Abstract
We describe a multistage approach to identify single nucleotide polymorphisms (SNPs) associated with neuroticism, a personality trait that shares genetic determinants with major depression and anxiety disorders. Whole genome association with 452 574 SNPs was performed on DNA pools from ~2000 individuals selected on extremes of neuroticism scores from a cohort of 88 142 people from southwest England. The most significant SNPs were then genotyped on independent samples to replicate findings. We were able to replicate association of one SNP within the PDE4D gene in a second sample collected by our laboratory and in a family-based test in an independent sample; however, the SNP was not significantly associated with neuroticism in two other independent samples. We also observed an enrichment of low P-values in known regions of copy number variations. Simulation indicates that our study had ~80% power to identify neuroticism loci in the genome with odds ratio (OR) > 2, and ~50% power to identify small effects (OR = 1.5). Since we failed to find any loci accounting for more than 1% of the variance, the heritability of neuroticism probably arises from many loci each explaining much less than 1%. Our findings argue the need for much larger samples than anticipated in genetic association studies and that the biological basis of emotional disorders is extremely complex.
Keywords: neuroticism, whole genome association, DNA pooling, personality, quantitative trait, PDE4D
Introduction
Ever since neuroticism was first proposed as personality trait that reflects a tendency toward states of negative affect,1 it has been included in nearly every theory of personality. Its importance as a psychological construct is further enhanced by its well documented correlation with common psychiatric disorders, that is, high levels of neuroticism predict the onset and subsequent episodes of major depression2–5 and are associated with symptoms of depression and anxiety in the general population.6–9 The correlation between anxiety, major depression and neuroticism is, in part, due to the presence of shared genetic factors,10–13 an observation that has spurred attempts to map the genetic basis of neuroticism.14–16 It is much easier to acquire the large samples necessary for genetic studies of neuroticism, which is assessed using a self-administered questionnaire, than it is to acquire large patient samples that require diagnostic interviews for accurate assessment of major depression and anxiety.
Genetic linkage analysis has had mixed success in mapping susceptibility loci for neuroticism. One study of extremely discordant and concordant siblings identified five loci that exceeded a 5% genome-wide significance threshold,14 but a second study, also using a selected sample, failed to identify any genome-wide significant signals.15 One explanation for the difficulties encountered is that the proportion of genetic variance attributable to an individual locus is extremely small, so that the linkage studies are underpowered. A recent survey of some 50 meta-analyses and 752 individual studies concluded that the typical effect sizes of individual genetic variants for complex disease ranged from 1.2 to 1.6.17
Whole genome association provides an alternative approach to detect small genetic effects. This is still an expensive option, but costs can be reduced by pooling equal amounts of DNA from each individual, separately for cases and controls, and estimating allele frequencies from the DNA pools (allelotyping). Even though allelotyping is replicated several times to increase accuracy, there is still a 10- to 100-fold decrease in cost compared to individual genotyping. DNA pooling can also be applied to quantitative traits in a method known as selective DNA pooling.18 Selective DNA pooling combines DNA pooling and selective genotyping, which is the genotyping of individuals only from the extreme of the distribution, with the most informative trait values. While DNA pooling offers a substantial reduction in genotyping cost, it possesses some disadvantages, primarily in the loss of information (genotype and haplotype distributions) and by introducing estimation errors. Recent studies have shown the feasibility and accuracy of estimating single-nucleotide polymorphism (SNP) allele frequencies in DNA pools using microarrays containing thousands of SNPs,19–23 which makes it possible to perform whole genome association studies in a cost-effective way.
We have collected a sample suitable for association studies of small genetic effects on neuroticism. We selected ~3600 individuals from a very large community cohort (n = 88 142) from southwest England, on the basis of the extremity of their neuroticism scores. Using a genomic control approach we have previously shown that stratification is unlikely to be a major confusion in the sample.24–26 Here we describe a multistage approach to identify SNPs associated with neuroticism in this as well as in replication samples. Although our initial sample was phenotyped for other personality dimensions, the sample used in this study was selected for having extreme neuroticism score and thus it is not suitable for the study of other dimensions.
Materials and methods
The sample
Individuals were selected from the patient registers of general practices in four counties in southwest England, that is, Oxfordshire, Gloucestershire, Somerset and Berkshire (as described by Fullerton et al.14 and Martin et al.27). We chose patients in rural practices, so that the sample is likely to be relatively genetically homogeneous. A total of more than 88 141 individuals completed the revised Eysenck Personality Questionnaire (EPQ). The neuroticism (N) scale of the EPQ consists of 23 questions scored on a two-point scale (0 or 1).28 N scores of the EPQ are correlated with similar measures on other personality measures such as the NEO PI-R N scale and the TPQ.29,30 The total scores were regressed on age and sex and the residuals were standardized so that each individual’s score is expressed in s.d. units.
From this population, two independent samples were selected for genotyping according to the extremeness of EPQ–N scores. The first population, originally selected for neuroticism linkage mapping, consists of one member of extremely concordant and discordant sibling pairs (as described by Fullerton et al.14 and Martin et al.27), while the second population, collected for genetic association, consists of singletons (as described by Willis-Owen et al.24). For the singletons, we received around 20%replies to our request of DNA samples. There was no difference in the distribution of N scores between subjects that replied and the ones that did not reply. We also received an equivalent rate of reply from high and low N extreme groups.
We screened a total of 3589 individuals; the first group of 2054 individuals was used in the whole genome scan with DNA pools and the second group of 1534 individuals was used as a replication sample. Out of the 2054 individuals (1038 high N and 1016 low N) that were included in the pools, 1301 were women and 753 were men. The replication sample consisted of 932 women and 601 men (831 high N and 702 low N). This study was approved by a research ethics committee in the UK, and appropriate informed consent was obtained.
DNA pools
DNA samples were collected and extracted from mouth swabs, quantified four times using the Pico-Green assay (Molecular Probes, Eugene, OR, USA) and tested by PCR. The samples were divided into eight groups according to N score (high and low) and gender (men and women). Equal amounts of DNA (150 ng) from each subject were mixed together to create 48 small pools, which on average were made from 43 individuals. Eight different pools were then created once by combining DNA from the small pools in proportion to the number of individuals in each pool. The eight pools are as follows: (1) men with high N score (n = 112), (2) men with low N score (n = 158), (3) men with very high N score (n = 245), (4) men with very low N score (n = 238), (5) women with high N score (n = 320), (6) women with low N score (n = 205), (7) women with very N high score (n = 340) and (8) women with very N low score (n = 436) (very high or low N scores are more than 1.5 s.d. from the mean score adjusted to age and sex (on average 2 s.d.); high and low N scores are between 1 and 1.5 s.d. from the mean score (on average 1.3 s.d.)).
We tested the accuracy of the pools before starting the experiment with the Affymetrix arrays by individually genotyping five SNPs and allelotyping the pools seven times using the Sequenom platform. Frequencies were averaged and corrected for unequal detection of the two alleles based on the ratios in individual heterozygotes.31 The pools were found to be very accurate, with an s.d. from the expected frequency of 0.016 for each of the eight large pools. The s.d. attributed to the pools (construction error) without the measurement error is estimated to be 0.012 for each pool and 0.0059 for frequency estimate for the high and low N groups with four pools in each. The observed frequencies were highly correlated with the expected frequencies for the small pools and, as expected, higher for the eight large pools (small pools: R2 = 98.8%; large pools: R2 = 99.7%).
Allelotyping with Affymetrix chips
We used the Affymetrix 100 and 500K mapping arrays sets. The arrays and the number of SNPs on each array are as follows: Nsp array with 255 931 SNPs, Sty with 238 304 SNPs, Hind with 57 244 SNPs and Xba with 58 960 SNPs. The total number of unique SNPs with dbSNP rs ID numbers is 577 889 (31 462 SNPs are present on more than one array). Out of those, 452 574 SNPs have a minor allele frequency above 5%.
According to the manufacturer’s protocol, 250 ng from each pool was digested with a restriction enzyme, ligated to an adaptor, amplified, fragmented, labeled and hybridized to the chip. Washing and staining was performed using the Fluidics Station 450 and scanned using the GeneChip Scanner 3000 7G. Each DNA pool was amplified and hybridized five times to different chips of the same type. The detection rate (MDR) for the 100K arrays was above 99% for all arrays. For the early access to 500K arrays the detection rate (MDR) was not available. Water, instead of DNA, was used as a negative control to test for contamination and the manufacturer’s individual reference DNA as a positive control to test for assay performance. Accuracy of genotyping call of the reference DNA was 99.8%.
