Abstract
Here we report the design, synthesis, and biochemical characterization of a new chemical tool, UNC0965. UNC0965 is a biotinylated version of our previously reported G9a chemical probe, UNC0638. Importantly, UNC0965 maintains high in vitro potency and is cell penetrant. The biotinylated tag of UNC0965 enables chemiprecipitation of G9a from whole cell lysates. Further, the cell penetrance of UNC0965 allowed us to explore the localization of G9a on chromatin both in vitro and in vivo.
Keywords: Chromatin, lysine methyltransferase, G9a, enzymes, medicinal chemistry
G9a is a lysine methyltransferase (KMT) responsible for catalyzing the mono- and dimethylation of histone H3 lysine 9 (H3K9).[1–6] Its activity is linked to multiple biological pathways and human diseases including cancer (reviewed in[7]). G9a is overexpressed in a number of tumors;[8] specifically, it is associated with increased metastasis and invasion of tumor cells in lung cancer.[9] Surprisingly, loss of G9a-dependent H3K9me2 (dimethylated H3K9) has also been observed in various cancer cell lines.[10] This discrepancy underscores the importance of further research into the biological role of G9a.
Chemical probes that demonstrate sufficient cellular potency and target selectivity can greatly accelerate understanding of protein function.[11, 12] Here, we report the design, synthesis, and biochemical characterization of such a chemical tool, UNC0965. UNC0965 is a biotinylated version of our previously reported G9a chemical probe UNC0638.[13] UNC0965 displays low nanomolar in vitro potency, is active in cellular assays, and selectively chemiprecipitates G9a from whole cell lysates. In addition, UNC0965 is a useful tool for exploring the localization of G9a on chromatin both in vitro, and in vivo.
We previously reported the X-ray cocrystal structure of the G9a-UNC0638-SAH (S-adenosyl-L-homocysteine) complex.[13] In this structure, UNC0638 adopted a conformation that buried the 7-(3-pyrrolidin-1-yl-)propoxy side chain in the lysine binding channel, and the isopropyl group of the 4-(N-isopropylpiperidin-4-ylamino) moiety was solvent exposed. We reiterated our previous efforts with EZH2[14] by appending a biotin tag to the solvent exposed region of UNC0638, which yielded UNC0965, a functionalized chemical tool for biological applications (Figure 1a). We hypothesized that a relatively long linker (~20 atoms from the piperidinyl nitrogen to the biotin carbonyl) would ensure that the biotin was at a sufficient distance from the surface of G9a to permit access to streptavidin-conjugated magnetic beads. This hypothesis was supported by docking studies using the G9a X-ray structure (Figure 1b). As expected, UNC0965 docked in a nearly identical orientation to UNC0638; with the 7-(3-pyrrolidin-1-yl)propoxy side chain embedded in the lysine binding channel, and the biotin solvent exposed, well away from the surface of the protein.
Figure 1.
Design of UNC0965. a) G9a-UNC0638-SAH co-crystal structure (PDB: 3RJW) was previously solved and demonstrated that the N-isopropyl group of UNC0638 is solvent exposed. We took advantage of this chemical handle and designed a biotinylated derivative, UNC0965. b) Docking of UNC0965 into the G9a crystal structure predicts a similar binding mode as UNC0638. Importantly, the biotin moiety of UNC0965 is solvent exposed and suitable for streptavidin capture.
