ABSTRACT
Proper cellular functioning requires that cellular machinery behave in a spatiotemporally regulated manner in response to global changes in nutrient availability. Mounting evidence suggests that one way this is achieved is through the establishment of physically defined gradients of O-GlcNAcylation (O-linked addition of N-acetylglucosamine to serine and threonine residues) and O-GlcNAc turnover. Because O-GlcNAcylation levels are dependent on the nutrient-responsive hexosamine signaling pathway, this modification is uniquely poised to inform upon the nutritive state of an organism. The enzymes responsible for O-GlcNAc addition and removal are encoded by a single pair of genes: both the O-GlcNAc transferase (OGT) and the O-GlcNAcase (OGA, also known as MGEA5) genes are alternatively spliced, producing protein variants that are targeted to discrete cellular locations where they must selectively recognize hundreds of protein substrates. Recent reports suggest that in addition to their catalytic functions, OGT and OGA use their multifunctional domains to anchor O-GlcNAc cycling to discrete intracellular sites, thus allowing them to establish gradients of deacetylase, kinase and phosphatase signaling activities. The localized signaling gradients established by targeted O-GlcNAc cycling influence many important cellular processes, including lipid droplet remodeling, mitochondrial functioning, epigenetic control of gene expression and proteostasis. As such, the tethering of the enzymes of O-GlcNAc cycling appears to play a role in ensuring proper spatiotemporal responses to global alterations in nutrient supply.
KEY WORDS: O-GlcNAc, O-GlcNAcylation, Metabolism, Post-translational modification, Signaling, Lipid droplets, Mitochondria, Epigenetics, Nuclear pores
Introduction
Hundreds of proteins are modified at the hydroxyl group of their serine and threonine residues by N-acetylglucosamine. This modification has been termed O-GlcNAcylation (Torres and Hart, 1984). The dynamic addition and removal of O-GlcNAc is governed by a single pair of enzymes, O-GlcNAc transferase (OGT) and β-N-acetylglucosaminidase C (O-GlcNAcase; OGA), respectively (Kreppel et al., 1997; Lubas et al., 1997). Both OGT and OGA are expressed in all tissue types, where they are necessary for either vertebrate development (OGT) or for the transition from the fetal stage to adulthood (OGA) (Shafi et al., 2000; Yang et al., 2012). Deregulation of O-GlcNAc cycling has been implicated in a variety of disease states, including cardiovascular disease, neurodegeneration, cancer and diabetes (Bond and Hanover, 2013; Hart et al., 2011; Ngoh et al., 2010; Zachara, 2012).
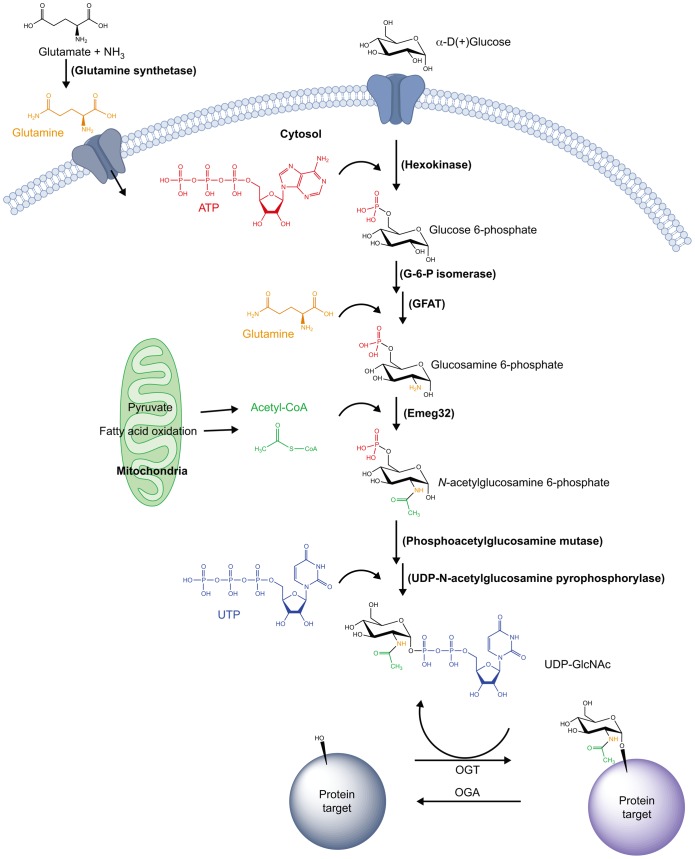
O-GlcNAc metabolism is widely believed to be part of a nutrient-sensing pathway termed the hexosamine biosynthetic pathway (HBP). This pathway is sensitive to carbohydrate, amino acid, fatty acid and ATP levels (Slawson et al., 2010) (Fig. 1). The substrate of OGT, uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), is the final product of the HBP, making O-GlcNAc addition ideal to reflect the nutritional state of the cell. Specifically, key components that feed into UDP-GlcNAc synthesis are derived from nutrients, such as the amino acid glutamine, acetyl-coenzyme A (acetyl-CoA), glucose and uridine. In this regard, the enzymes of the HBP join the signaling proteins AMP activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR), which act as sensors of energy charge and amino acid levels, respectively.
Fig. 1.
Generation of the OGT substrate, UDP-GlcNAc – an overview of the hexosamine biosynthetic pathway. The hexosamine biosynthetic pathway (HBP) integrates many extracellular physiological inputs, including nutritional intake, with the metabolism of key carbohydrate, amino acid, nucleotide and fatty acid components, to produce the OGT substrate UDP-GlcNAc. O-GlcNAc cycling, the dynamic addition to and removal of O-GlcNAc on serine or threonine residues of a plethora of OGT and OGA targets, allows for the regulation of important downstream cellular processes in a nutrient-dependent manner. The enzymes involved are given in brackets.
O-GlcNAcylation is often considered to be analogous to protein phosphorylation. However, unlike protein phosphorylation, whereby phosphate is added to and removed from proteins by many different kinases and phosphatases, protein O-GlcNAcylation is performed by the products of a single pair of human genes (Nolte and Muller, 2002; Shafi et al., 2000). Hundreds of proteins with a range of functions have been shown to be O-GlcNAcylated (Copeland et al., 2008), suggesting that many pathways exist by which specific substrates are modified to ensure proper downstream cellular responses. The specific pattern of protein O-GlcNAcylation in the cell is determined in part by O-GlcNAc levels, the subcellular localization of OGT and OGA, and the different interaction partners of OGT and OGA either at specific cellular locations, during specific times, or in specific cell types. Because disease states are associated with deregulated O-GlcNAc cycling (Bond and Hanover, 2013; Hart et al., 2011; Ngoh et al., 2010; Zachara, 2012), the factors that ensure that OGT and OGA are able to recognize their substrates and achieve substrate specificity must be tightly regulated.
Owing to its nutrient dependence, the presence or absence of an O-GlcNAc modification on a protein could initiate a cascade that is intended to ‘direct’ the cell to augment processes that are associated with nutrient conservation, such as a reduction in cell proliferation and attenuation of nutrient storage and cell growth. The question remains as to how a balance is achieved between the apparently competing functions of OGA and OGT to modulate highly specific spatiotemporal responses. One possibility is that under certain conditions, such as those outlined in Fig. 2A, O-GlcNAcylation of proteins might be influenced in a global manner (Fig. 2A). Concurrently, a more fine-tuned mechanism might govern protein O-GlcNAcylation to ensure both global and local cellular response to changes in UDP-GlcNAc levels, which would be important both under conditions of very low or very high UDP-GlcNAc, or under conditions of UDP-GlcNAc homeostasis. Based on recent findings in the field, we propose that this could be achieved by interaction of OGA and OGT with specific binding partners that tether them to precise intracellular locations or organelles to allow them to evoke localized responses, as has been reported in the past for other enzyme systems (Görlich et al., 1996; Griffin et al., 2011; Izaurralde et al., 1997; Pratt et al., 2005). This local gradient of OGA or OGT might therefore establish a threshold of activity: enzyme activity is ‘on’ when the target proteins are close to the tethered enzyme or ‘off’ with increasing distance, allowing for sensitive and specific responses to UDP-GlcNAc levels (Fig. 2B). Such a mechanism would allow the cell to translate information about the nutritional state into highly specialized changes in cellular responses. Interestingly, a study using fluorescence resonance energy transfer (FRET) to detect changes in the spatiotemporal dynamics of the O-GlcNAc modification in response to signal-inducing stimuli suggested that O-GlcNAc cycling does in fact exhibit compartment-specific dynamics (Carrillo et al., 2011). Specifically, Carrillo and colleagues observed a rapid increase in the O-GlcNAcylation of a FRET-based sensor specifically at the plasma membrane and nucleus, but not elsewhere in the cytoplasm, following serum-stimulated signal transduction in Cos7 cells.