Data analysis of DNA pools
An allele frequency estimate was calculated based on the relative intensities of the two alleles (know as the relative allele score; RAS) for each SNP and for each of the eight pools.32 The frequency estimate in each pool is the average across the five measurements. Frequency estimates for high and low N were obtained from the average frequency in the four pools belonging to each category. An SNP-specific variance was estimated by calculating the variance of the five measurements for each DNA pool across the eight pools . The allele frequencies from HapMap Centre d’Etude du Polymorphisme Humain (CEPH) trios were used as an estimate for the allele frequencies in the population (P̂). We modified the standard chi-squared statistic to include the error variance introduced by the DNA pooling procedure. The test statistic is expressed as:
where PH is the frequency in high N pools and PL in low N pools, NH is the number of individuals in high N pools and NL is the number of individuals in low N pools and npH and npL are the number of measurements for high and low N pools. P-values were calculated using a chi-squared distribution for all SNPs with a minor allele frequency above 0.05. We did not attempt to produce a test statistic with desirable statistical properties. The log P values calculated based on this statistic do not correspond to the expected type I error and were only used to rank and prioritize the SNPs for individual genotyping. The empirical distribution of the pools’ log P values was assessed using a simulation with a null effect (see below).
Simulation
To estimate a genome-wide significance threshold, we generated 1000 panels of 4000 chromosomes by random sampling with the replacement from the phased HapMap data (n = 120 from unrelated individuals in CEU).33 For each panel we calculated the significance of allele frequency differences between two groups of 2000 chromosomes for all SNPs with a minor allele frequency greater than 0.05, included in the 500 and 100K Affymetrix arrays. The most significant value in each panel was recorded.
For each SNP in the DNA pooling simulation the allele frequency in the HapMap CEPH sample was used. To simulate the errors introduced by the DNA pooling procedure we first generated a random normal deviate from the true HapMap frequency using the estimated construction error variance. We then assigned a number of measurements to each simulated pool with an error based on the SNP-specific measurement error and assumed a normal distribution.
To evaluate the power of individual genotyping and DNA pooling we designated 2556 random SNPs from the HapMap (whether they were included in the arrays or not) to be the causal variant with an effect size, measured in terms of odds ratios (ORs), of 1.2, 1.3, 1.5, 1.7, 2 and 3. All the SNPs in HapMap phase II with minor allele frequency (MAF) > 0.05, in the CEPH population,33 had an equal chance (0.1%) of being the causal variant. Based on the frequency of the causal SNP, we assigned the allele frequencies to two groups with a frequency difference that corresponded to the effect sizes. We sampled phased chromosomes with different probabilities based on the allele frequencies. We calculated the significance of the allelic χ2 test at the causal SNP and for SNPs in a 100 kb window surrounding the causal SNP that are included in the Affymetrix arrays. For the DNA pooling we introduced two sources of error, that is, construction error and measurement error, as described above. Similar to our study design, we simulated 20 measurements for each of the two groups for all SNPs in the 100 kb window. We calculated the significance of each SNP in the same way used for the real allelotyping data. For the individual genotyping simulation we recorded the minimum P-value across all SNPs in the window. For the DNA pooling we simulated a design where all SNPs with log P > 4 (the negative logarithm, base 10, of the P-value) were individually genotyped. To simulate this design, we recorded the minimum P-value of the individual genotypes across all SNPs in the window that exceeded a log P of 4 in the DNA pooling simulation.
Individual genotyping
Individual genotyping was performed using the Sequenom MassArray platform according to the manufacturer’s instructions (SEQUENOM, San Diego, CA, USA). The DNA samples in the pools were genotyped using the iPLEX assay with up to 29 SNPs multiplexed together. The replication sample was genotyped using the homogenous MassEXTEND (hME) assay. Chips were read using the Bruker Autoflex Mass Spectrometer system (Bruker-Sequenom, San Diego, CA, USA). As a quality control for the iPLEX assay we genotyped 69 HapMap CEPH DNA samples with known genotypes. The concordance level between the genotypes was on average 99.4%. Hardy–Weinberg (HW) equilibrium was tested on all SNPs, but was not used automatically to exclude SNPs because all samples used are selected from the extreme of the population and might show deviation from HW in SNPs associated with the trait. To check the genotyping quality of SNP rs702543, we compared 634 genotypes obtained using the HME and iPLEX assays. The concordance level was 100%. Haplotypes were analyzed based on a score statistic using ‘haplo.stats’ package for R.34
Results
Sensitivity and specificity of DNA pooling
The design and interpretation of the DNA pooling experiment require an assessment of the method’s likely power, that is, its sensitivity and specificity. If we were to genotype all 2000 individuals individually, we would be able to detect an effect explaining 1, 0.5 and 0.1% (ORs of 1.72, 1.47 and 1.19) of the additive genetic variance with power of 0.94, 0.34 and 0.001, respectively (assuming an alpha of 1.66 × 10−7 (300 000 independent tests, a complete linkage disequilibrium (LD) between the marker and the causative SNP and a trait increasing allele frequency of 0.2)). If we test only 20 markers, for example, in a replication sample, our power is 1, 0.96 and 0.19 for 1, 0.5 and 0.1% effects, respectively.
To investigate the pooling strategy we simulated the design of the study in which we used eight different pools, each allelotyped five times (that is, 20 measurements for each group). Using samples of CEPH individuals genotyped as part of the HapMap project,33 we established that we could accurately simulate the pooling experiment (see Supplementary material for a full description). We first simulated a situation with a null effect and found that the genome-wide 5% threshold for individual genotyping is 1.7 × 10−7 (log P = 6.77), which is equivalent to a 5% threshold after applying a Bonferroni correction for 294 071 independent tests. Then, by simulation, we determined the power to detect an association by genotyping directly the simulated causal SNP, by individual genotyping of SNPs on the Affymetrix arrays (500 and 100K with MAF> 0.05) and using a DNA pooling strategy. We designed a random SNP from the HapMap to be a causal SNP and we looked at SNPs in the array within a 100 kb window. By simulating an SNP from the HapMap as a causal SNP we include in our simulation the effect of the coverage of the arrays (the LD between the SNPs on the array and SNPs in the genome, represented by HapMap data) and the possibility that more than one SNP on the array is in LD with the causal SNP.
We looked at two study designs. In the first design, we assessed our ability to detect an SNP that exceeds the 5% genome-wide threshold (log P = 6.77). In the second design, we assessed what would happen when we test in a second sample all SNPs that exceed a log P of 3 in the analysis of the first sample. We calculated the power using different effect sizes of OR between 1.2 and 3 (Table 1). For small effect sizes, that is, ORs of 1.2, 1.3 and 1.5, the power to exceed the 5% genome-wide threshold using the DNA pooling strategy is 0, 3 and 31%, respectively. This is around 75% of the power achieved by individual genotyping, but incurs only 2% of the cost (40 × 4 arrays relative to 2000 × 4). For the study design that includes testing all log P > 3 SNPs in a second sample, power is the probability of inclusion in the list for replication without taking into account the power of the second sample to replicate the findings. The power of DNA pooling with the second design is 4, 16 and 47% for ORs of 1.2, 1.3 and 1.5, respectively. For this design and with these ORs the reduction in power ranges between 41 and 79% compared to individual genotyping.
Table 1.
Power of DNA pooling strategy compared to individual genotyping
| OR | Functional SNP (%) |
Individual genotyping (%) |
DNA pooling (%) |
|---|---|---|---|
| 5% threshold | |||
| 1.2 | 0 | 0 | 0 |
| 1.3 | 6 | 4 | 3 |
| 1.5 | 51 | 41 | 31 |
| 1.7 | 80 | 68 | 56 |
| 2 | 93 | 82 | 72 |
| 3 | 99 | 90 | 85 |
| Log P >3 | |||
| 1.2 | 19 | 19 | 4 |
| 1.3 | 53 | 49 | 16 |
| 1.5 | 88 | 80 | 47 |
| 1.7 | 97 | 90 | 66 |
| 2 | 99 | 92 | 76 |
| 3 | 100 | 95 | 87 |
Abbreviations: OR, odds ratio; SNP, single-nucleotide polymorphism; 5% threshold, power to detect the association with significance above the 5% genome-wide threshold; log P > 3, percentage of cases with log P > 3, in a design that includes testing the log P > 3 SNPs in a replication sample.
