-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–57. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 31.Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, et al. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. Journal of Clinical Oncol. 2012;30:282–90. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 32.Quackenbush J. Microarray data normalization and transformation. Nature Genetics. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 33.Zien A, Aigner T, Zimmer R, Lengauer T. Centralization: a new method for the normalization of gene expression data. Bioinformatics. 2001;17:323–31. doi: 10.1093/bioinformatics/17.suppl_1.s323. [DOI] [PubMed] [Google Scholar]
- 34.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.