Fig. 2.
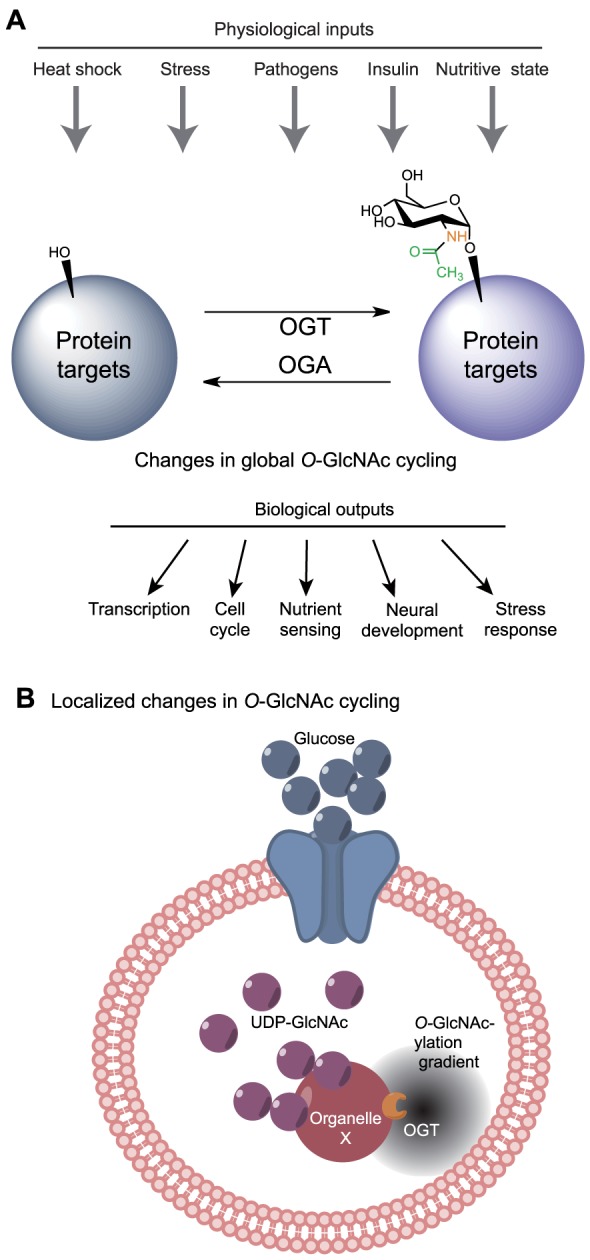
Global and local responses to changes in UDP-GlcNAc levels. (A) The cell can respond to certain extracellular physiological inputs with global changes in intracellular UDP-GlcNAc levels and subsequent changes in total protein O-GlcNAcylation levels to modulate proper downstream cellular responses. (B) The tethering of OGA or OGT at specific cellular locations could establish local gradients of protein O-GlcNAcylation, and thus potentially introduce gradients of protein activity (illustrated by the gray ‘cloud’) that allow for highly specific spatiotemporal responses to the overall UDP-GlcNAc levels, with consequent cellular responses.
In this Commentary, we will present in detail the localization and properties of the different isoforms of OGT and OGA, as well as the specific interactions they undergo with a wide variety of cellular components. This information can provide insight into how the cell is able to translate changes in nutritional status into distinct metabolic responses as a means to maintain cellular homeostasis. Furthermore, we will examine the mechanisms by which the modulation of protein O-GlcNAcylation alters cellular metabolism and touch upon how the deregulation of these signaling pathways, either genetically or through altered nutritional conditions, might result in certain disease states.
Domain structure and localization of OGT and OGA isoforms
O-GlcNAc transferase
In mammals, the gene coding for OGT is found on the X chromosome (Shafi et al., 2000), and several alternatively spliced transcripts encoding distinct OGT isoforms have been identified, including isoforms that localize to the nucleocytoplasm (ncOGT) and to the mitochondria (mOGT) (Love et al., 2003), as well as a short isoform (sOGT) that shows nucleocytoplasmic localization (Hanover et al., 2003; Nolte and Muller, 2002; Shafi et al., 2000). More recently, a genetically unrelated enzyme with O-GlcNAc transferase activity, extracellular OGT (eOGT), has also been described (Sakaidani et al., 2012; Sakaidani et al., 2011). The ncOGT isoform is ∼116 kDa and is encoded by all 23 exons of the OGT gene (Hanover et al., 2003), whereas mOGT is ∼103 kDa and is encoded by exons 5 to 23 (Love et al., 2003), and sOGT is ∼78 kDa and encoded by exons 10 to 23 (Hanover et al., 2003; Nolte and Muller, 2002; Shafi et al., 2000) (Fig. 3A). The newly described eOGT utilizes UDP-GlcNAc but performs its function in the endomembrane system; it has been less extensively characterized than the three OGT isoforms, to which it exhibits no apparent homology (Sakaidani et al., 2011). As such, our discussion of OGT will focus on the similarities and differences between ncOGT, mOGT and sOGT.
Fig. 3.
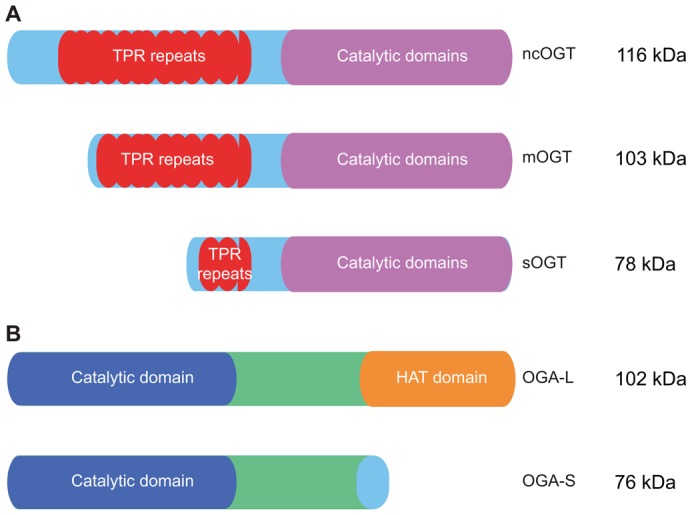
OGT and OGA isoforms and domain structure. (A) The three splice isoforms of OGT, ncOGT, mOGT and sOGT, possess variable N-termini but identical catalytic domains. (B) The two splice isoforms of OGA, OGA-L and OGA-S, differ at their C-termini. OGA-L possesses a putative HAT domain, whereas OGA-S lacks this domain and instead contains a unique 15-amino-acid long C-terminal extension.
In general, the structural domains of the three differentially spliced OGT isoforms can be divided into regions that include a variable N-termini and tetratricopeptide repeat (TPR) domain, a linker region and a multidomain catalytic region (Kreppel et al., 1997; Lazarus et al., 2011; Lubas et al., 1997) (Fig. 3A). Each isoform possesses an identical catalytic domain, and variation between the isoforms occurs primarily in the length and sequence of their unique N-termini. For example, mOGT possesses a mitochondrial-targeting sequence in its variable N-terminal region upstream of the TPR domain (Love et al., 2003). The isoforms also differ in their TPR domains, and contain between 2.5 and 12.5 TPR motifs, consisting of 34 amino acid repeats each, that are thought to have a role in protein–protein interactions (Goebl and Yanagida, 1991; Iyer et al., 2003; Liu et al., 1999). Specifically, ncOGT possesses 12.5 TPR motifs, whereas mOGT has 9.5 motifs and sOGT only 2.5 motifs (Hanover et al., 2003). The TPR domain of ncOGT has been crystallized and exists as a superhelix with an amphipathic center groove that is lined with Asn residues (Blatch and Lässle, 1999; Jínek et al., 2004). The TPR domain bears some resemblance to the importin-α family of nuclear transport receptors (Jínek et al., 2004). The characteristics of this groove might have a role in determining substrate specificity for a variety of OGT substrates, although a specific substrate sequence motif is not apparent (Blatch and Lässle, 1999; Hanover et al., 2003; Iyer et al., 2003; Lazarus et al., 2006). Consequently, the differences in the length of the TPR domains in each OGT isoform has been suggested to modulate the substrate specificity among the different isoforms (Lubas and Hanover, 2000).