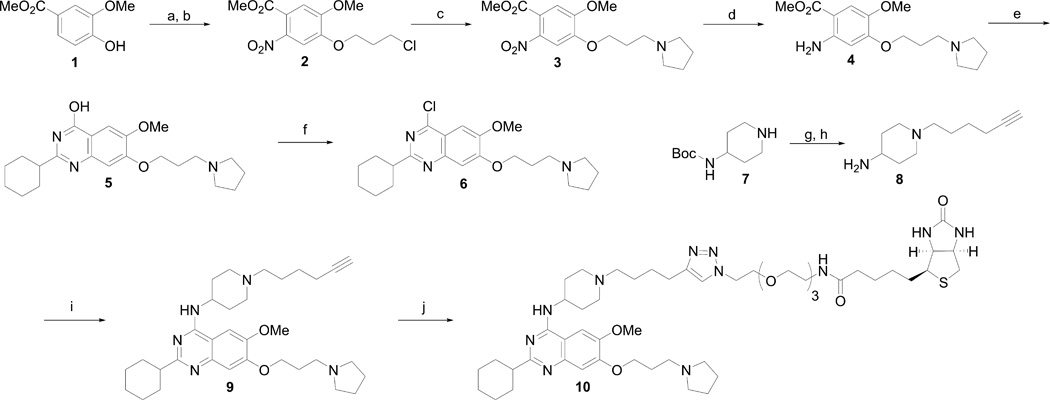
The synthesis of UNC0965 is outlined in Scheme 1. Compound 6 was prepared from the commercially available starting material (compound 1) according to the synthetic route developed previously.[13, 15–17] Compound 8 was synthesized from commercially available tert-butyl piperidin-4-ylcarbamate (7) in two straightforward steps. Nucleophilic aromatic substitution of compound 6 with compound 8 gave compound 9, which is a UNC0638 derivative containing a terminal alkyne. We used this terminal alkyne to perform a copper catalyzed click reaction with commercially available biotin-PEG3-azide to give compound 10, UNC0965.
Scheme 1.
Synthesis of UNC0965. Reagents and Conditions: (a) 1-Chloro-3-iodopropane, K2CO3, CH3CN, reflux; (b) HNO3, Ac2O, 0 °C to RT, 75% over two steps; (c) pyrrolidine, K2CO3, NaI, cat. tetrabutylammonium iodide, CH3CN, reflux, 81%; (d) Fe dust, NH4OAc, EtOAc-H2O, reflux, 63%; (e) 4 N HCl, dioxane, cyclohexanecarbonitrile; (f) N,N-diethylaniline, POCl3, reflux, 47% over two steps; (g) hex-5-yn-1-yl 4-methylbenzenesulfonate, K2CO3, DMF, 60 °C, 88%; (h) TFA, DCM, rt, 99%; (i) 1-(hex-5-yn-1-yl)piperidin-4-amine·2TFA (8), K2CO3, DMF, 60 °C, 81%; (j) biotin-PEG3-azide, cat. Cu(CH3CN)4PF6, DCM, rt, 92%.
We next confirmed that UNC0965 binds G9a and inhibits its catalytic activity. We used a scintillation proximity assay as previously reported[18] to determine the IC50 of UNC0965 (Figure 2a). This assay is a direct measurement of the G9a catalyzed transfer of a tritiated methyl group from 3H-SAM (S-adenosyl-L-methionine) to the histone peptide substrate. UNC0965 showed potent inhibition, with an IC50 value of < 2.5 nM. This strong inhibition of the G9a catalytic activity implied that UNC0965 would bind the protein tightly enough to efficiently chemiprecipitate G9a from cell lysates.
Figure 2.
In vitro characterization of UNC0965. a) UNC0965 shows potent inhibition (IC50 < 2.5 nM) of G9a catalytic activity in an in vitro scintillation proximity assay. Experiments were performed in duplicate. b) Streptavidin-conjugated magnetic beads pre-coated with UNC0965 selectively chemiprecipitate G9a from HEK 293T cell lysates. G9a western blot indicates that UNC0965-treated beads (lane 3) chemiprecipitate G9a compared to DMSO-treated beads (lane 2). This interaction can be blocked or retained, by pretreating lysate with UNC0638 (lane 4) or its negative control UNC1152 (lane 5), respectively. The large (G9aL) and small (G9aS) isoforms of G9a are indicated. The asterisk (*) denotes a background band. These data are representative of three biological replicates.