Pooling and individual genotyping
We screened 2054 extreme scoring individuals in eight DNA pools for an association with neuroticism (Figure 1). We calculated an association log P value for 452 574 SNPs (with MAF > 5%) based on the frequency differences between high and low N pools, the SNP allele frequency and the SNP-specific measurement error. SNPs with MAF below 0.05 were excluded because of the limited power to detect a significant association and the decrease in the accuracy of DNA pooling with rare variants. We ranked the SNPs based on the log P values and used a log P>4 as the cutoff for individual genotyping. We excluded 78 SNPs because the allele frequency estimates had been biased by an outlier in one of the pools. Four hundred and forty SNPs were considered for individual genotyping. Out of 440 SNPs, the assays of 405 SNPs were successfully designed and were used to genotype a subset of the sample (759 individuals). We calculated the significance for allele frequency differences between high and low N for sex-averaged and sexspecific frequencies for each SNP. Forty-two SNPs (10%) had a P-value between 0.05 and 0.01, and 28 SNPs (7%) had a P-value < 0.01. We genotyped the whole sample with 110 SNPs that showed sex-specific or sex-averaged P-values < 0.1. No SNP had a P-value that remained significant after correcting for multiple testing (300 000 tests). Eighteen SNPs had a P-value <10−3, three SNPs had a P-value < 10−4 and two had a P-value < 10−5. As expected, our results using DNA pooling (with a log P > 4 as cutoff) produced a significant deficiency in P-values < 10−3 compared to what would be expected by chance if all SNPs were individually genotyped (~300 expected by chance). However, we do not see a significant deficiency of P-values < 10−5 (~3 expected by chance), which indicates that our method would have potentially picked SNPs with genome-wide significant values after Bonferroni correction for multiple tests (P-values < 0.05/300 000 = 1.67 × 10−7). The highest scoring SNPs and their associated genes are shown in Table 2.
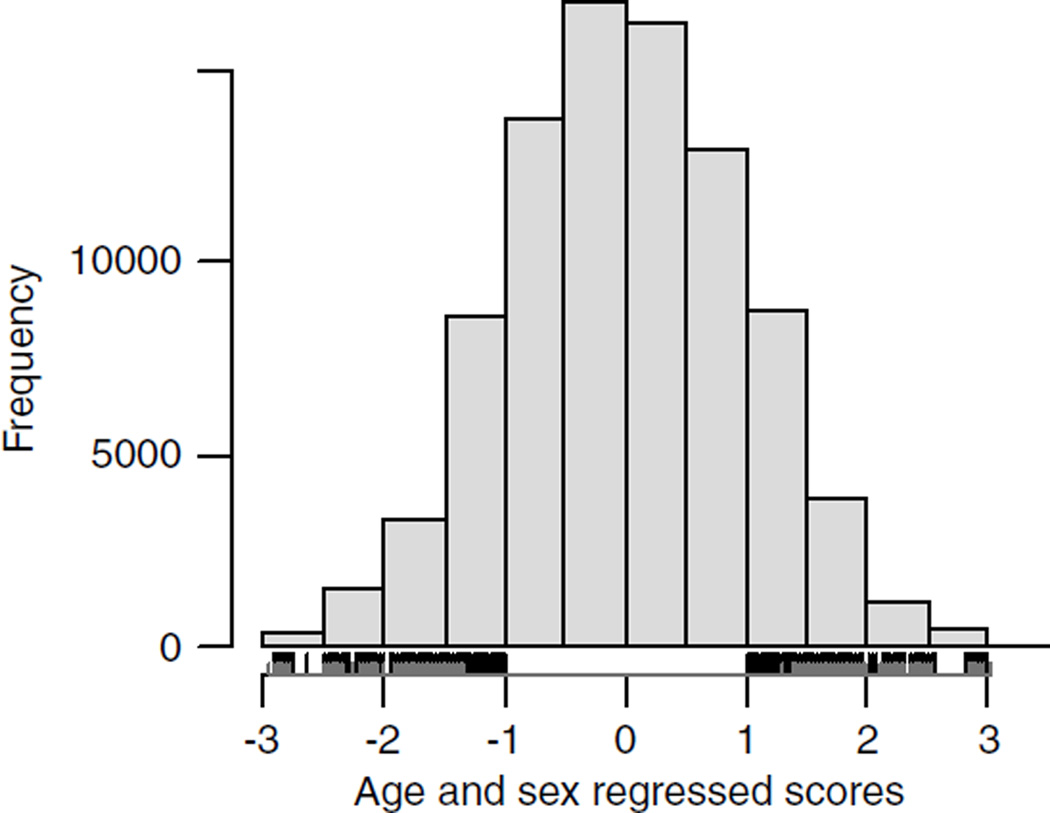
Figure 1.
Distribution of N scores as standardized residuals of the sum scores after regression on age and sex (88 141 individuals). The selection of individuals with extreme score is presented below the histogram. The black ticks mark the sample used in the DNA pools, and the gray represents the sample used in the replication sample.
Table 2.
Results of individual genotyping
| SNP | DNA pooling ranka |
P-values for allele frequencies differences | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP identifier |
Geneb | Chrc | Position | Genotypingd | Replicatione | |||||
| All | M | F | All | M | F | |||||
| rs322239 | PTN | 7 | 136413862 | 12 | 4.8E–06 | 1.1E–04 | 5.7E–03 | 8.8E–01 | 7.4E–01 | 6.5E–01 |
| rs4841017 | 8 | 8614503 | 53 | 6.5E–06 | 2.3E–05 | 1.5E–02 | 2.3E–01 | 3.7E–01 | 4.1E–01 | |
| rs17385253 | 1 | 34564102 | 21 | 3.3E–05 | 7.7E–03 | 1.5E–03 | 2.8E–01 | 1.3E–01 | 8.5E–01 | |
| rs9329165f | 8 | 8614052 | 160 | 3.8E–05 | 3.0E–05 | 4.9E–02 | 2.1E–01 | 9.4E–01 | 9.5E–02 | |
| rs10483573 | 14 | 46932227 | 258 | 5.6E–05 | 3.6E–03 | 3.2E–03 | 6.1E–01 | 7.8E–01 | 3.9E–01 | |
| rs10504830 | CNBD1 | 8 | 88448321 | 115 | 1.1E–04 | 6.8E–04 | 2.2E–02 | 5.7E–01 | 1.4E–01 | 6.8E–01 |
| rs13290746g | 9 | 34858377 | 146 | 1.5E–04 | 3.6E–02 | 1.6E–03 | 3.8E–01 | 2.8E–01 | 7.8E–01 | |
| rs9959800 | 18 | 52115138 | 48 | 1.7E–04 | 1.1E–02 | 6.0E–03 | 1.4E–01 | 8.0E–01 | 3.8E–02 | |
| rs10797812 | 1 | 179716254 | 159 | 2.4E–04 | 2.2E–02 | 4.1E–03 | 5.1E–01 | 2.6E–01 | 7.2E–02 | |
| rs4875610 | CSMD1 | 8 | 3165318 | 430 | 4.1E–04 | 2.6E–02 | 6.3E–03 | 6.5E–01 | 4.7E–01 | 9.9E–01 |
| rs2123315f | 4 | 18642838 | 136 | 4.3E–04 | 2.0E–01 | 3.8E–03 | 9.6E–02 | 5.1E–01 | 1.3E–01 | |
| rs702543 | PDE4D | 5 | 58878531 | 184 | 8.2E–04 | 9.8E–04 | 9.5E–02 | 7.7E–04 | 2.7E–01 | 5.9E–04 |
| rs7666238 | 4 | 142453368 | 315 | 9.0E–04 | 2.1E–03 | 6.9E–02 | 6.1E–01 | 1.8E–01 | 7.9E–02 | |
| rs9431663 | TRIM67 | 1 | 227620956 | 211 | 1.2E–03 | 8.2E–01 | 1.2E–04 | 9.7E–01 | 8.6E–02 | 1.6E–01 |
| rs7594674g | 2 | 67191611 | 432 | 2.6E–03 | 4.3E–01 | 6.5E–04 | 3.9E–01 | 4.0E–01 | 7.9E–02 | |
| rs873989 | DAB1 | 1 | 58268619 | 148 | 3.5E–03 | 8.2E–01 | 1.3E–04 | 9.3E–01 | 4.9E–01 | 5.2E–01 |
| rs401897 | C20orf32 | 20 | 54463165 | 394 | 5.6E–03 | 6.5E–04 | 3.4E–01 | 5.2E–01 | 8.3E–01 | 4.4E–01 |
| rs201997 | ADAM18 | 8 | 39703325 | 186 | 6.4E–03 | 2.7E–04 | 5.3E–01 | 1.2E–01 | 6.0E–02 | 6.4E–01 |
| rs1452788 | TCF4 | 18 | 51268302 | 384 | 6.5E–03 | 6.7E–01 | 9.3E–04 | 5.7E–01 | 3.9E–01 | 9.7E–01 |
Abbreviation: SNPs, single-nucleotide polymorphisms.