As mentioned above, the catalytic domain of all three OGT isoforms is identical, and several crystal structures of OGT variants that are in a complex with UDP alone, with UDP and peptide substrate or with sugar donor analogs have been solved (reviewed in Vocadlo, 2012). These structures have demonstrated that when in a complex with UDP alone, OGT adopts a ‘closed’ conformation, with interactions between TPR10 and 11 and a helix of the catalytic domain acting as a ‘latch’ to stabilize this conformation (Lazarus et al., 2011). In the structure of OGT in complex with both UDP and peptide substrate, a wide cleft appears to be formed through a hinge-like rotation between TPR12 and 13, opening the active site for peptide substrate binding (Lazarus et al., 2011). Based on these crystal structures and kinetic experiments, Lazarus and co-workers proposed a sequential bi-bi kinetic mechanism for OGT action whereby UDP-GlcNAc binds prior to the protein substrate, although the exact mechanism of glycosyl transfer, including the identity of the catalytic residue, remains to be determined (Lazarus et al., 2011; Schimpl et al., 2012b). To examine the ternary complex with both UDP and modified peptide substrate bound, crystal structures were solved using a very slowly hydrolyzed UDP-GlcNAc analog, UDP-5SGlcNAc, in which the endocyclic oxygen atom is replaced by sulfur (Lazarus et al., 2012). These structures suggest that OGT promotes catalysis using an electrophilic migration mechanism (Lazarus et al., 2012). This ternary complex also demonstrates that the substrate interaction interface is large (Lazarus et al., 2012), suggesting that the binding of different protein substrates is likely to cause global structural changes in the active center of OGT, which are reflected in the wide range of affinities observed for UDP-GlcNAc (from 1 µm to over 20 µm) (Shen et al., 2012).
In addition, the cell cycle regulator host cell factor 1 (HCF-1) is an evolutionarily conserved epigenetic regulator that undergoes an unusual proteolytic processing event to generate two stably associated fragments that regulate different stages of the cell cycle (Hanover, 2011; Mazars et al., 2010). Interestingly, recent reports have demonstrated that OGT is involved in the proteolytic maturation of HFC-1, with cleavage of HCF-1 being carried out in the same active site where protein glycosylation occurs and with UPD-GlcNAc used as a co-substrate during the cleavage reaction (Capotosti et al., 2011; Daou et al., 2011; Hanover, 2011; Lazarus et al., 2013). It is not yet clear if there are other substrates that might be cleaved by OGT.
O-GlcNAcase
First identified as meningioma-expressed antigen 5 (MGEA5), the gene encoding OGA is located on chromosome 10 in humans (Comtesse et al., 2001; Heckel et al., 1998). Alternative gene splicing yields two isoforms, a long isoform (OGA-L) and a short isoform (OGA-S). OGA-L is found predominantly in the cytoplasm (Comtesse et al., 2001), whereas OGA-S exhibits nuclear (Comtesse et al., 2001) and lipid-droplet-associated localization (Keembiyehetty et al., 2011). Both isoforms possess an identical N-terminal hyaluronidase domain, but differ at their C-termini (Fig. 3B). The OGA-L isoform contains a C-terminal extension with similarity to known histone acetyl transferase (HAT) domains (Comtesse et al., 2001; Schultz and Pils, 2002), whereas OGA-S has a unique 15-amino-acid-long C-terminal extension. Despite the similarity of the OGA-L C-terminus to known HAT domains, it is unclear whether the protein functions as a HAT (Butkinaree et al., 2008; He et al., 2014; Toleman et al., 2004). OGA-L is encoded by 16 exons, and produces a 916-amino-acid protein product (Gao et al., 2001). OGA-S is a splice variant of the OGA gene wherein the 5′ splice site of intron 11 is skipped, leading to an extended exon 10 that utilizes an alternate stop codon relative to OGA-L (Comtesse et al., 2001). As a result, OGA-S is produced as a truncated 677-amino-acid species. In OGA-L, a linker region between the N- and C-terminal domains contains a caspase 3 cleavage site that is cleaved during apoptosis (Butkinaree et al., 2008). In in vitro experiments, OGA-S exhibits lower enzyme activity than OGA-L (Kim et al., 2006; Macauley and Vocadlo, 2009), although it is unclear whether this reflects their relative activities in an intact cell.
Because efforts to crystallize human OGA have not yet been successful, work to understand the catalytic mechanism of OGA has been based on structural studies of bacterial homologs (Dennis et al., 2006; He et al., 2010; Rao et al., 2006) and on mechanistic studies that use the human variant and synthetic OGA substrates and inhibitors (Çetinbaş et al., 2006; Greig et al., 2009; Macauley et al., 2005a; Macauley et al., 2005b; Whitworth et al., 2007). These studies suggest that the catalytic mechanism in human OGA-L requires two key catalytic aspartate residues, Asp174 and Asp175, and proceeds via a two-step substrate-assisted mechanism. Additional studies have focused on determining the factors that are important for substrate recognition by OGA, including determination of the residues of OGA involved and the features of the substrate that contribute to catalysis (Schimpl et al., 2012a; Schimpl et al., 2010; Shen et al., 2012). The results of these studies suggest that it is either contacts between OGA and the sugar on the protein substrate that modulate OGA recognition (Shen et al., 2012), or contacts between OGA and both the sugar and the peptide backbone of the protein substrate (Schimpl et al., 2012a). The individual amino acid side chains appear not to play a role in substrate binding and recognition.
The spatiotemporal effects of targeted O-GlcNAc cycling
Overall, it is likely that the unique characteristics of the various OGA and OGT isoforms provide a means for the differential recognition of specific protein substrates in a highly regulated manner. Further, the specific localization patterns and physical properties of these isoforms are likely to allow them to interact with specific protein substrates in a nutritionally regulated manner. Identifying these interactions might provide insight into how the cell is able to communicate its nutritional status and elicit appropriate responses in order to maintain cellular homeostasis. Moreover, they might offer an explanation for the disease states that arise when homeostasis cannot be achieved owing to dietary or genetic factors. Specific examples of targeted O-GlcNAc cycling and the subsequent cellular response are discussed below.
Lipid droplet remodeling – OGA-S, perilipin-2 and the proteasome
Lipid droplets are cellular organelles responsible for the storage and hydrolysis of neutral lipids. Lipid droplets were originally considered to be inert organelles; however, more recently, they have emerged as key players in various aspects of cellular homeostasis, including lipid and energy metabolism and beyond (reviewed in Guo et al., 2009). Furthermore, their study has garnered interest because excessive lipid storage in lipid droplets can be linked to the development of obesity, diabetes and atherosclerosis. A better understanding of the complex and dynamic mechanism by which lipid droplets are formed and remodeled will provide further insight into the regulation of lipid and energy metabolism.
