Abstract
In 2002–2003, the American College of Medical Informatics (ACMI) undertook a study of the future of informatics training. This project capitalized on the rapidly expanding interest in the role of computation in basic biological research, well characterized in the National Institutes of Health (NIH) Biomedical Information Science and Technology Initiative (BISTI) report. The defining activity of the project was the three-day 2002 Annual Symposium of the College. A committee, comprised of the authors of this report, subsequently carried out activities, including interviews with a broader informatics and biological sciences constituency, collation and categorization of observations, and generation of recommendations. The committee viewed biomedical informatics as an interdisciplinary field, combining basic informational and computational sciences with application domains, including health care, biological research, and education. Consequently, effective training in informatics, viewed from a national perspective, should encompass four key elements: (1) curricula that integrate experiences in the computational sciences and application domains rather than just concatenating them; (2) diversity among trainees, with individualized, interdisciplinary cross-training allowing each trainee to develop key competencies that he or she does not initially possess; (3) direct immersion in research and development activities; and (4) exposure across the wide range of basic informational and computational sciences. Informatics training programs that implement these features, irrespective of their funding sources, will meet and exceed the challenges raised by the BISTI report, and optimally prepare their trainees for careers in a field that continues to evolve.
The 1999 report of the Biomedical Information Science and Technology Initiative (BISTI), issued by the National Institutes of Health (NIH), signaled a new era in biomedical informatics.1 The report stated categorically that: “… the impact of computer technology is so extensive that it is no longer possible to think about [the biomedical] mission [of NIH] without computers.” The BISTI report documented the critical current and future role of computation in genomics and proteomics, and the full sweep of modern biological science. It also envisioned a program of National Centers of Excellence in Biomedical Computing that embraced both research and training.
The BISTI report and related rising expectations for application of information technology to basic biological research have generated fundamental questions for the informatics community. Will bioinformatics become an area of science largely separate from traditional clinical informatics, independently growing (and perhaps reinventing) its own body of knowledge? Or will the two fields gradually converge through actions that consolidate common interests and scientific challenges—jointly approaching a research agenda that connects the genotype and phenotype? A critical challenge related to these questions is how to educate and train future professionals for careers in these evolving fields. Decisions made now about the support and structure of informatics training programs and their curricula will shape the next generation of scientists and determine the future of the field(s). Should the NIH, other government agencies, and private foundations develop plans to support bioinformatics training outside the current framework of informatics training (ongoing since the early 1980s)? Alternatively, should funding agencies seek to promote more integrated approaches to education, with each program seen as a variation on a common theme? These questions cannot be answered until common themes, if any, have been articulated.
The American College of Medical Informatics (ACMI), stimulated by the above questions, obtained support from the National Library of Medicine (NLM) to study and report on future biomedical informatics training. While ACMI previously considered issues related to the future of biomedical informatics as a field, 2 the current study focused more on the challenges of training for future careers in bioinformatics. ACMI created a task force, comprised of the coauthors of this report, to coordinate and carry out the study. The Task Force included eight ACMI Fellows, one postdoctoral fellow in a biomedical informatics training program, and two staff members. The defining activity of the study was the 2002 ACMI Symposium, held February 14–17 in Palm Springs. Among the 36 ACMI Fellows attending the 2002 symposium were, for the first time, individuals elected to the College who consider themselves to be primarily bioinformaticians (as opposed to clinical informaticians, clinicians, computer scientists, biomedical librarians, etc.). In addition, the College invited a representative from industry with experience in bioinformatics to join the symposium as a guest. Study activities following the symposium included several discussions among members of the Task Force, a series of interviews with ten prominent individuals in bioinformatics who did not attend the retreat, and a plenary session panel presented at the November 2002 meeting of the American Medical Informatics Association.
The central study result characterizes effective training in biomedical informatics along several dimensions. Programs with the defined features will prepare their trainees well, irrespective of the source of funding that supports a program or its trainees. The current ACMI report hopes to influence current and future directors of biomedical informatics training and agencies that support such training to create, endorse, implement, and evolve toward programs with the defined characteristics.