SNPs within a copy number variation region are in bold.
Rank of SNPs based on log P results from the DNA pooling experiment.
SNP associate gene based on NCBI dbSNP.
Chromosome.
Results from individual genotyping the sample in the pools.
Results from genotyping the replication sample.
Genotyping distribution deviates from Hardy–Weinberg equilibrium in sample included in the pools (P-value < 0.0026).
Genotyping distribution deviates from Hardy–Weinberg equilibrium in both samples using two different SNP genotyping platforms (iPLEX and HME).
Haplotype analysis
To examine the possibility that multiple SNPs, each of small effect, might lie within the same gene, we searched for genes or intervals of 50 kb or less containing one SNP with a log P greater than 4 (based on the pooling analysis) and at least one other SNP with a log P greater than 2. Since LD would explain the presence of clusters of SNPs with high log P values, we excluded the pairs of SNPs whose r2 values (from the HapMap project) were greater than 0.6. We identified 81 SNPs from 34 regions for individual genotyping. Nineteen clustered SNPs showed a P-value < 0.05 and eight SNPs a P-value < 0.01 (Table 3). From each region we selected three SNPs (or two when three could not be found) with the most significant P-values for haplotype analysis since, in some situations, haplotype analysis is statistically more powerful.35 We compared the minimum P-value obtained from the single SNP analysis and the minimum P-value obtained from the haplotype analysis (global and haplotype-specific analyses). The minimum P-value from the haplotype analysis was substantially lower (< 0.5) in only two cases than that obtained from single marker analysis. The analysis of three SNP haplotypes (rs702543, rs17782374 and rs35277) of the PDE4D gene gave a simulated P-value for the maximum haplotype-specific score statistics of 0.00022 relative to a minimum P-value of 0.00082 for the single SNP analysis. The analysis of two SNP haplotypes (rs17225638 and rs11177914) at the RAB3IP gene gave a simulated P-value for the global statistics of 0.0069 relative to a minimum P-value of 0.020 for the single SNP analysis.
Table 3.
Haplotype analysis
| Genea | Chrb | Position | SNP1 | SNP2 | SNP3 | SNP min Pc | Global sim Pd | Max-stat sim Pe |
|---|---|---|---|---|---|---|---|---|
| LOC284661 | 1 | 4370062 | rs6679220 | rs349410 | 4.2E–02 | 2.1E–01 | 1.9E–01 | |
| DAB1 | 1 | 58268619 | rs873989 | rs1213659 | rs1202835 | 1.3E–04 | 1.5E–02 | 3.9E–02 |
| NRXN1 | 2 | 50761056 | rs7576283 | rs1037426 | rs930295 | 1.0E–01 | 7.2E–01 | 7.3E–01 |
| CTNNA2 | 2 | 80715942 | rs216617 | rs3770369 | rs7609261 | 2.9E–03 | 3.6E–02 | 1.2E–01 |
| DPP10 | 2 | 116252992 | rs10496506 | rs11123306 | 2.1E–01 | 5.4E–01 | 8.2E–01 | |
| LRP1B | 2 | 140847651 | rs13021003 | rs2046563 | rs17551974 | 1.5E–02 | 8.6E–02 | 2.9E–02 |
| FHIT | 3 | 60694463 | rs17361653 | rs1882899 | rs1716741 | 1.3E–03 | 8.6E–04 | 1.9E–02 |
| NA | 3 | 153382923 | rs641025 | rs989920 | rs325738 | 2.6E–03 | 2.6E–03 | 8.7E–03 |
| NA | 3 | 162481199 | rs7652267 | rs1478565 | 8.3E–02 | 1.3E–01 | 1.4E–01 | |
| NLGN1 | 3 | 175393516 | rs6806890 | rs6779246 | rs7623402 | 1.6E–02 | 7.2E–02 | 1.1E–02 |
| NA | 4 | 116523708 | rs1112531 | rs963251 | 5.1E–02 | 3.6E–01 | 6.4E–01 | |
| PDE4D | 5 | 58878531 | rs702543 | rs17782374 | rs35277 | 8.2E–04 | 4.7E–04 | 2.2E–04 |
| NA | 6 | 33080668 | rs592625 | rs7743563 | rs2523666 | 6.9E–02 | 4.2E–01 | 1.9E–01 |
| NA | 6 | 120286484 | rs10484965 | rs10499094 | rs9320695 | 7.3E–03 | 1.3E–01 | 3.7E–01 |
| LAMA2 | 6 | 129281463 | rs7776116 | rs9321142 | rs6928495 | 1.1E–03 | 3.4E–02 | 5.9E–03 |
| PARK2 | 6 | 162539612 | rs6904579 | rs3019449 | rs17649761 | 1.6E–03 | 4.1E–02 | 1.9E–02 |
| LOC442275 | 6 | 164461465 | rs206694 | rs2764649 | 4.3E–02 | 8.1E–02 | 5.0E–02 | |
| CREB5 | 7 | 28296787 | rs4722801 | rs177582 | 3.7E–02 | 9.0E–02 | 1.3E–01 | |
| NA | 7 | 95721578 | rs2724041 | rs2524976 | 6.9E–03 | 6.5E–02 | 1.0E–01 | |
| RELN | 7 | 102871243 | rs362698 | rs17321820 | rs1541329 | 2.4E–02 | 3.0E–01 | 2.6E–01 |
| PTN | 7 | 136413862 | rs322239 | rs322323 | rs322236 | 4.8E–06 | 8.0E–05 | 1.0E–05 |
| NA | 8 | 8614503 | rs4841017 | rs9329165 | rs6601729 | 6.5E–06 | 1.9E–04 | 1.7E–04 |
| CSMD1 | 8 | 3178277 | rs4395910 | rs10503232 | rs4875610 | 4.1E–04 | 8.5E–04 | 6.2E–04 |
| NRG1 | 8 | 32336767 | rs1487152 | rs7002732 | rs12114401 | 4.6E–04 | 4.1E–04 | 3.0E–03 |
| NA | 8 | 108959180 | rs1389976 | rs10505119 | 1.9E–02 | 7.9E–02 | 3.2E–02 | |
| CSMD3 | 8 | 114031134 | rs10505196 | rs4876501 | rs10505174 | 2.2E–02 | 2.0E–01 | 4.4E–01 |
| PTPRD | 9 | 8481878 | rs6477311 | rs1359114 | rs10511494 | 1.2E–02 | 7.5E–02 | 3.1E–02 |
| ASTN2 | 9 | 116859364 | rs2418446 | rs10513281 | rs10983238 | 3.4E–02 | 2.2E–01 | 4.1E–01 |
| TMEM16C | 11 | 26402767 | rs10834976 | rs1381181 | rs10834971 | 4.4E–03 | 1.4E–02 | 2.7E–02 |
| BDNF | 11 | 27715568 | rs985205 | rs2049045 | 1.7E–01 | 4.3E–01 | 3.6E–01 | |
| DLG2 | 11 | 84283401 | rs17147761 | rs891773 | rs10501548 | 4.4E–02 | 3.7E–01 | 4.9E–01 |
| CNTN5 | 11 | 98413115 | rs10501893 | rs10501898 | rs10501903 | 4.1E–02 | 4.9E–02 | 6.2E–02 |
| GRIP1 | 12 | 65150558 | rs4913307 | rs12314237 | rs12581890 | 1.2E–02 | 1.1E–02 | 2.0E–02 |
| RAB3IP | 12 | 68480702 | rs17225638 | rs11177914 | 2.0E–02 | 6.9E–03 | 1.5E–02 | |
| LOC284058 | 17 | 41550514 | rs10514901 | rs17631303 | rs2532276 | 9.4E–03 | 6.7E–02 | 2.2E–02 |
| TCF4 | 18 | 51268302 | rs1452788 | rs12326693 | rs9957668 | 9.3E–04 | 2.8E–01 | 3.2E–01 |
| NA | 18 | 52115138 | rs9959800 | rs1229587 | 1.7E–04 | 1.5E–03 | 1.4E–03 | |
| GRIK1 | 21 | 30306168 | rs2832533 | rs2832438 | 5.4E–03 | 1.0E–02 | 1.0E–02 | |
| ERG | 21 | 38888946 | rs17194186 | rs2836502 | rs2836365 | 2.2E–02 | 2.4E–02 | 1.8E–02 |
Abbreviation: NA, not available.