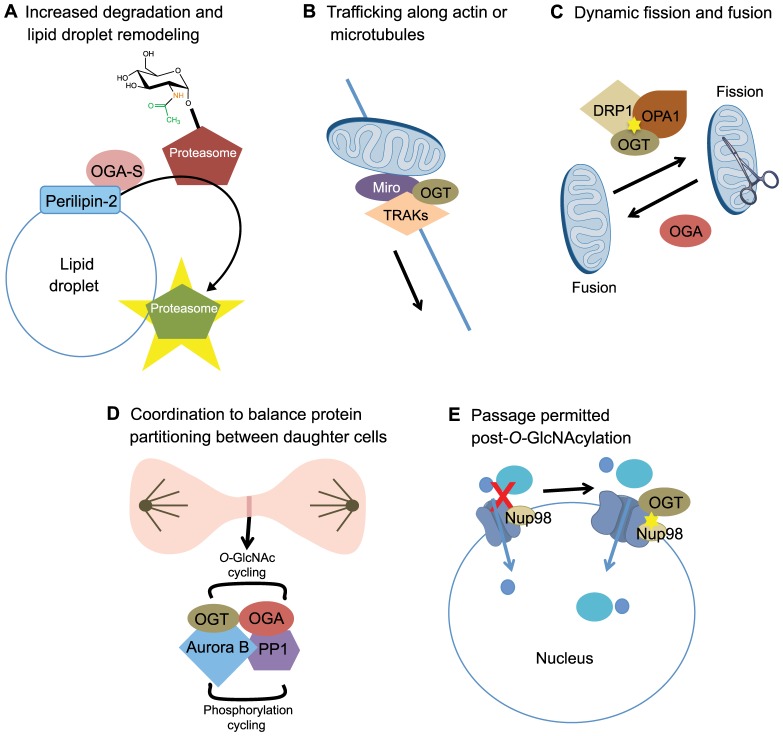
Interestingly, OGA-S has been shown to be targeted to the surface of nascent lipid droplets, where it colocalizes with perilipin-2, a protein known to be actively involved in lipid droplet formation (Fig. 4A) (Bickel et al., 2009; Brasaemle, 2007; Londos et al., 1999). In addition, it is known that the activity of the 26S subunit of the proteasome is inhibited by O-GlcNAc modification (Zhang et al., 2007); this is interesting because proteasomal activity is required at the surface of lipid droplets for lipid droplet remodeling (Ohsaki et al., 2006). Furthermore, the activity of OGA-S is enhanced by lipid accumulation (Keembiyehetty et al., 2011). Therefore, a high concentration of OGA-S on the surface of lipid droplets could lead to the removal of the O-GlcNAc modification from the proteasome in the immediate area surrounding the lipid droplet, resulting in a local increase in proteasomal activity and protein remodeling on the lipid droplet surface. Moreover, it has been demonstrated that reduction of OGA-S levels results in the stabilization of proteins that are associated with lipid droplets (Keembiyehetty et al., 2011), suggesting that there might be a complex feedback system between proteasomal activity and the level of OGA-S that could have a role in regulating lipid droplet maturation. Potentially, this is a mechanism by which the cell regulates lipid storage and mobilization, with OGA-S uniquely suitable to link carbohydrate and lipid metabolism.
Fig. 4.
O-GlcNAc cycling and localized cellular remodeling. (A) Lipid droplet remodeling. The targeted localization of OGA-S by perilipin-2 at the surface of lipid droplets results in the removal of O-GlcNAc from the proteasome and its subsequent activation. Proteasomal activation at the surface of lipid droplets increases the degradation of surface localized proteins, resulting in lipid droplet remodeling. (B) Mitochondrial trafficking. OGT binds to TRAKs and has a role in the trafficking of mitochondria along actin or microtubule networks, potentially either directly through TRAK binding, via TRAK O-GlcNAcylation, or both. (C) Mitochondrial fission and fusion. OGT O-GlcNAcylates DRP1 and OPA1 (indicated by the yellow star), which results in increased mitochondrial fission. Overexpression of OGA reverses this effect and promotes mitochondrial fusion. (D) Cytokinesis. During cytokinesis, both OGT and OGA are found in a transient complex with both Aurora B and the phosphatase PP1. It has been shown that the localization of OGT in this complex is dependent on Aurora B. These pairs of enzymes with opposing activities somehow coordinate their activities in a presumably complex manner to ensure proper protein partitioning into daughter cells and completion of cytokinesis. (E) Nuclear pore remodeling. O-GlcNAcylation of Nup98 by OGT (yellow star) can relax the ‘sieve’-like structure of the nuclear pore, allowing the passage of larger molecules not permitted prior to Nup98 O-GlcNAcylation.
Mitochondrial trafficking – OGT, Miro, the TRAKs and kinesins
Mitochondria are trafficked along both actin and microtubule networks (Ligon and Steward, 2000a; Ligon and Steward, 2000b; Morris and Hollenbeck, 1995; Rube and van der Bliek, 2004), and the regulation of mitochondrial trafficking is particularly important during embryonic development and for the regulation of apoptosis, as well as in neurons, where mitochondria often need to be transported over long distances (Detmer and Chan, 2007). In Drosophila, mitochondrial trafficking is regulated by a protein complex containing the mitochondrial Rho GTPase Miro and the kinesin-binding protein Milton (Guo et al., 2005; Stowers et al., 2002). This complex also includes the kinesin-binding adaptor proteins TRAK1 (OIP106 in humans) and TRAK2 (OIP98 in humans, also known as Grif-1) (Brickley and Stephenson, 2011), which, interestingly, have been found to bind to OGT (Iyer et al., 2003). It is therefore possible that TRAK O-GlcNAcylation either modulates their function directly, allows the TRAKs to recruit OGT to Miro or kinesins, or alters both the function and recruitment roles of the TRAKs (Fig. 4B). Interestingly, using co-immunoprecipitation experiments, Iyer et al. (Iyer et al., 2003) have shown that OIP106 interacts with both OGT and RNA polymerase II (RNA Pol II) in vivo and forms a RNA-Pol-II–OIP106–OGT ternary complex. The authors suggest that, in addition to a role in localized regulation of mitochondrial trafficking through its interaction with OGT, OIP106 might also target OGT to transcriptional complexes where it could modify transcriptional machinery components (i.e. RNA Pol II and transcription factors). As the levels of UDP-GlcNAc are influenced by input from a number of nutritional pathways (Fig. 1), including those that are necessary for mitochondrial function, it is possible that the O-GlcNAcylation state of regulatory proteins might communicate nutritional status (i.e. the availability of mitochondrial substrates, to modulate mitochondrial mobility) (Wells et al., 2003). Because mitochondrial trafficking is particularly important in neurons (Schwarz, 2013), its deregulation could result in neurological disease. It would be particularly interesting to address whether OGT activity is involved in modulating mitochondrial transport through O-GlcNAcylation of Miro, the TRAKs or other kinesins, as this could provide a link between nutritional status and the development of neurological pathologies (MacAskill and Kittler, 2010).
Mitochondrial fission versus fusion – O-GlcNAcylation, OPA1 and DRP1
In addition to proper trafficking, the regulated and dynamic remodeling of mitochondria is also essential for appropriate mitochondrial function (Kane and Youle, 2010; Westermann, 2010). One protein that plays a prominent role in mitochondrial fission is the GTPase dynamin-related protein 1 (DRP1, also known as DNM1L) (Chang and Blackstone, 2010). Although primarily a cytoplasmic protein (Smirnova et al., 2001), Gawlowski and colleagues have demonstrated that, in cardiac cells, DRP1 translocates to the mitochondria upon O-GlcNAcylation by OGT (Gawlowski et al., 2012). Furthermore, they have shown that O-GlcNAcylation increases the amount of DRP1 that is in an active GTP-bound state. Following the O-GlcNAcylation and mitochondrial translocation of DRP1, Gawlowski and colleagues observed increased mitochondrial fragmentation with reduced membrane potential. In addition, they found that high glucose levels both increase the O-GlcNAcylation of optical atrophy 1 (OPA1), a mitochondrial fusion-related protein, and decrease either OPA1 expression or protein stability in neonatal cardiac myocytes, resulting in the same phenotype as observed upon increased DRP1 O-GlcNAcylation. Furthermore, they determined that overexpression of OGA was able to restore proper mitochondrial structure, suggesting that the localized activity of OGT or OGA results in the subsequent fragmentation (fission) or restoration (fusion) of mitochondria, respectively (Fig. 4C). Perhaps this is a mechanism by which the cell communicates its nutritive state for the maintenance of proper mitochondrial function, for example in preparation for cellular division, in which a localized fragmentation or fission of mitochondria is required to ensure that mitochondria are allocated properly, or to help maximize the capacity for oxidative phosphorylation, through fusion, when the cell is under low nutrient conditions. As such, the role of O-GlcNAc cycling in mitochondrial trafficking and fission–fusion could have particularly important implications in the development of mitochondrial dysfunction observed in diabetes.