Study Methods and Activities
The 2002 ACMI Symposium addressed the following specific questions derived from the overall goal of the study:
What skill sets are needed in the next generation of biomedical informaticians, using “biomedical” in the broad sense of the word?
Training programs can be organized in many ways. Given the skill sets identified from question 1, what are the strengths and drawbacks of various approaches to training that address these skill sets?
What kinds of training can and should take place in the NIH BISTI centers that cannot take place in the current NLM-funded programs?
Several plenary presentations and panels were organized to frame the issues and provide a forum for discussion of the future of informatics training. On the initial day of the symposium, a keynote address was provided by Russ Altman, MD, PhD, with the theme of “The Biological Data Explosion: Creating New Pressures on Informatics Training.” Drs. Ted Shortliffe and Isaac Kohane offered comments as discussants. Subsequent small-group deliberations addressed the issues raised in the session, with focus on question 1 relating to essential skill sets for informatics training. The conclusions of each group were transferred to posters that were on display for all participants the following morning.
After the poster session to open the second day of the meeting, Perry Miller, MD, PhD gave his keynote presentation, “From Genomics to Clinical Informatics,” with discussant presentations by Drs. Mark Boguski and Gregory Cooper. A wide-ranging discussion focused on the importance of integration between clinical informatics and bioinformatics. Discussion within the same four small groups as the first day addressed the full set of questions guiding the symposium. Again, each group prepared posters for display on the next and final day of the meeting. The second day ended with a panel discussion on “Tribalism and Culture,” with Drs. Charles Friedman, Bo Saxberg, Larry Hunter, and William Hersh as panelists.
The third day focused on the theme of “Training for a Practice Discipline” and, as such, addressed a series of more tactical issues relating to the design of training in biomedical informatics.
Following the symposium, five members of the Task Force interviewed ten prominent individuals in the field of bioinformatics. While several ACMI bioinformaticians participated in the 2002 ACMI Symposium, the Task Force wanted to obtain a more diverse vision of bioinformatics research and training, beyond ACMI per se. The interviewees selected to provide this additional perspective included four directors of bioinformatics-related programs at the NIH and six university-based individuals who are currently directing funded BISTI centers or are otherwise prominent in bioinformatics research and training. The interviews were “semistructured” in that all interviewees were asked the same basic set of open-ended questions, but the conversations were otherwise unconstrained. Interviewees were asked to offer their personal definitions of bioinformatics and computational biology, their views of evolving professional roles required to sustain this new area of science in which biology meets computing, and their views about different programs that support training in these fields.
The Task Force considered the observations and recommendations from the ACMI Symposium and the interviews during several discussions, leading to generation of Task Force findings and recommendations regarding the future of training in biomedical informatics.
The Landscape of Informatics
The Task Force adopted a practical view of informatics that shaped its recommended approach to training. The practice of informatics, most generally, requires the presence of two components: (1) a set of skills and methodologic tools derived from knowledge of the basic informational and computing sciences; and (2) knowledge, experience, and activity in one or more application domains. The coexistence of, and interactions between, these key components gives meaning and significance to informatics as a field. The basic sciences relevant to informatics include, but are not restricted to, computer science, information and telecommunication science, cognitive science, statistics, decision science, and management/organizational science. Application domains for informatics can literally be any area of human endeavor supportable by information technology, but for biomedical informatics are typically constrained to biomedicine. In this sense, biomedical informatics is the union of the basic informational and computing sciences listed above, with biomedicine as an application domain. Biomedicine is a broad application domain spanning all health professional practice (including public health and bioimaging); basic biological research; clinical research; education of future and current health professionals; and the administration of practice, research, and education. It follows from these definitions that biomedical informatics is the umbrella discipline that embraces a range of subdisciplines defined by specific application areas. Within the framework of this report, then, the term bioinformatics refers to the union of the basic informational and computing sciences with biological research as a specific application domain. The authors acknowledge that different constituencies use the term bioinformatics in ways different from the one proposed.