UNC0965 was coupled to streptavidin-conjugated magnetic beads to chemiprecipitate G9a from HEK 293T whole cell extracts (Figure 2b). Beads pre-incubated in the presence of DMSO (1%) did not pull down G9a (Figure 2b, lane 2). UNC0965-coated streptavidin-conjugated beads effectively pulled down G9a, while pre-incubation of the lysate with UNC0638 blocked this interaction (Figure 2b, lanes 3 and 4). Additionally, UNC1152, a negative control of UNC0638 that does not significantly inhibit G9a catalytic activity (compound 26a in[18]), was unable to block the ability of UNC0965-coated beads to chemiprecipitate G9a (Figure 2b, lanes 3 and 5). Therefore, UNC0965 chemiprecipitates G9a from cell lysates, which further confirms that the biotin moiety of UNC0965 does not interfere with its binding to G9a. Future applications for this chemiprecipitation technique could include proteomic analysis of precipitated proteins to identify G9a-binding partners.
To further characterize this chemical tool, we investigated whether UNC0965 could capture G9a from formaldehyde crosslinked chromatin. Chromatin is a highly dynamic structure, and an effective barrier to gene transcription. As such, it is frequently remodeled to create or prevent access of transcription factors to underlying DNA sequences. Traditional antibody-based chromatin immunoprecipitation (ChIP) was developed as a tool to provide a snapshot of protein-DNA interactions in the dynamic chromatin environment (reviewed in[19–21]). In a standard ChIP assay, a crosslinking reagent such as formaldehyde is used to covalently attach proteins to DNA, the cells are lysed, and the chromatin is fragmented by sonication or enzymatic digestion. An antibody is then used to immunoprecipitate a protein of interest from the chromatin extract and the associated DNA sequences are analyzed by methods such as quantitative PCR (qPCR) or next-generation sequencing (ChIP-seq). We hypothesized that UNC0965 could substitute for an antibody to precipitate G9a protein in a method referred to as chemical inhibitor chromatin precipitation (chem-ChIP).[22]
Use of a functionalized chemical tool for chromatin precipitation has distinct advantages over an antibody. First, such chemical tools can be easily synthesized at low cost, and eliminate the batch-to-batch variability common with antibody reagents. Second, the biotin tag can be designed to be solvent exposed for chemical tools that have a well-defined mode of binding. This design has a potential added advantage over traditional ChIP, where crosslinking can prevent access of an antibody to the protein of interest. Third, chemical tools can be cell permeable, which allows cell treatment prior to crosslinking. This in vivo chem-ChIP method would be particularly well suited for proteins that are difficult to precipitate after crosslinking due to antibody epitope masking. Therefore, we tested the ability of UNC0965 to precipitate G9a from chromatin extracted from crosslinked cells (in vitro chem-ChIP), and chromatin generated from cells treated with the compound prior to crosslinking (in vivo chem-ChIP) and compared these results to the normal antibody method.
Using traditional antibody ChIP, we identified four chromosomal regions in HEK 293T cells (Supplemental Table S1) that showed a significant fold change in G9a occupancy over background when compared to a negative control region near the GAPDH gene (Figure 3a). We performed in vitro chem-ChIP using the same protocol used for antibody ChIP. In place of the G9a antibody and IgG control, we used streptavidin-conjugated magnetic beads pre-incubated with UNC0965 and DMSO, respectively. We then examined the identified G9a occupied chromosomal regions by qPCR (Figure 3b). The UNC0965 in vitro chem-ChIP showed a greater than 5-fold increase over DMSO at each chromosomal region. The difference between the G9a antibody ChIP signal and the IgG signal was less significant (p < 0.5 to p < 0.01) (Figure 3a), than the signal for UNC0965 in vitro chem-ChIP compared to DMSO (p < 0.01 to p < 0.001). Thus, UNC0965 effectively captures G9a in an in vitro ChIP assay, and in this case with a better G9a signal over background than the antibody ChIP.
Figure 3.