Cell division and cytokinesis – O-GlcNAc cycling, Aurora B and PP1
During cell division, many proteins need to be targeted to distinct cellular locations to accurately regulate mitotic progression and ensure that each daughter cell receives the appropriate complement of cellular material and a complete set of chromosomes. One such protein is Aurora B, a kinase essential for normal mitotic progression (Carmena and Earnshaw, 2003), that exhibits cell-cycle-dependent localization and is found to be targeted to the midbody during cytokinesis, where its activity is necessary for the successful completion of cytokinesis. Disruption of either Aurora B expression or activity has been shown to increase polyploidy in cells (Adams et al., 2001; Nguyen et al., 2005; Ota et al., 2002; Tatsuka et al., 1998). Interestingly, both OGT and OGA are also found at the midbody during cytokinesis in a transient complex with Aurora B and protein phosphatase 1 (PP1) (Fig. 4D) (Slawson et al., 2008), a phosphatase whose activity opposes that of Aurora B (Emanuele et al., 2008; Goto et al., 2006; Sugiyama et al., 2002). Furthermore, Aurora B activity appears to be necessary for the recruitment of OGT to the midbody, as its localization is lost upon reduction of Aurora B activity (Slawson et al., 2008). It appears that the combined activities of the members of this complex regulate the post-translational modification status of mitotic substrates, such as that of the cytoskeletal intermediate filament protein vimentin, in a highly localized manner (Slawson et al., 2008). It is possible that the pattern of modifications achieved by these pairs of enzymes plays a role in ensuring the proper segregation of the substrate proteins into the daughter cells, in the case of vimentin potentially by inducing the disassembly of vimentin filaments (Izawa and Inagaki, 2006). It is not yet clear how a balance between the activities of these pairs of enzymes with opposing actions is achieved to accomplish this task.
Nuclear pore structure and permeability – O-GlcNAcylation and Nup98
Nuclear pore complexes (NPCs) are important for controlling the passage of material from the cytoplasm into the nucleus and are composed of nucleoporins (Nups). A common motif in the Nups is the phenylalanine-glycine (FG) motif that consists of up to 50 repetitive FG sequences (Starr and Hanover, 1991). It has been suggested that the FG–FG contacts between Nups creates a ‘sieve’-like structure in the pore cavity (Ribbeck and Görlich, 2001) through which small molecules can easily pass into the nucleus, but which excludes species larger than 5 nm (Mohr et al., 2009). Nuclear transport receptors (NTRs) provide a means for import and export of molecules larger than this cutoff. Interestingly, the FG-repeat domains of the Nups are also binding sites for NTRs, further emphasizing the importance of the repeats in creating the NPC barrier. It has also been suggested that FG-repeat domains function in modulating nuclear entry and exit, and that the binding of NTRs to Nup FG-repeats replaces FG–FG contacts, resulting in a local disruption of the sieve structure and subsequent nuclear entrance of molecules with higher molecular mass (Frey and Görlich, 2007; Frey et al., 2006; Labokha et al., 2013). Interestingly, it has been shown that a subset of FG-containing Nups are modified by O-GlcNAc (Hanover et al., 1987), and one of these is Nup98 (Powers et al., 1995). Recently, using Nup98-derived hydrogels, Labokha and co-workers demonstrated that O-GlcNAc modification of Nup98 relieves the strict sieving property of the hydrogel, thereby allowing passage of larger molecules that could not be accommodated prior to Nup98 O-GlcNAcylation (Fig. 4E) (Labokha et al., 2013). Because other Nups are also known to be O-GlcNAcylated, it would be particularly interesting to determine whether the targeted or localized addition and removal of the O-GlcNAc moiety allows the cell to dynamically regulate the permeability of specific nuclear pore complexes in response to nutritional cues, which could have a profound effect on cellular homeostasis.
Transcriptional repression through interaction of OGT with Sin3
The packaging of DNA into chromatin is an essential mechanism by which transcription is regulated. The transcription of specific genes can be up- or down-regulated through post-translational modification of histones that alter both protein interactions and chromatin structure (Kouzarides, 2007). In addition, it has been proposed that a complex ‘histone code’ exists: specific combinations of multiple histone modifications result in the activation or repression of particular genes, thus allowing for the fine-tuning of gene expression in order to appropriately respond to a variety of environmental and nutritional cues (Jenuwein and Allis, 2001; Strahl and Allis, 2000; Turner, 2000).
One of these histone modifications is the acetylation of specific lysine residues, which alters chromatin structure and thus the state of activation of specific genes (Kuo and Allis, 1998). Consequently, enzymes that either acetylate or deacetylate histones, HATs and histone deacetylases (HDACs), respectively, play an integral role in determining gene expression profiles. In general, HDACs can be found in a diverse array of corepressor complexes that allow for the repression of distinct subsets of genes (Burke and Baniahmad, 2000; Ng and Bird, 2000). One of these complexes is the Sin3-containing HDAC corepressor complex. Furthermore, Sin3 has been shown to interact physically with and be O-GlcNAcylated by OGT (Fig. 5), and this interaction results in the repression of genes through a pathway that might be independent of HDAC activity (Yang et al., 2002). Potentially, the localized activity of Sin3 and OGT, together with the O-GlcNAc modification, could act synergistically to repress specific subsets of regulatory genes, as has been seen with, for example, the retinoblastoma tumor suppressor protein (Siddiqui et al., 2003).
Fig. 5.
O-GlcNAc cycling and transcriptional regulation. The proteins Sin3 (shown on the left) and TETs (shown on the right) might interact with OGT at targeted locations to repress or activate, respectively, the transcription of a specific subset of genes in response to nutritional cues. Simultaneously, the putative histone acetyl transferase (HAT) domain of OGA might regulate its binding to specific histone tail marks, allowing OGA, in conjunction with histone acetylation, to activate gene transcription.
In addition, as noted above, OGA-L has a predicted HAT domain that is similar to that of the histone acetyltransferase GCN5. Despite the debate over the ability of OGA to act as a HAT (Butkinaree et al., 2008; He et al., 2014), owing to this similarity, it is possible that this domain could facilitate the association of OGA-L with histone proteins. Potentially, a specific pattern of histone modification could recruit OGA at certain locations and times, for example, cell cycle stages, to remove the O-GlcNAc modification and, in conjunction with histone acetylation, activate gene transcription (Fig. 5).
Transcriptional activation through interaction of OGT with TET proteins
As mentioned above, post-translational modification of histones is one way in which transcriptional activity can be regulated. Another mechanism of regulating transcriptional activity is through direct modification of DNA itself. It is known that DNA methylation on cytosine by DNA methyltransferases, termed 5-methylcytosine or 5-mC, generally results in gene repression (Cedar and Bergman, 2009; Suzuki and Bird, 2008). Proteins from the ten-eleven translocation (TET) family (TET1, TET2 and TET3) catalyze the conversion of 5-mC to cytosine 5-hydroxymethylation or 5-hmC, and much work is being done to better understand the roles of this mark (Ito et al., 2010; Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Recently, it has been shown that OGT interacts physically with and is recruited to chromatin by the TET proteins (Chen et al., 2013; Deplus et al., 2013b; Vella et al., 2013) (Fig. 5). The interaction of OGT with the TETs plays a complicated role in regulating transcription, which is just beginning to be understood, although it appears that these interactions are important for TET-dependent gene activation. Specifically, it has been shown that OGT positively regulates TET1 protein levels and hmC levels on TET1 target genes (Shi et al., 2013). Furthermore, OGT and TET1 are found together at promoters near CpG-rich transcription start sites genome wide, where low levels of DNA methylation are observed (Vella et al., 2013). This might mean TET1 and OGT together play a role in regulating CpG island methylation. Interestingly, TET2 levels positively influence histone 2B (H2B) Ser112 O-GlcNAcylation levels, independently of TET2 enzymatic activity (Chen et al., 2013). This suggests OGT recruitment by TET2 can increase O-GlcNAcylation of specific chromatin substrates, influencing the transcriptional activation of specific subsets of genes. OGT and TET2 and TET3 have also been found to localize to promoters where they appear to play a role in H3K4me3 enrichment by promoting binding of the SET1/COMPASS H3K4 methyltransferase SETD1A, and subsequent transcriptional activation (Deplus et al., 2013a). The results of these studies are particularly interesting considering that under other conditions, for example, while interacting with the Sin3 HDAC complex, OGT results in transcriptional repression of a distinct subset of genes. Thus, it is possible that changes in the nutritional status can simultaneously translate into the up- and down-regulation of distinct subsets of genes to ensure appropriate transcriptional profiles are achieved (Fig. 5).