SNPs within a copy number variation region are in bold.
SNP associate gene based on NCBI dbSNP.
Chromosome.
Minimum P-value from the single SNP analysis.
Simulation P-value for the global score statistic.
Simulation P-value for the maximum score statistic.
Distribution of P-values obtained in the DNA pooling stage in linkage regions
To enhance the power to detect association in regions where we had prior information for a genetic effect, we took into account evidence from a previous linkage scan that used the same population analyzed here by association.27 Analysis of allele sharing in discordant and concordant sib pairs previously identified five significant quantitative trait loci (QTLs) for neuroticism, on 1q, 4q, 7p, 12q and 13q. We tested whether the distribution of P-values obtained in the DNA pooling stage was higher than expected in the QTL regions.
We calculated the total proportion of log P values above 2, 3, 4 and 5 in the 1-LOD score intervals for the five significant peaks. The proportion of log P values greater than 2, 3, 4 and 5 was also calculated in a set of five regions of the same size chosen at random from across the genome, and this process was repeated 1000 times. We found no significant enrichment in the 1-LOD score intervals of the QTL regions, when analyzed together. However, when we tested individual regions (comparing the proportion of log P values above a certain value in each 1-LOD score interval compared to random selected region), we found a nominally significant (P = 0.022) enrichment in log P > 2 values in the 1-LOD interval on chromosome 1q. This result becomes non-significant when corrected for multiple testing.
Copy number variants
We noticed that some SNPs with high log P values in the DNA pooling experiment are clustered in regions of known copy number variation (CNV). In the DNA pooling experiment we calculated the association score using the relative intensities of the probes, which might be influenced by CNVs. Hence, a cluster of high log P values could be a result of different frequencies of CNVs in cases and controls rather than of SNP allele frequency differences. We tested the distribution of log P values in regions of known CNVs and random regions of the same size (in the same way as we investigated the distribution of log P values in the linkage regions). We observed a positive enrichment of log P values in CNVs obtained from The Database of Genomic Variants, http://projects.tcag.ca/variation/ (not including the data from Redon et al.36) (P = 0.0085, 0.017, 0.024 and 0.031 for log P of 2, 3, 4 and 5). We also tested the distribution of log P values in CNVs reported recently.36 We examined CNVs identified using the 500K EA arrays with a frequency of greater than 5%. The positive enrichment of log P values using this dataset was even more significant (P = 0.017, 0.003, 0.002 and P < 0.001 for log P of 2, 3, 4 and 5).
We considered whether this result could be artifactual. Within CNVs, the amount of DNA added to each pool might be more variable than in other regions, so we expect first that there will be more variation in allele frequency estimates in these regions and second that the differences between groups (as measured by the log P values) will be less when frequencies are obtained by individual genotyping.
Neither expectation was realized. Variation between DNA pools at SNPs within CNVs was not significantly different from that seen at SNPs else-where in the genome, and the enrichment of high log P values at CNVs was also seen in some cases in individually genotyped SNPs. These results indicate that the explanation for the enrichment may be related to the biology of neuroticism.
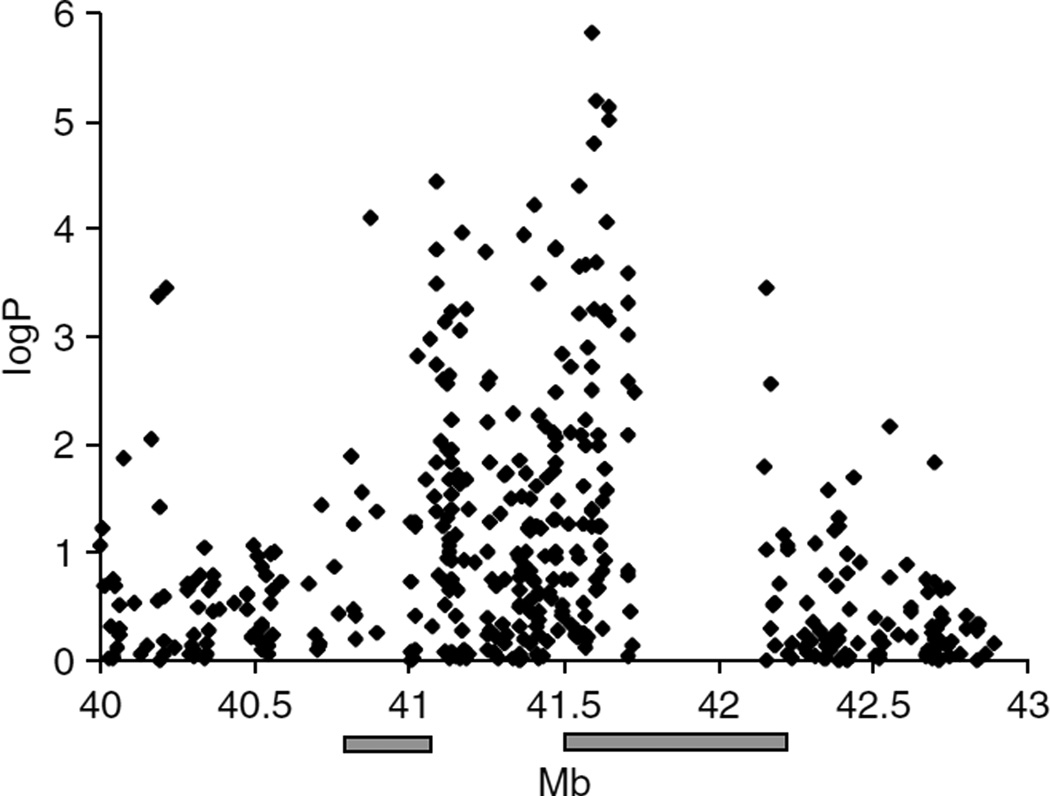
We examined each CNV to identify those contributing most to the observed enrichment of log P values. A region on chromosome 17q21.31 is the CNV region with the highest enrichment of high log P values (Figure 2). The enrichment of log P values in other CNVs is no longer significant when chromosome 17 is excluded from the analysis (minimum P = 0.087 for log P = 5). Individual genotyping of the sample used in the pools with an SNP (rs10514901) within the CNV region showed a modest association with neuroticism (P=0.0094).
Figure 2.
DNA pooling association results in a region of chromosome 17q21.31. Results of the DNA pooling are presented in log P values (vertical axis). The gray bars show the positions in megabases (Mb) of copy number variations (CNVs) on 17q21.3.36
Replication
We attempted to replicate the results of the most significant SNPs in an independent sample. Nineteen SNPs with a P-value < 0.001 (sex-specific or sex-averaged) were genotyped on a sample that consisted of 933 women and 601 men (all in the 10% low or high tail of the N distribution) (Figure 1). Only one SNP showed a statistically significant association in the replication. The significant SNP, rs702543, had a P-value of 0.00081 in the original sample, a P-value of 0.00078 (Bonferroni corrected P-value with 19 tests = 0.015) in the replication sample and a combined P-value = 2 × 10−6. In both the samples the frequency of the A allele was increased in the high Ns (frequency in the combined sample = 0.61) relative to the low Ns (frequency = 0.55) with an OR of 1.27 (95% confidence interval for the OR (95% CI) is 1.15– 1.4) and proportion of variance explained of 0.64% in an ANOVA model. Using the age and sex regressed N score to analyze the replication sample we obtain a P-value of 0.00065 for the additive model (note that because the scores are so extreme in the selected samples, this result is almost identical to the value obtained by applying a χ2 test: P = 0.00078). The ORs for men (OR = 1.30 in the combined sample) and women (OR = 1.25) were not statistically different in the pooling sample, the replication sample or the combined sample.