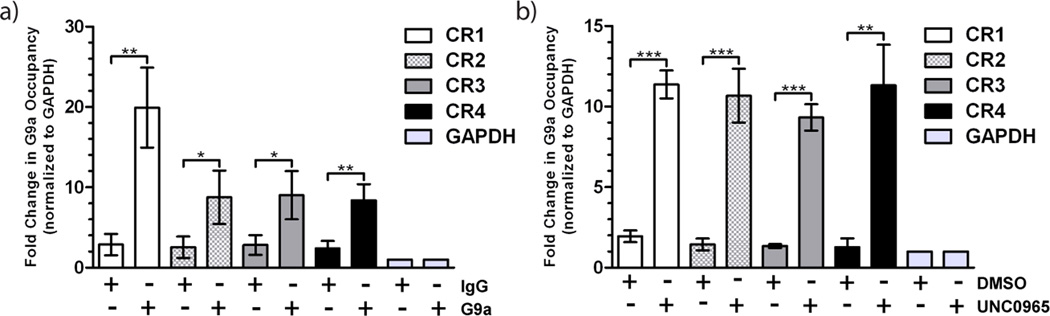
G9a occupancy on chromatin can be determined using antibody ChIP or in vitro chem-ChIP with UNC0965. a) ChIP using G9a antibody or IgG control followed by qPCR. b) In vitro chem-ChIP with UNC0965-coated streptavidin-conjugated magnetic beads followed by qPCR. PCR primer sets correspond to chromosomal regions (CR) occupied by G9a as well as a GAPDH control region that shows no G9a occupancy. Chromosomal regions assayed include: CR1 (white), CR2 (checkered), CR3 (grey), and CR4 (black), and GAPDH (light grey). Error bars represent the standard deviation of three biological replicates. Statistical significance between DMSO and UNC0965 chem-ChIP for each region was determined by an unpaired t-test and indicated by asterisks (* = p < .05, ** = p < .01, *** = p < .001).
As noted earlier, unlike antibodies, cell permeable small-molecule tools can be used to pre-bind the target in cells prior to crosslinking. We assessed cellular activity of UNC0965 using an in-cell western assay reported previously.[13] Treatment of cells for 48 hours with increasing concentrations of UNC0965 resulted in a decrease in H3K9me2 (IC50 = 3.3 ± 1.2 µM), with low cell toxicity (EC50 > 50 µM) (Figure 4a). While UNC0965 has lower cell permeability compared to UNC0638,[13] it shows sufficient inhibition of H3K9me2 to be useful for cell-based assays.
Figure 4.
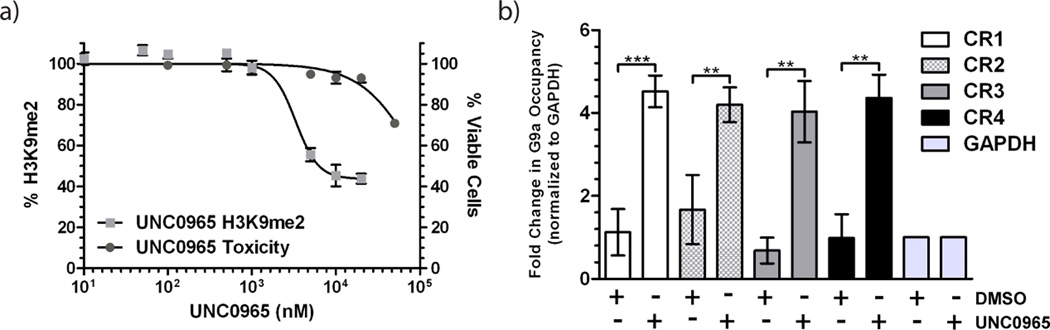
UNC0965 is cell penetrant and can be used for in vivo chem-ChIP assays. a) In-cell western assay of MDA-MB-231 cells treated with UNC0965 and assayed for total cellular H3K9me2 levels. UNC0965 reduces cellular levels of H3K9me2 (light grey squares; IC50 = 3.3 ± 1.2 µM) and is not toxic in the compound concentration range tested (grey circles; EC50 > 50 µM). Error bars represent the standard deviation of 3 biological replicates. These data confirm that UNC0965 is cell penetrant. b) In vivo chem-ChIP. HEK 293T cells were treated with UNC0965 (5 µM) or DMSO (0.05%) for 15 minutes, crosslinked with formaldehyde, and then chromatin was isolated. ChIP was performed using streptavidin-conjugated magnetic beads followed by qPCR. PCR primer sets correspond to chromosomal regions (CR) occupied by G9a as well as a GAPDH control region that shows no G9a occupancy. Chromosomal regions assayed include: CR1 (white), CR2 (checkered), CR3 (grey), and CR4 (black), and GAPDH (light grey). Error bars represent the standard deviation of three biological replicates. Statistical significance between DMSO and UNC0965 chem-ChIP for each region was determined by an unpaired t-test and indicated by asterisks (** = p < .01, *** = p < .001).