Conclusions and perspectives
Although, in this Commentary, we have explored a few examples of the localization-dependent effects of O-GlcNAc cycling, the O-GlcNAc field is in its infancy and much remains to be discovered. As such, many more interaction partners responsible for OGA and OGT tethering and their functional relevance in the cell will likely be uncovered in the future. For example, although it has not yet been demonstrated directly that the protein myosin phosphatase target subunit 1 (MYPT1), a regulatory subunit of PP1, can recruit OGT to a particular cellular location, there are a number of observations that suggest that localized interactions between MYPT1 and OGT might be involved in ensuring that events take place accurately and at the correct cellular location during mitosis and cytokinesis, as has been demonstrated for Aurora B: (1) MYPT1 plays an integral and highly spatiotemporal role in mitosis and cytokinesis, (2) disruptions in O-GlcNAc cycling also disrupt mitosis and cytokinesis (Sakabe and Hart, 2010), and (3) MYPT1 interacts with OGT (Cheung et al., 2008). A similar mechanism in targeting OGA or OGT might also take places during Polycomb-mediated repression, the specialized silencing of genes that must be tightly regulated in a spatiotemporal manner. In fact, it has been shown in both Drosophila and mouse embryonic stem cells that OGT plays an essential role in Polycomb-mediated repression that is crucial for proper development and pluripotency (Gambetta et al., 2009; Myers et al., 2011; Sinclair et al., 2009). However, although it has been shown that OGT O-GlcNAcylates the Polycomb group member Polyhomeotic in Drosophila (Gambetta et al., 2009), the molecular details of its role in Polycomb-mediated repression remain incompletely defined and might involve a similar targeting mechanism to that described above for the other cellular processes that require localized OGT and OGA activity. Further efforts to elucidate the interaction partners that target OGT and OGA to particular locations for specific cellular functions will be pivotal to fully elucidate the pathways that communicate between lipid, amino acid, nucleotide and sugar metabolism. This research is anticipated to provide crucial insights that will allow us to both better understand the various disease states that can arise from such metabolic defects.
Footnotes
Competing interests
The authors declare no competing interests.
Funding
The work of our laboratory is funded from the intramural program of the National Institutes of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health. Deposited in PMC for release after 12 months.
References
- Adams R. R., Maiato H., Earnshaw W. C., Carmena M. (2001). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153, 865–880 10.1083/jcb.153.4.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel P. E., Tansey J. T., Welte M. A. (2009). PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta 1791, 419–440 10.1016/j.bbalip.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch G. L., Lässle M. (1999). The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939 [DOI] [PubMed] [Google Scholar]
- Bond M. R., Hanover J. A. (2013). O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 33, 205–229 10.1146/annurev--nutr--071812--161240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle D. L. (2007). Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48, 2547–2559 10.1194/jlr.R700014--JLR200 [DOI] [PubMed] [Google Scholar]
- Brickley K., Stephenson F. A. (2011). Trafficking kinesin protein (TRAK)-mediated transport of mitochondria in axons of hippocampal neurons. J. Biol. Chem. 286, 18079–18092 10.1074/jbc.M111.236018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke L. J., Baniahmad A. (2000). Co-repressors 2000. FASEB J. 14, 1876–1888 10.1096/fj.99--0943rev [DOI] [PubMed] [Google Scholar]
- Butkinaree C., Cheung W. D., Park S., Park K., Barber M., Hart G. W. (2008). Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J. Biol. Chem. 283, 23557–23566 10.1074/jbc.M804116200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotosti F., Guernier S., Lammers F., Waridel P., Cai Y., Jin J., Conaway J. W., Conaway R. C., Herr W. (2011). O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144, 376–388 10.1016/j.cell.2010.12.030 [DOI] [PubMed] [Google Scholar]
- Carmena M., Earnshaw W. C. (2003). The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4, 842–854 10.1038/nrm1245 [DOI] [PubMed] [Google Scholar]
- Carrillo L. D., Froemming J. A., Mahal L. K. (2011). Targeted in vivo O-GlcNAc sensors reveal discrete compartment-specific dynamics during signal transduction. J. Biol. Chem. 286, 6650–6658 10.1074/jbc.M110.191627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H., Bergman Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10, 295–304 10.1038/nrg2540 [DOI] [PubMed] [Google Scholar]
- Çetinbaş N., Macauley M. S., Stubbs K. A., Drapala R., Vocadlo D. J. (2006). Identification of Asp174 and Asp175 as the key catalytic residues of human O-GlcNAcase by functional analysis of site-directed mutants. Biochemistry 45, 3835–3844 10.1021/bi052370b [DOI] [PubMed] [Google Scholar]
- Chang C. R., Blackstone C. (2010). Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 1201, 34–39 10.1111/j.1749--6632.2010.05629.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Chen Y., Bian C., Fujiki R., Yu X. (2013). TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 10.1038/nature11742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. D., Sakabe K., Housley M. P., Dias W. B., Hart G. W. (2008). O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem. 283, 33935–33941 10.1074/jbc.M806199200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comtesse N., Maldener E., Meese E. (2001). Identification of a nuclear variant of MGEA5, a cytoplasmic hyaluronidase and a beta-N-acetylglucosaminidase. Biochem. Biophys. Res. Commun. 283, 634–640 10.1006/bbrc.2001.4815 [DOI] [PubMed] [Google Scholar]
- Copeland R. J., Bullen J. W., Hart G. W. (2008). Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am. J. Physiol. 295, E17–E28 10.1152/ajpendo.90281.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou S., Mashtalir N., Hammond-Martel I., Pak H., Yu H., Sui G., Vogel J. L., Kristie T. M., Affar B. (2011). Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc. Natl. Acad. Sci. USA 108, 2747–2752 10.1073/pnas.1013822108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis R. J., Taylor E. J., Macauley M. S., Stubbs K. A., Turkenburg J. P., Hart S. J., Black G. N., Vocadlo D. J., Davies G. J. (2006). Structure and mechanism of a bacterial beta-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol. 13, 365–371 10.1038/nsmb1079 [DOI] [PubMed] [Google Scholar]
- Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E. et al. (2013a). TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E. et al. (2013b). TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 10.1038/emboj.2012.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer S. A., Chan D. C. (2007). Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8, 870–879 10.1038/nrm2275 [DOI] [PubMed] [Google Scholar]
- Emanuele M. J., Lan W., Jwa M., Miller S. A., Chan C. S., Stukenberg P. T. (2008). Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181, 241–254 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S., Görlich D. (2007). A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130, 512–523 10.1016/j.cell.2007.06.024 [DOI] [PubMed] [Google Scholar]
- Frey S., Richter R. P., Görlich D. (2006). FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- Gambetta M. C., Oktaba K., Müller J. (2009). Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 10.1126/science.1169727 [DOI] [PubMed] [Google Scholar]
- Gao Y., Wells L., Comer F. I., Parker G. J., Hart G. W. (2001). Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 10.1074/jbc.M010420200 [DOI] [PubMed] [Google Scholar]
- Gawlowski T., Suarez J., Scott B., Torres-Gonzalez M., Wang H., Schwappacher R., Han X., Yates J. R., 3rd, Hoshijima M., Dillmann W. (2012). Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-β-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J. Biol. Chem. 287, 30024–30034 10.1074/jbc.M112.390682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M., Yanagida M. (1991). The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 16, 173–177 10.1016/0968--0004(91)90070--C [DOI] [PubMed] [Google Scholar]
- Görlich D., Panté N., Kutay U., Aebi U., Bischoff F. R. (1996). Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15, 5584–5594 [PMC free article] [PubMed] [Google Scholar]
- Goto H., Kiyono T., Tomono Y., Kawajiri A., Urano T., Furukawa K., Nigg E. A., Inagaki M. (2006). Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 8, 180–187 10.1038/ncb1350 [DOI] [PubMed] [Google Scholar]
- Greig I. R., Macauley M. S., Williams I. H., Vocadlo D. J. (2009). Probing synergy between two catalytic strategies in the glycoside hydrolase O-GlcNAcase using multiple linear free energy relationships. J. Am. Chem. Soc. 131, 13415–13422 10.1021/ja904506u [DOI] [PubMed] [Google Scholar]