We attempted to replicate rs702543 in other populations. We investigated three populations: (1) a sample selected from 15 027 Australian twins and 11 389 of their family members. Unrelated individuals with neuroticism, anxiety and depression scores in the upper or lower deciles were genotyped (n=761)37; (2) unrelated individuals from the Virginia Adult Twin Study of Psychiatric and Substance Use Disorders.38 We used a sample of 1022 individuals selected from 9270 twins for a study of candidate genes on anxiety, depression and neuroticism; (3) a sample of 417 unrelated individuals from the Netherlands twin family for a study of anxious depression.39 Individuals were selected from a total of 13 717 individuals from 3344 families on the basis of extreme factor scores. The factor score included a neuroticism assessment, using the Amsterdamse Biografische Vragenlijst (ABV), a self-report personality instrument similar in content to the EPQ.40,41
We divided the samples from all populations into two groups based on N scores. We analyzed the allele and genotype frequencies in high and low N in each population and also in the combined sample (Table 4). The combined sample from the three populations showed a non-significant trend for an excess of the A allele in high N-score group (OR = 1.05, 95% CI = 0.92–1.19, P = 0.46). The allelic effect of SNP rs702543 estimated from the combination of the three populations and our replication sample in terms of OR is 1.13 (95% CI = 1.03–1.24, P=0.012).
Table 4.
Genotypes in replication samples
| Sample | High N | Low N | OR | P allele | P genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | A%a | N | AA | AG | GG | A% | N | ||||
| Pool sample | 348 | 428 | 156 | 60.3 | 932 | 291 | 468 | 197 | 54.9 | 956 | 1.25 | 0.00082 | 0.0035 |
| Our replication | 289 | 316 | 123 | 61.4 | 728 | 197 | 305 | 133 | 55.0 | 635 | 1.3 | 0.00077 | 0.0029 |
| Australian | 129 | 163 | 57 | 60.3 | 349 | 144 | 196 | 71 | 58.9 | 411 | 1.06 | 0.57 | 0.85 |
| Dutch | 75 | 84 | 31 | 62.0 | 190 | 85 | 106 | 36 | 61.0 | 227 | 1.03 | 0.82 | 0.88 |
| Virginia | 83 | 132 | 38 | 58.9 | 253 | 261 | 379 | 129 | 58.6 | 769 | 1.01 | 0.90 | 0.69 |
| Three groups | 287 | 379 | 126 | 60.2 | 792 | 490 | 681 | 236 | 59.0 | 1407 | 1.05 | 0.46 | 0.76 |
| All replications | 576 | 695 | 249 | 60.8 | 1520 | 687 | 986 | 369 | 57.8 | 2042 | 1.13 | 0.012 | 0.029 |
Abbreviation: OR, odds ratio.
Frequency of the A allele.
We also used family based analysis to test for the association of rs702543 with neuroticism in the Netherlands and Australian full family samples. There was no evidence for association in the Netherlands family sample (N= 1148, wave 6, analyzed using quantitative transmission-disequilibrium test (QTDT), P = 0.88). The Australian family sample was analyzed using the age and sex transformed scores42 based on the full 23-item EPQ-R N scale and the 10-item Kessler Psychological Distress scale43 (K10), a measure of current non-specific psychological distress in the anxiety–depression spectrum. Individuals were considered ‘affected’ for EPQ-R and K10 if their residual scores were in the top 5, 10, 15 and 20 percentile of available residuals. Consistent with the findings in the original sample, an overtransmission of the A allele to affected individuals was observed for all percentiles. The most significant transmission distortion was observed when the individuals in the 20 percentile were treated as affected (analyzed using UNPHASED v3.03, EPQ P-value = 0.040, K10 P-value = 0.0059).
Discussion
We have performed the first whole genome analysis of neuroticism, using pooled DNA. We show that, by using eight pools with five replicates of each, we retain between 21 and 94% (depending on the effect size and threshold used) of the power from genotyping all 452 574 SNPs individually, equivalent to a 50-fold saving in the costs. We identified one SNP rs702543 from an analysis of ~2000 individuals selected from the neuroticism extremes of 88 142 people, which we were able to replicate in a separate sample from the same cohort. The SNP was genotyped in three other laboratories, on related, but not identical, phenotypes. In each case the direction of allelic effects was the same as in our sample, but the test reached statistical significance in only one sample, that is, only in one method of analysis (family based analysis). Our study raises a number of issues for whole genome association studies in general and the genetic basis of neuroticism in particular.
We deal first with the performance of the pooling strategy. Our estimates of the power of our DNA pooling strategy were derived by simulation using the measurement error across all SNPs from both 500 and 100K arrays. However, there is a 3.2-fold difference between the variances of the repeated measurements for the two types of arrays (mean variance for 100K= 0.0017, and for 500K= 0.0055). This difference in measurement accuracy is probably due to the smaller number of features per SNP and the reduction in feature size in the 500K array relative to the 100 K.21 It means that the measurements in the 500K arrays should be at least triplicated to achieve the genotype accuracy obtained with the 100K arrays.
DNA pooling allows substantial savings in genotyping costs (see, for example, references44–46) and there have been successes using DNA pooling in the analysis of a relatively small number of markers, for example, in schizophrenia47,48 multiple sclerosis,49 and serum high-density lipoprotein cholesterol levels.50,51 However, in the two cases where whole genome association has been attempted (that is, in memory52 and mild mental impairment53), the yields have been low, that is, in both cases only one locus was considered significant.
Our results are comparable, in that we have found just one locus of small effect. Failure to replicate the finding in all samples is not unexpected, given the differences between the cohorts, in both phenotype and recruitment. No sample used exactly the same phenotype. We used the 23-item Eysenck N scale, while the US and Australian sample used the short (12-item) scale and the Dutch sample used the Amsterdamse Biografische Vragenlijst, which is based on the Eysenck scale but is not identical. However, differences in the measures are likely to be outweighed by differences in the recruitment strategy. Our sample represents the extremes of about 61 367 unrelated people, the Dutch from 3444 families, the Australian from around 7500 families and the US from 9270 twins.
Second, we consider the lessons for the design of whole genome association studies. Assuming that the genetic basis of neuroticism is typical of behavior, and also typical of other complex traits, our results reinforce the findings that the ORs of most complex trait loci are less than 1.6.54 Our linkage study on the same cohort failed to find any large effect loci (explaining more than 5% of the variance), and given that we had approximately 50% power to detect an OR of 1.5 (which is equivalent to a locus explaining 0.5–1% of the variance) and failed to find any loci accounting for more than 1% of the variance, it seems likely that the 40% additive genetic variance of neuroticism arises from many loci explaining much less than 1%. This means that to obtain adequate power in whole genome association studies, much larger samples will be needed than anticipated.
Using association analysis, we failed to identify any genes under the five significant linkage peaks that were obtained using the sib pairs from the same population-based study of personality (although it should be noted that there was a nominally significant enrichment in log P > 2 values in the 1-LOD interval on chromosome 1q). Our significant SNP is located on chromosome 5, a region that was not indicated by the linkage analysis. Since linkage analysis is robust to allelic heterogeneity at a locus, this may indicate that rare variants are a major contributor to the heritability of neuroticism. The linkage signal could also be due to the co-localization under the linkage peaks of variants in different genes, each of too insufficient individual effect to be detected by association. Other possible factors that could contribute to the lower power of our study to identify the genes under the linkage peaks include the effect of incomplete genome coverage (low LD between the SNPs typed and the causal variants) and a high false-negative rate of the association analysis.
Unlike the linkage intervals that did not show a significant enrichment of high log P values, we did see the enrichment in known CNVs. Chromosomal abnormalities have been reported in patients with schizophrenia, bipolar disorder and major depression,55,56 but it is not clear to what extent CNVs contribute to variation in behavior. It is noteworthy that CNVs on chromosome 17 (enriched for high log P values) contain two genes involved in behavior, that is, the Tau gene (MAPT) and the corticotropin-releasing hormone receptor-1 (CRHR1). CRHR1 has been shown to mediate anxiety-related behavior and hormonal adaptation to stress.57
Finally, our findings are important for the understanding of the genetic basis of neuroticism and the associated psychiatric disorders of anxiety and depression. The rs702543 SNP is located in the phosphodiesterase 4D, cAMP-specific (PDE4D) gene, in an intron between exon D3 and D8. The HapMap database (CEU, Release #21) shows one SNP (rs702542) in complete correlation with rs702543, and two other SNPs that are partially correlated, that is, rs296410 (r2 = 0.5) and rs40216 (r2 = 0.56). All three SNPs lie within a 65.3-kb haplotype block on chromosome 5 (58 872 707–58 937 990 bp). The G allele of rs702543 creates a putative cAMP response element (CRE) for CREB2/c-jun heterodimer (TGACGTTA), while the A allele destroys it.58,59 The transcription of different isoforms of PDE4D was shown to be regulated by cAMP levels. For example, a CRE (TGACGTT) in the promoter of an isoform of PDE4D (PDE4D5) was shown to be involved in the cAMP responsiveness of the PDE4D5 promoter.60
Several other lines of evidence support the involvement of the PDE4D gene in susceptibility to neuroticism and depression. A PDE4-specific inhibitor rolipram has antidepressant effects on animals and patients with major depression.61–63 PDE4D knockout mice show antidepressant-like behavior, which is further increased by the antidepressants desipramine and fluoxetine but not by rolipram. This suggests that the PDE4D subtype is an essential mediator of the antidepressant effects of rolipram. PDE4D expression is increased in mouse cerebral cortex by the repeated treatment with desipramine, fluoxetine and rolipram and in the hippocampus by fluoxetine and rolipram. Recently, variants in two genes encoding PDEs were found to be associated with the diagnosis of major depression, and one of them (PDE11A) was also found to be associated with remission in response to antidepressants.64 Together, these observations indicate that the PDE4D gene is likely to be involved in susceptibility to neuroticism and the associated psychiatric disorders of major depression and anxiety.