For the in vivo chem-ChIP assay, we wanted to pulse the cells with the compound, rather than treat for several days, in order to minimize any effects of G9a inhibition on its chromatin localization. Therefore, we grew HEK 293T cells to 80% confluency, and then exposed the cells to 5 µM UNC0965 or 0.05% (volume equivalent) DMSO for 15 minutes prior to formaldehyde crosslinking. We then performed the standard antibody ChIP protocol (see Supplemental Methods) except the precipitation step was performed using streptavidin-conjugated magnetic beads. Cells treated with UNC0965 showed a significant average change in G9a occupancy over DMSO-treated cells (p < 0.01) (Figure 4b), which was very comparable to the in vitro chem-ChIP results (Figure 3b). These data demonstrate that even with compound pre-treatment of the cells, the biotin moiety on UNC0965 remains solvent exposed after formaldehyde crosslinking, confirming that UNC0965 is a useful chemical tool for in vivo chem-ChIP studies and could be used in additional cellular applications.
Our success with UNC0965 and G9a highlights methods to convert highly selective chemical probes into extremely valuable tools that may aide in understanding biological questions. The in vivo chem-ChIP technique could be an alternative method for targets that have no suitable ChIP-grade antibody due to masking of the appropriate epitope after crosslinking. In fact, use of a labeled chemical tool could prevent the need for overexpression of a tagged protein. We noted that the signal to noise ratio was lower for the antibody ChIP compared to both the in vitro and in vivo chem-ChIP assays. Thus, it would be interesting to compare genome-wide occupancy of G9a between all three methods to determine how the overall signal for UNC0965 chem-ChIP compares to that of traditional ChIP.
Experimental Section
In vitro chem-ChIP
Chromatin was prepared from HEK 293T cells as described for the antibody ChIP. Each ChIP contained chromatin from approximately 8x106 cells. Streptavidin-conjugated magnetic beads (Dynabeads® M-280 streptavidin, Life Technologies #11205D, 250 µg per chemiprecipitation) were washed 3 times with 500 µL TBST (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Tween-20), diluted in 100 µL TBST, and 4 µL of 10 mM UNC0965 was added, followed by rotation on an end-over-end rotator for 1 hour at room temperature to saturate the beads. After binding, the beads were washed with TBST (500 µL×3) to remove excess unbound compound. Chromatin was then added to the beads and the mixture was placed on a rotator at 4°C overnight. The next morning, tubes were removed from the rotator, and supernatant was aspirated from the beads. The beads were washed (500 µL×3) with ChIP wash buffer (0.1% SDS, 1% TritonX-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris, pH=8.0) followed by one wash with 500 µL of ChIP final wash buffer (1% SDS, 1% TritonX-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl, pH 8.0). During each wash, the sample was rotated at room temperature for 3 minutes. The washed beads were resuspended in 200 µL TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0) with 20 µg proteinase K, and the contents were transferred into a PCR tube. The mixture was incubated at 55°C, for 30 minutes followed by 2 hours at 80°C to reverse the crosslinks. The DNA in the collected supernatant was then purified as described in Active Motif ChIP-IT® High Sensitivity kit #53040.
In vivo chem-ChIP
HEK 293T cells were grown to 80% confluency on 15 cm plates as described. Cells were treated with 5 µM UNC0965 for 15 minutes prior to formaldehyde crosslinking. Chromatin was prepared as described for the antibody ChIP. Each chem-ChIP represented approximately 8x106 cells. Streptavidin-conjugated magnetic beads (Dynabeads® M-280 streptavidin, Life Technologies #11205D, 500 µg per chemiprecipitation) were washed 3 times with 500 µL TBST (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Tween-20). Chromatin was then added to the beads and the mixture was placed on a rotator at 4°C overnight. Wash and DNA purification steps were performed as described in the method for in vitro chem-ChIP, above.