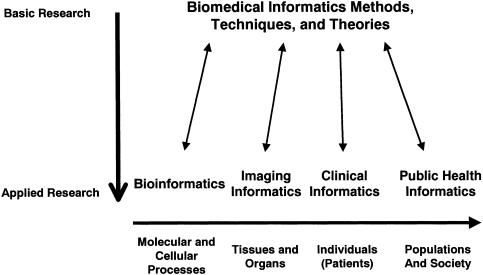
▶ illustrates how four (of many) informatics subdisciplines arise when the methods, techniques, and theories drawn from the basic informational and computational sciences interact with specific application areas. Each of the illustrated application areas is directed at problems existing at a particular level of scale, such as the molecular/cellular level in the case of bioinformatics. From the perspective of informatics, research activities are more “basic,” to the extent that they are directed at methods, techniques, and theories that transcend the application areas. Research activities are more applied, to the extent that they address issues specific to an application area.
Figure 1.
The relationship between biomedical informatics as a core scientific discipline and its diverse array of application domains that span biological science, imaging, clinical practice, public health, and others not illustrated. From: Shortliffe EH. JBI status report [editorial]. J Biomed Inform. 2002;35:279–80. Reprinted with the author's permission.
It also follows from this definition that the informational/computing sciences and the application domain have coequal and essential status in informatics. Indeed, it is the assembly of individuals, each having expertise both in these basic sciences and in one or more relevant application domains, that gives centers and departments of informatics their fundamental and unique character. Informatics centers are multidisciplinary along two axes: (1) the multiplicity of basic fields (each an academic discipline) that serve as components of its scientific foundation, and (2) the multiplicity of domains to which information processing resources and other “tools” developed in informatics centers can be applied.
According to this view, informatics should not be seen as a specialization of the application domains of clinical medicine, molecular biology, or educational psychology. Clinicians, biologists, and educators who “use computers as a component of their professional work” are not practicing informatics unless they have mastered one or more of the foundational information/computing sciences and are directly applying their knowledge of these sciences to their domain of choice.
The authors of this report strongly endorse the use of biomedical informatics as an umbrella term that names the core discipline while also denoting the union of the information and computer sciences with domains including clinical practice, biomedical research, imaging, public health, and health professions education. We believe that use of “biomedical informatics” as the overarching term and “bioinformatics” in a more focused sense will lead over time to the clearest portrayal of the field and, as such, will promote effective communication among the varied constituencies that work in the field. Our specific recommendations for training in biomedical informatics, that follow, will build on this view of the field.
Elements of Effective Training
The study asserts, as a central finding, that the core purpose of biomedical informatics training programs should be to prepare individuals capable of making major contributions to the creation and evaluation of computational tools with application to biomedical or clinical research, health care practice, and education. The most sophisticated and important tools will be those that integrate across diverse domains of application and build the future capacity to handle the increasing explosion of biological/clinical data. It should be emphasized that the creation of effective tools that implement innovative methods is itself a scientific activity. This is because of the perpetual need for new and better conceptual and mathematical models to empower these tools, the countless open questions regarding the optimal design and methods to develop and deploy these tools, and the ongoing imperative to understand through empirical studies the effectiveness of tools that have been deployed and how to improve them. Major contributions can be made to any of these three activities: modeling, development and deployment, and empirical studies. Few trainees in biomedical informatics will emerge with the full repertoire of skills necessary to make contributions to all three activities. Nonetheless, the capacity to train individuals who can make creative contributions to tool building is a defining aspect of a biomedical informatics training program.
The Task Force proposes four essential elements for training in biomedical informatics, reflecting the views offered above. Excellent training in computer science (absent specific attachment to a domain) or in application domains such as molecular biology (absent attachment to the foundational information/computing sciences) can occur without these elements. However, our model of excellent training in biomedical informatics requires them.
Integrated Curriculum
The strongest training programs are those that integrate the basic informational/computing sciences with appropriate application domains. Integration is simultaneously the biggest challenge to create effective training environments in biomedical informatics and the greatest potential source of benefit. Integrated curricula offer courses and other educational experiences that explicitly relate the basic informational/computing sciences to problems in the relevant domain(s). In this light, an index of the quality of a training curriculum is the fraction of the total training experience that explicitly combines basic science (computational) and domain (biological or clinical) issues. Information/computer science courses that are domain-independent (such as introductory graduate-level courses offered in a computer science department) and domain-oriented courses that do not invoke issues of information or computing (such as introductory graduate-level courses in molecular biology) may be important components of a training curriculum, but they do not contribute directly to integration. A highly integrated curriculum will also bring educational benefits that transcend the specific course content that is mastered by the students. For example, integrated curricula provide experiences that help trainees learn to collaborate in a multidisciplinary setting, to integrate data of varied types and from varied sources, and to weave together varying modes of idea expression and varying modes of thinking.