Supplementary Material
Acknowledgments
Funding for this work was provided by the Wellcome Trust. SS is supported by the European Molecular Biology Organization.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Eysenck HJ. The Biological Basis of Personality. Springfield, IL: Thomas; 1967. [Google Scholar]
- 2.Angst J, Clayton P. Premorbid personality of depressive, bipolar and schizophrenic patients with special reference to suicidal issues. Compr Psychiatry. 1986;27:511–532. doi: 10.1016/0010-440x(86)90055-6. [DOI] [PubMed] [Google Scholar]
- 3.Boyce P, Parker G, Barnett B, Cooney M, Smith F. Personality as a vulnerability factor to depression. Br J Psychiatry. 1991;159:106–114. doi: 10.1192/bjp.159.1.106. [DOI] [PubMed] [Google Scholar]
- 4.Hirschfeld RM, Klerman GL, Lavori P, Keller MB, Griffith P, Coryell W. Premorbid personality assessments of first onset of major depression. Arch Gen Psychiatry. 1989;46:345–350. doi: 10.1001/archpsyc.1989.01810040051008. [DOI] [PubMed] [Google Scholar]
- 5.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. A longitudinal twin study of personality and major depression in women. Arch Gen Psychiatry. 1993;50:853–862. doi: 10.1001/archpsyc.1993.01820230023002. [DOI] [PubMed] [Google Scholar]
- 6.Bienvenu OJ, Samuels JF, Costa PT, Reti IM, Eaton WW, Nestadt G. Anxiety and depressive disorders and the five-factor model of personality: a higher- and lower-order personality trait investigation in a community sample. Depress Anxiety. 2004;20:92–97. doi: 10.1002/da.20026. [DOI] [PubMed] [Google Scholar]
- 7.Cox BJ, MacPherson PS, Enns MW, McWilliams LA. Neuroticism and self-criticism associated with posttraumatic stress disorder in a nationally representative sample. Behav Res Ther. 2004;42:105–114. doi: 10.1016/s0005-7967(03)00105-0. [DOI] [PubMed] [Google Scholar]
- 8.Jylha P, Isometsa E. The relationship of neuroticism and extraversion to symptoms of anxiety and depression in the general population. Depress Anxiety. 2006;23:281–289. doi: 10.1002/da.20167. [DOI] [PubMed] [Google Scholar]
- 9.Jylha P, Isometsa E. Temperament, character and symptoms of anxiety and depression in the general population. Eur Psychiatry. 2006;21:389–395. doi: 10.1016/j.eurpsy.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Hettema JM, Neale MC, Myers JM, Prescott CA, Kendler KS. A population-based twin study of the relationship between neuroticism and internalizing disorders. Am J Psychiatry. 2006;163:857–864. doi: 10.1176/ajp.2006.163.5.857. [DOI] [PubMed] [Google Scholar]
- 11.Kendler KS, Gardner CO, Prescott CA. Toward a comprehensive developmental model for major depression in women. Am J Psychiatry. 2002;159:1133–1145. doi: 10.1176/appi.ajp.159.7.1133. [DOI] [PubMed] [Google Scholar]
- 12.Kendler KS, Gardner CO, Prescott CA. Toward a comprehensive developmental model for major depression in men. Am J Psychiatry. 2006;163:115–124. doi: 10.1176/appi.ajp.163.1.115. [DOI] [PubMed] [Google Scholar]
- 13.Kendler KS, Gatz M, Gardner CO, Pedersen NL. Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry. 2006;63:1113–1120. doi: 10.1001/archpsyc.63.10.1113. [DOI] [PubMed] [Google Scholar]
- 14.Fullerton J, Cubin M, Tiwari H, Wang C, Bomhra A, Davidson S, et al. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. Am J Hum Genet. 2003;72:879–890. doi: 10.1086/374178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nash MW, Huezo-Diaz P, Williamson RJ, Sterne A, Purcell S, Hoda F, et al. Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum Mol Genet. 2004;13:2173–2182. doi: 10.1093/hmg/ddh239. [DOI] [PubMed] [Google Scholar]
- 16.Neale BM, Sullivan PF, Kendler KS. A genome scan of neuroticism in nicotine dependent smokers. Am J Med Genet B Neuropsychiatr Genet. 2005;132:65–69. doi: 10.1002/ajmg.b.30095. [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol. 2006;164:609–614. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 18.Darvasi A, Soller M. Selective DNA pooling for determination of linkage between a molecular marker and a quantitative trait locus. Genetics. 1994;138:1365–1373. doi: 10.1093/genetics/138.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butcher LM, Meaburn E, Liu L, Fernandes C, Hill L, Al-Chalabi A, et al. Genotyping pooled DNA on microarrays: a systematic genome screen of thousands of SNPs in large samples to detect QTLs for complex traits. Behav Genet. 2004;34:549–555. doi: 10.1023/B:BEGE.0000038493.26202.d3. [DOI] [PubMed] [Google Scholar]
- 20.Craig I, Meaburn E, Butcher L, Hill L, Plomin R. Single-nucleotide polymorphism genotyping in DNA pools. Methods Mol Biol. 2005;311:147–164. doi: 10.1385/1-59259-957-5:147. [DOI] [PubMed] [Google Scholar]
- 21.Kirov G, Nikolov I, Georgieva L, Moskvina V, Owen MJ, O’Donovan MC. Pooled DNA genotyping on Affymetrix SNP genotyping arrays. BMC Genomics. 2006;7:27. doi: 10.1186/1471-2164-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macgregor S, Visscher PM, Montgomery G. Analysis of pooled DNA samples on high density arrays without prior knowledge of differential hybridization rates. Nucleic Acids Res. 2006;34:e55. doi: 10.1093/nar/gkl136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meaburn E, Butcher LM, Schalkwyk LC, Plomin R. Genotyping pooled DNA using 100K SNP microarrays: a step towards genomewide association scans. Nucleic Acids Res. 2006;34:e27. doi: 10.1093/nar/gnj027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willis-Owen SA, Turri MG, Munafo MR, Surtees PG, Wainwright NW, Brixey RD, et al. The serotonin transporter length polymorphism, neuroticism, and depression: a comprehensive assessment of association. Biol Psychiatry. 2005;58:451–456. doi: 10.1016/j.biopsych.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 25.Willis-Owen SA, Shifman S, Copley RR, Flint J. DCNP1: a novel candidate gene for major depression. Mol Psychiatry. 2006;11:121–122. doi: 10.1038/sj.mp.4001747. [DOI] [PubMed] [Google Scholar]
- 26.Willis-Owen SA, Fullerton J, Surtees PG, Wainwright NW, Miller S, Flint J. The Val66Met coding variant of the brain-derived neurotrophic factor (BDNF) gene does not contribute toward variation in the personality trait neuroticism. Biol Psychiatry. 2005;58:738–742. doi: 10.1016/j.biopsych.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Martin N, Goodwin G, Fairburn C, Wilson R, Allison D, Cardon LR, et al. A population based study of personality in 34 000 sib-pairs. Twin Res. 2000;3:310–315. [PubMed] [Google Scholar]
- 28.Eysenck HJ, Eysenck SBG, editors. Manual of the Eysenck Personality Questionnaire. San Diego, CA: Educational and Industrial Testing Service; 1975. [Google Scholar]
- 29.Aluja A, García O, Garcia LF. A comparative study of Zuckerman’s three structural models for personality through the NEO-PI-R: ZKPQ-III-R, EPQ-RS and Goldberg’s 50-bipolar adjectives. Pers Individ Dif. 2002;33:713–725. [Google Scholar]
- 30.De Fruyt F, Van De Wiele L, Van Heeringen C. Cloninger’s psychobiological model of temperament and character and the five-factor model of personality. Pers Individ Dif. 2000;29:441–452. [Google Scholar]
- 31.Shifman S, Pisante-Shalom A, Yakir B, Darvasi A. Quantitative technologies for allele frequency estimation of SNPs in DNA pools. Mol Cell Probes. 2002;16:429–434. doi: 10.1006/mcpr.2002.0440. [DOI] [PubMed] [Google Scholar]