- Griffin E. E., Odde D. J., Seydoux G. (2011). Regulation of the MEX-5 gradient by a spatially segregated kinase/phosphatase cycle. Cell 146, 955–968 10.1016/j.cell.2011.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Macleod G. T., Wellington A., Hu F., Panchumarthi S., Schoenfield M., Marin L., Charlton M. P., Atwood H. L., Zinsmaier K. E. (2005). The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron 47, 379–393 10.1016/j.neuron.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Guo Y., Cordes K. R., Farese R. V., Jr, Walther T. C. (2009). Lipid droplets at a glance. J. Cell Sci. 122, 749–752 10.1242/jcs.037630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover J. A. (2011). A versatile sugar transferase makes the cut. Cell 144, 321–323 10.1016/j.cell.2011.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover J. A., Cohen C. K., Willingham M. C., Park M. K. (1987). O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem. 262, 9887–9894 [PubMed] [Google Scholar]
- Hanover J. A., Yu S., Lubas W. B., Shin S. H., Ragano-Caracciola M., Kochran J., Love D. C. (2003). Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch. Biochem. Biophys. 409, 287–297 10.1016/S0003--9861(02)00578--7 [DOI] [PubMed] [Google Scholar]
- Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011). Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 10.1146/annurev--biochem--060608--102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Macauley M. S., Stubbs K. A., Vocadlo D. J., Davies G. J. (2010). Visualizing the reaction coordinate of an O-GlcNAc hydrolase. J. Am. Chem. Soc. 132, 1807–1809 10.1021/ja9086769 [DOI] [PubMed] [Google Scholar]
- He Y., Roth C., Turkenburg J. P., Davies G. J. (2014). Three-dimensional structure of a Streptomyces sviceus GNAT acetyltransferase with similarity to the C-terminal domain of the human GH84 O-GlcNAcase. Acta Crystallogr. D Biol. Crystallogr. 70, 186–195 10.1107/S1399004713029155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel D., Comtesse N., Brass N., Blin N., Zang K. D., Meese E. (1998). Novel immunogenic antigen homologous to hyaluronidase in meningioma. Hum. Mol. Genet. 7, 1859–1872 10.1093/hmg/7.12.1859 [DOI] [PubMed] [Google Scholar]
- Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., Zhang Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 10.1038/nature09303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S. P., Akimoto Y., Hart G. W. (2003). Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J. Biol. Chem. 278, 5399–5409 10.1074/jbc.M209384200 [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Kutay U., von Kobbe C., Mattaj I. W., Görlich D. (1997). The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16, 6535–6547 10.1093/emboj/16.21.6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa I., Inagaki M. (2006). Regulatory mechanisms and functions of intermediate filaments: a study using site- and phosphorylation state-specific antibodies. Cancer Sci. 97, 167–174 10.1111/j.1349--7006.2006.00161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D. (2001). Translating the histone code. Science 293, 1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Jínek M., Rehwinkel J., Lazarus B. D., Izaurralde E., Hanover J. A., Conti E. (2004). The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 11, 1001–1007 10.1038/nsmb833 [DOI] [PubMed] [Google Scholar]
- Kane L. A., Youle R. J. (2010). Mitochondrial fission and fusion and their roles in the heart. J. Mol. Med. 88, 971–979 10.1007/s00109--010--0674--6 [DOI] [PubMed] [Google Scholar]
- Keembiyehetty C. N., Krzeslak A., Love D. C., Hanover J. A. (2011). A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome. J. Cell Sci. 124, 2851–2860 10.1242/jcs.083287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. J., Kang D. O., Love D. C., Hanover J. A. (2006). Enzymatic characterization of O-GlcNAcase isoforms using a fluorogenic GlcNAc substrate. Carbohydr. Res. 341, 971–982 10.1016/j.carres.2006.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693–705 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kreppel L. K., Blomberg M. A., Hart G. W. (1997). Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 10.1074/jbc.272.14.9308 [DOI] [PubMed] [Google Scholar]
- Kriaucionis S., Heintz N. (2009). The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 10.1126/science.1169786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. H., Allis C. D. (1998). Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20, 615–626 [DOI] [PubMed] [Google Scholar]
- Labokha A. A., Gradmann S., Frey S., Hülsmann B. B., Urlaub H., Baldus M., Görlich D. (2013). Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 32, 204–218 10.1038/emboj.2012.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus B. D., Love D. C., Hanover J. A. (2006). Recombinant O-GlcNAc transferase isoforms: identification of O-GlcNAcase, yes tyrosine kinase, and tau as isoform-specific substrates. Glycobiology 16, 415–421 10.1093/glycob/cwj078 [DOI] [PubMed] [Google Scholar]
- Lazarus M. B., Nam Y., Jiang J., Sliz P., Walker S. (2011). Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 10.1038/nature09638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus M. B., Jiang J., Gloster T. M., Zandberg W. F., Whitworth G. E., Vocadlo D. J., Walker S. (2012). Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol. 8, 966–968 10.1038/nchembio.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus M. B., Jiang J., Kapuria V., Bhuiyan T., Janetzko J., Zandberg W. F., Vocadlo D. J., Herr W., Walker S. (2013). HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 342, 1235–1239 10.1126/science.1243990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon L. A., Steward O. (2000a). Movement of mitochondria in the axons and dendrites of cultured hippocampal neurons. J. Comp. Neurol. 427, 340–350 [DOI] [PubMed] [Google Scholar]
- Ligon L. A., Steward O. (2000b). Role of microtubules and actin filaments in the movement of mitochondria in the axons and dendrites of cultured hippocampal neurons. J. Comp. Neurol. 427, 351–361 [DOI] [PubMed] [Google Scholar]
- Liu F. H., Wu S. J., Hu S. M., Hsiao C. D., Wang C. (1999). Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J. Biol. Chem. 274, 34425–34432 10.1074/jbc.274.48.34425 [DOI] [PubMed] [Google Scholar]
- Londos C., Brasaemle D. L., Schultz C. J., Segrest J. P., Kimmel A. R. (1999). Perilipins, ADRP, and other proteins that associate with intracellular neutral lipid droplets in animal cells. Semin. Cell Dev. Biol. 10, 51–58 10.1006/scdb.1998.0275 [DOI] [PubMed] [Google Scholar]
- Love D. C., Kochan J., Cathey R. L., Shin S. H., Hanover J. A. (2003). Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J. Cell Sci. 116, 647–654 10.1242/jcs.00246 [DOI] [PubMed] [Google Scholar]
- Lubas W. A., Hanover J. A. (2000). Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J. Biol. Chem. 275, 10983–10988 10.1074/jbc.275.15.10983 [DOI] [PubMed] [Google Scholar]
- Lubas W. A., Frank D. W., Krause M., Hanover J. A. (1997). O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272, 9316–9324 10.1074/jbc.272.14.9316 [DOI] [PubMed] [Google Scholar]
- MacAskill A. F., Kittler J. T. (2010). Control of mitochondrial transport and localization in neurons. Trends Cell Biol. 20, 102–112 10.1016/j.tcb.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Macauley M. S., Vocadlo D. J. (2009). Enzymatic characterization and inhibition of the nuclear variant of human O-GlcNAcase. Carbohydr. Res. 344, 1079–1084 10.1016/j.carres.2009.04.017 [DOI] [PubMed] [Google Scholar]
- Macauley M. S., Stubbs K. A., Vocadlo D. J. (2005a). O-GlcNAcase catalyzes cleavage of thioglycosides without general acid catalysis. J. Am. Chem. Soc. 127, 17202–17203 10.1021/ja0567687 [DOI] [PubMed] [Google Scholar]
- Macauley M. S., Whitworth G. E., Debowski A. W., Chin D., Vocadlo D. J. (2005b). O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 280, 25313–25322 10.1074/jbc.M413819200 [DOI] [PubMed] [Google Scholar]
- Mazars R., Gonzalez-de-Peredo A., Cayrol C., Lavigne A. C., Vogel J. L., Ortega N., Lacroix C., Gautier V., Huet G., Ray A. et al. (2010). The THAP-zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase: a link between DYT6 and DYT3 dystonias. J. Biol. Chem. 285, 13364–13371 10.1074/jbc.M109.072579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D., Frey S., Fischer T., Güttler T., Görlich D. (2009). Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 28, 2541–2553 10.1038/emboj.2009.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R. L., Hollenbeck P. J. (1995). Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J. Cell Biol. 131, 1315–1326 10.1083/jcb.131.5.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S. A., Panning B., Burlingame A. L. (2011). Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 108, 9490–9495 10.1073/pnas.1019289108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Bird A. (2000). Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25, 121–126 10.1016/S0968--0004(00)01551--6 [DOI] [PubMed] [Google Scholar]