Supplementary Material
Acknowledgements
The authors thank Dr. Stephen V. Frye for critical reading of the manuscript and Dr. Yan Xiong for proofreading the experimental section. The research described here was supported by the grant R01GM103893 from the National Institute of General Medical Sciences of the National Institutes of Health, the University Cancer Research Fund and Carolina Partnership from University of North Carolina at Chapel Hill, the V Foundation for Cancer Research, and the Structural Genomics Consortium, a registered charity (number 1097737) that receives funds from the Canada Foundation for Innovation, Eli Lilly Canada, GlaxoSmithKline, the Ontario Ministry of Economic Development and Innovation, the Novartis Research Foundation, Pfizer, AbbVie, Takeda, Janssen, Boehringer Ingelheim and the Wellcome Trust.
Footnotes
Supplemental Methods
Methods for chemical synthesis, in vitro scintillation proximity assay, chemiprecipitation of G9a, western blotting, antibody ChIP, quantitative PCR, in-cell western, and cell toxicity assays are described in the supplemental information.
References
- 1.Jenuwein T, Laible G, Dorn R, Reuter G. Cell. Mol. Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 3.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 4.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. J Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 5.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinkai Y, Tachibana M. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL. J Biol. Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen MW, Hua KT, Kao HJ, Chi CC, Wei LH, Johansson G, Shiah SG, Chen PS, Jeng YM, Cheng TY, Lai TC, Chang JS, Jan YH, Chien MH, Yang CJ, Huang MS, Hsiao M, Kuo ML. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 10.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Nat. Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 12.Frye SV. Nat. Chem. Biol. 2010;6:159–161. doi: 10.1038/nchembio.296. [DOI] [PubMed] [Google Scholar]
- 13.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, Dong A, Tempel W, Wang SC, Chen X, Chau I, Mangano TJ, Huang XP, Simpson CD, Pattenden SG, Norris JL, Kireev DB, Tripathy A, Edwards A, Roth BL, Janzen WP, Garcia BA, Petronis A, Ellis J, Brown PJ, Frye SV, Arrowsmith CH, Jin J. Nat. Chem. Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, Liu F, Gao C, Huang XP, Kuznetsova E, Rougie M, Jiang A, Pattenden SG, Norris JL, James LI, Roth BL, Brown PJ, Frye SV, Arrowsmith CH, Hahn KM, Wang GG, Vedadi M, Jin J. ACS Chem. Biol. 2013;8:1324–1344. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu F, Barsyte-Lovejoy D, Allali-Hassani A, He Y, Herold JM, Chen X, Yates CM, Frye SV, Brown PJ, Huang J, Vedadi M, Arrowsmith CH, Jin J. J Med. Chem. 2011;54:6139–6150. doi: 10.1021/jm200903z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, Kozieradzki I, Senisterra G, Chau I, Siarheyeva A, Kireev DB, Jadhav A, Herold JM, Frye SV, Arrowsmith CH, Brown PJ, Simeonov A, Vedadi M, Jin J. J Med. Chem. 2009;52:7950–7953. doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, Dong A, Senisterra G, Chau I, Siarheyeva A, Norris JL, Kireev DB, Jadhav A, Herold JM, Janzen WP, Arrowsmith CH, Frye SV, Brown PJ, Simeonov A, Vedadi M, Jin J. J Med. Chem. 2010;53:5844–5857. doi: 10.1021/jm100478y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, Barsyte-Lovejoy D, Li F, Xiong Y, Korboukh VK, Huang X, Allali-Hassani A, Janzen WP, Roth BL, Frye SV, Arrowsmith CH, Brown PJ, Vedadi M, Jin J. J Med. Chem. 2013 doi: 10.1021/jm401480r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo MH, Allis CD. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 20.Collas P. Mol. Biotechnol. 2010;45:87–100. doi: 10.1007/s12033-009-9239-8. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Ubreva J, Ballestar E. Methods Mol Biol. 2014;1094:309–318. doi: 10.1007/978-1-62703-706-8_24. [DOI] [PubMed] [Google Scholar]
- 22.Bradner JE, Young AR. Nature Biotechnology, in press [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.