- 32.Meaburn E, Butcher LM, Liu L, Fernandes C, Hansen V, Al-Chalabi A, et al. Genotyping DNA pools on microarrays: tackling the QTL problem of large samples and large numbers of SNPs. BMC Genomics. 2005;6:52. doi: 10.1186/1471-2164-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaid DJ. Evaluating associations of haplotypes with traits. Genet Epidemiol. 2004;27:348–364. doi: 10.1002/gepi.20037. [DOI] [PubMed] [Google Scholar]
- 36.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirk KM, Birley AJ, Statham DJ, Haddon B, Lake RI, Andrews JG, et al. Anxiety and depression in twin and sib pairs extremely discordant and concordant for neuroticism: prodromus to a linkage study. Twin Res. 2000;3:299–309. doi: 10.1375/136905200320565274. [DOI] [PubMed] [Google Scholar]
- 38.Hettema JM, An SS, Neale MC, Bukszar J, van den Oord EJ, Kendler KS, et al. Association between glutamic acid decarboxylase genes and anxiety disorders, major depression, and neuroticism. Mol Psychiatry. 2006;11:752–762. doi: 10.1038/sj.mp.4001845. [DOI] [PubMed] [Google Scholar]
- 39.Boomsma DI, Beem AL, van den Berg M, Dolan CV, Koopmans JR, Vink JM, et al. Netherlands twin family study of anxious depression (NETSAD) Twin Res. 2000;3:323–334. doi: 10.1375/136905200320565300. [DOI] [PubMed] [Google Scholar]
- 40.Middeldorp CM, Birley AJ, Cath DC, Gillespie NA, Willemsen G, Statham DJ, et al. Familial clustering of major depression and anxiety disorders in Australian and Dutch twins and siblings. Twin Res Hum Genet. 2005;8:609–615. doi: 10.1375/183242705774860123. [DOI] [PubMed] [Google Scholar]
- 41.Rettew DC, Vink JM, Willemsen G, Doyle A, Hudziak JJ, Boomsma DI. The genetic architecture of neuroticism in 3301 Dutch adolescent twins as a function of age and sex: a study from the Dutch twin register. Twin Res Hum Genet. 2006;9:24–29. doi: 10.1375/183242706776403028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jardine R, Martin NG, Henderson AS. Genetic covariation between neuroticism and the symptoms of anxiety and depression. Genet Epidemiol. 1984;1:89–107. doi: 10.1002/gepi.1370010202. [DOI] [PubMed] [Google Scholar]
- 43.Kessler R, Mroczek D. Final Version of the Psychological Distress Scale. Technical Note. Ann Arbor, MI: Institute for Social Research, University of Michigan; 1994. [Google Scholar]
- 44.Sham P, Bader JS, Craig I, O’Donovan M, Owen M. DNA pooling: a tool for large-scale association studies. Nat Rev Genet. 2002;3:862–871. doi: 10.1038/nrg930. [DOI] [PubMed] [Google Scholar]
- 45.Mohlke KL, Erdos MR, Scott LJ, Fingerlin TE, Jackson AU, Silander K, et al. High-throughput screening for evidence of association by using mass spectrometry genotyping on DNA pools. Proc Natl Acad Sci USA. 2002;99:16928–16933. doi: 10.1073/pnas.262661399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barratt BJ, Payne F, Rance HE, Nutland S, Todd JA, Clayton DG. Identification of the sources of error in allele frequency estimations from pooled DNA indicates an optimal experimental design. Ann Hum Genet. 2002;66:393–405. doi: 10.1017/S0003480002001252. [DOI] [PubMed] [Google Scholar]
- 47.Norton N, Williams HJ, Williams NM, Spurlock G, Zammit S, Jones G, et al. Mutation screening of the Homer gene family and association analysis in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2003;120:18–21. doi: 10.1002/ajmg.b.20032. [DOI] [PubMed] [Google Scholar]
- 48.Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A, et al. A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet. 2002;71:1296–1302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawcer S, Maranian M, Setakis E, Curwen V, Akesson E, Hensiek A, et al. A whole genome screen for linkage disequilibrium in multiple sclerosis confirms disease associations with regions previously linked to susceptibility. Brain. 2002;125:1337–1347. doi: 10.1093/brain/awf143. [DOI] [PubMed] [Google Scholar]
- 50.Bansal A, van den Boom D, Kammerer S, Honisch C, Adam G, Cantor CR, et al. Association testing by DNA pooling: an effective initial screen. Proc Natl Acad Sci USA. 2002;99:16871–16874. doi: 10.1073/pnas.262671399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinds DA, Seymour AB, Durham LK, Banerjee P, Ballinger DG, Milos PM, et al. Application of pooled genotyping to scan candidate regions for association with HDL cholesterol levels. Hum Genomics. 2004;1:421–434. doi: 10.1186/1479-7364-1-6-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 53.Butcher LM, Meaburn E, Knight J, Sham PC, Schalkwyk LC, Craig IW, et al. SNPs, microarrays and pooled DNA: identification of four loci associated with mild mental impairment in a sample of 6000 children. Hum Mol Genet. 2005;14:1315–1325. doi: 10.1093/hmg/ddi142. [DOI] [PubMed] [Google Scholar]
- 54.Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol. 2006;164:609–614. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 55.Lee JA, Lupski JR. Genomic rearrangements and gene copynumber alterations as a cause of nervous system disorders. Neuron. 2006;52:103–121. doi: 10.1016/j.neuron.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 56.Wilson GM, Flibotte S, Chopra V, Melnyk BL, Honer WG, Holt RA. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum Mol Genet. 2006;15:743–749. doi: 10.1093/hmg/ddi489. [DOI] [PubMed] [Google Scholar]
- 57.Smith GW, Aubry JM, Dellu F, Contarino A, Bilezikjian LM, Gold LH, et al. Corticotropin releasing factor receptor 1-deficient mice display decreased anxiety, impaired stress response, and aberrant neuroendocrine development. Neuron. 1998;20:1093–1102. doi: 10.1016/s0896-6273(00)80491-2. [DOI] [PubMed] [Google Scholar]
- 58.Benbrook DM, Jones NC. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- 59.Benbrook DM, Jones NC. Different binding specificities and transactivation of variant CRE’s by CREB complexes. Nucleic Acids Res. 1994;22:1463–1469. doi: 10.1093/nar/22.8.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Le Jeune IR, Shepherd M, Van Heeke G, Houslay MD, Hall IP. Cyclic AMP-dependent transcriptional up-regulation of phosphodiesterase 4D5 in human airway smooth muscle cells. Identification and characterization of a novel PDE4D5 promoter. J Biol Chem. 2002;277:35980–35989. doi: 10.1074/jbc.M204832200. [DOI] [PubMed] [Google Scholar]
- 61.Fleischhacker WW, Hinterhuber H, Bauer H, Pflug B, Berner P, Simhandl C, et al. A multicenter double-blind study of three different doses of the new cAMP-phosphodiesterase inhibitor rolipram in patients with major depressive disorder. Neuropsychobiology. 1992;26:59–64. doi: 10.1159/000118897. [DOI] [PubMed] [Google Scholar]
- 62.Scott AI, Perini AF, Shering PA, Whalley LJ. In-patient major depression: is rolipram as effective as amitriptyline? Eur J Clin Pharmacol. 1991;40:127–129. doi: 10.1007/BF00280065. [DOI] [PubMed] [Google Scholar]
- 63.Zhang HT, Huang Y, Jin SL, Frith SA, Suvarna N, Conti M, et al. Antidepressant-like profile and reduced sensitivity to rolipram in mice deficient in the PDE4D phosphodiesterase enzyme. Neuropsychopharmacology. 2002;27:587–595. doi: 10.1016/S0893-133X(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 64.Wong ML, Whelan F, Deloukas P, Whittaker P, Delgado M, Cantor RM, et al. Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc Natl Acad Sci USA. 2006;103:15124–15129. doi: 10.1073/pnas.0602795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.