- Ngoh G. A., Facundo H. T., Zafir A., Jones S. P. (2010). O-GlcNAc signaling in the cardiovascular system. Circ. Res. 107, 171–185 10.1161/CIRCRESAHA.110.224675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. G., Chinnappan D., Urano T., Ravid K. (2005). Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property. Mol. Cell. Biol. 25, 4977–4992 10.1128/MCB.25.12.4977--4992.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte D., Muller U. (2002). Human O-GlcNAc transferase (OGT): genomic structure, analysis of splice variants, fine mapping in Xq13.1. Mamm. Genome. 13, 62–64 [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Cheng J., Fujita A., Tokumoto T., Fujimoto T. (2006). Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol. Biol. Cell 17, 2674–2683 10.1091/mbc.E05--07--0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T., Suto S., Katayama H., Han Z. B., Suzuki F., Maeda M., Tanino M., Terada Y., Tatsuka M. (2002). Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 62, 5168–5177 [PubMed] [Google Scholar]
- Powers M. A., Macaulay C., Masiarz F. R., Forbes D. J. (1995). Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 128, 721–736 10.1083/jcb.128.5.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt S. J., Epple H., Ward M., Feng Y., Braga V. M., Longmore G. D. (2005). The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J. Cell Biol. 168, 813–824 10.1083/jcb.200406083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F. V., Dorfmueller H. C., Villa F., Allwood M., Eggleston I. M., van Aalten D. M. (2006). Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 25, 1569–1578 10.1038/sj.emboj.7601026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Görlich D. (2001). Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20, 1320–1330 10.1093/emboj/20.6.1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rube D. A., van der Bliek A. M. (2004). Mitochondrial morphology is dynamic and varied. Mol. Cell. Biochem. 256-257, 331–339 10.1023/B:MCBI.0000009879.01256.f6 [DOI] [PubMed] [Google Scholar]
- Sakabe K., Hart G. W. (2010). O-GlcNAc transferase regulates mitotic chromatin dynamics. J. Biol. Chem. 285, 34460–34468 10.1074/jbc.M110.158170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaidani Y., Nomura T., Matsuura A., Ito M., Suzuki E., Murakami K., Nadano D., Matsuda T., Furukawa K., Okajima T. (2011). O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat. Commun. 2, 583 10.1038/ncomms1591 [DOI] [PubMed] [Google Scholar]
- Sakaidani Y., Ichiyanagi N., Saito C., Nomura T., Ito M., Nishio Y., Nadano D., Matsuda T., Furukawa K., Okajima T. (2012). O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochem. Biophys. Res. Commun. 419, 14–19 10.1016/j.bbrc.2012.01.098 [DOI] [PubMed] [Google Scholar]
- Schimpl M., Schüttelkopf A. W., Borodkin V. S., van Aalten D. M. (2010). Human OGA binds substrates in a conserved peptide recognition groove. Biochem. J. 432, 1–7 10.1042/BJ20101338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimpl M., Borodkin V. S., Gray L. J., van Aalten D. M. (2012a). Synergy of peptide and sugar in O-GlcNAcase substrate recognition. Chem. Biol. 19, 173–178 10.1016/j.chembiol.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimpl M., Zheng X., Borodkin V. S., Blair D. E., Ferenbach A. T., Schüttelkopf A. W., Navratilova I., Aristotelous T., Albarbarawi O., Robinson D. A. et al. (2012b). O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nat. Chem. Biol. 8, 969–974 10.1038/nchembio.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Pils B. (2002). Prediction of structure and functional residues for O-GlcNAcase, a divergent homologue of acetyltransferases. FEBS Lett. 529, 179–182 10.1016/S0014--5793(02)03322--7 [DOI] [PubMed] [Google Scholar]
- Schwarz T. L. (2013). Mitochondrial trafficking in neurons. Cold Spring Harb. Perspect. Biol. 5, a011304 10.1101/cshperspect.a011304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., Marth J. D. (2000). The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97, 5735–5739 10.1073/pnas.100471497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D. L., Gloster T. M., Yuzwa S. A., Vocadlo D. J. (2012). Insights into O-linked N-acetylglucosamine ([0-9]O-GlcNAc) processing and dynamics through kinetic analysis of O-GlcNAc transferase and O-GlcNAcase activity on protein substrates. J. Biol. Chem. 287, 15395–15408 10.1074/jbc.M111.310664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F. T., Kim H., Lu W., He Q., Liu D., Goodell M. A., Wan M., Songyang Z. (2013). Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J. Biol. Chem. 288, 20776–20784 10.1074/jbc.M113.460386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui H., Solomon D. A., Gunawardena R. W., Wang Y., Knudsen E. S. (2003). Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol. Cell. Biol. 23, 7719–7731 10.1128/MCB.23.21.7719--7731.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D. A., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., Vocadlo D. J., Brock H. W., Honda B. M. (2009). Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. USA 106, 13427–13432 10.1073/pnas.0904638106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C., Lakshmanan T., Knapp S., Hart G. W. (2008). A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol. Biol. Cell 19, 4130–4140 10.1091/mbc.E07--11--1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C., Copeland R. J., Hart G. W. (2010). O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 35, 547–555 10.1016/j.tibs.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr C. M., Hanover J. A. (1991). A common structural motif in nuclear pore proteins (nucleoporins). Bioessays 13, 145–146 10.1002/bies.950130309 [DOI] [PubMed] [Google Scholar]
- Stowers R. S., Megeath L. J., Górska-Andrzejak J., Meinertzhagen I. A., Schwarz T. L. (2002). Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36, 1063–1077 10.1016/S0896--6273(02)01094--2 [DOI] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D. (2000). The language of covalent histone modifications. Nature 403, 41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- Sugiyama K., Sugiura K., Hara T., Sugimoto K., Shima H., Honda K., Furukawa K., Yamashita S., Urano T. (2002). Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene 21, 3103–3111 10.1038/sj.onc.1205432 [DOI] [PubMed] [Google Scholar]
- Suzuki M. M., Bird A. (2008). DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476 10.1038/nrg2341 [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L. et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 10.1126/science.1170116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuka M., Katayama H., Ota T., Tanaka T., Odashima S., Suzuki F., Terada Y. (1998). Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 58, 4811–4816 [PubMed] [Google Scholar]
- Toleman C., Paterson A. J., Whisenhunt T. R., Kudlow J. E. (2004). Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J. Biol. Chem. 279, 53665–53673 10.1074/jbc.M410406200 [DOI] [PubMed] [Google Scholar]
- Torres C. R., Hart G. W. (1984). Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- Turner B. M. (2000). Histone acetylation and an epigenetic code. Bioessays 22, 836–845 [DOI] [PubMed] [Google Scholar]
- Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K. et al. (2013). Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell 49, 645–656 10.1016/j.molcel.2012.12.019 [DOI] [PubMed] [Google Scholar]
- Vocadlo D. J. (2012). O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr. Opin. Chem. Biol. 16, 488–497 10.1016/j.cbpa.2012.10.021 [DOI] [PubMed] [Google Scholar]
- Wells L., Vosseller K., Hart G. W. (2003). A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell. Mol. Life Sci. 60, 222–228 10.1007/s000180300017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B. (2010). Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 11, 872–884 10.1038/nrm3013 [DOI] [PubMed] [Google Scholar]
- Whitworth G. E., Macauley M. S., Stubbs K. A., Dennis R. J., Taylor E. J., Davies G. J., Greig I. R., Vocadlo D. J. (2007). Analysis of PUGNAc and NAG-thiazoline as transition state analogues for human O-GlcNAcase: mechanistic and structural insights into inhibitor selectivity and transition state poise. J. Am. Chem. Soc. 129, 635–644 10.1021/ja065697o [DOI] [PubMed] [Google Scholar]
- Yang X., Zhang F., Kudlow J. E. (2002). Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80 10.1016/S0092--8674(02)00810--3 [DOI] [PubMed] [Google Scholar]
- Yang Y. R., Song M., Lee H., Jeon Y., Choi E. J., Jang H. J., Moon H. Y., Byun H. Y., Kim E. K., Kim D. H. et al. (2012). O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell 11, 439–448 10.1111/j.1474--9726.2012.00801.x [DOI] [PubMed] [Google Scholar]
- Zachara N. E. (2012). The roles of O-linked β-N-acetylglucosamine in cardiovascular physiology and disease. Am. J. Physiol. 302, H1905–H1918 10.1152/ajpheart.00445.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Hu Y., Huang P., Toleman C. A., Paterson A. J., Kudlow J. E. (2007). Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. J. Biol. Chem. 282, 22460–22471 10.1074/jbc.M702439200 [DOI] [PubMed] [Google Scholar]