Cross-training
The roles of professionals in biomedical informatics should ultimately relate to issues of human health and disease. This should happen either directly, through contributions to biological research or health care, or indirectly, for example, through improvement of health professional education. As such, effective training programs must produce individuals with knowledge and skills in the basic information/computer sciences and in at least one biomedical/health domain area. Because individuals often enter training programs with significantly stronger backgrounds in one side of this equation, maximally effective training programs must have the capability of individualizing the training experience to provide the elements that each trainee does not possess. Programs that admit trainees who, as a group, have backgrounds exclusively in informational/computing science or relevant application domains create less attractive training environments than programs that provide training on both sides to individuals coming from both sides. Trainees with strong information/computer science backgrounds offer perspectives that are highly complementary to the perspectives of trainees with strong domain backgrounds (clinicians, biologists, educators). The interplay between these different types of trainees realizes, within the architecture of the program itself, the interaction between the computational sciences and application domains that is the essence of informatics.
We also take the position that training programs emphasizing multiple application domains (such as clinical medicine and basic biomedical science) offer superior environments to those that emphasize one application domain exclusively. Colocated applied research across the spectrum, illustrated in ▶, allows important cross-fertilization between trainees and faculty whose work emphasizes different domains, inevitably comingling the professional cultures that have grown up around these domains. For example, interactions between trainees and faculty who are oriented to clinical applications and those who are oriented to biological applications will engender creative experiences around important cross-domain issues such as genotypic–phenotypic studies (e.g., pharmacogenomics) while also promoting mutual respect and understanding among individuals who work in these varied domains.
Immersion in the Research and Development Work of Informatics
Effective training programs are situated in environments with ongoing faculty-directed research and development activities in which trainees are directly involved. Project work is a required, not optional, part of the training experience. Curricula that consist entirely of courses are not sufficient for optimal training. Whereas trainees may undertake independent projects in their own areas of specific interest during the later stages of training, project work at the early stages of training best occurs in faculty laboratories as part of the ongoing work of those laboratories. Training programs with more of an applied than a basic research focus would situate these trainee projects in information system deployment settings, but these projects would still be undertaken with some significant level of faculty supervision and would avoid placing trainees on the critical path for development of specific information services within an institution. Effective training programs combine didactic experiences such as coursework with direct immersion in research and development. There is no a priori ideal balance of didactics and project work, but extremes in either direction are not desirable.
Incorporation of Core Skills from the Fields Related to the Information and Computational Sciences
Competence and capacity for the development, implementation, and evaluation of informatics tools require a curriculum addressing a broad set of computational topics. At a general level, these topics include representation, modeling, data analysis, systems (biological, computational, organizational), and decision making. Comprehensive training in biomedical informatics—whether applied to genomics, clinical care, or any other application domain—should therefore include sufficient exposure for some level of competence in most, if not all, of the areas listed below. The challenge and the craft of designing informatics curricula are seen in the need to balance emphasis of these fields with that required for mastery of one or more application domains.
Algorithms
Basic mathematics, including calculus
Cognitive/human factors and interfaces
Data structures
Database design
Evaluation/research methods
Information retrieval
Knowledge representation
Modeling
Networking/architecture
Ontology/vocabulary
Probability/statistics
Programming languages
Simulation
Software engineering
BISTI and the Quality of Informatics Training
With publication of the BISTI report in 1999 and the subsequent proliferation of informatics training programs sponsored by various federal agencies and private foundations, the Task Force emphasizes that the qualities of strong biomedical informatics training programs are independent of the sources that fund these programs. Many training programs fund students' support using multiple resources, including research awards, training grants, students' personal resources (self-pay tuition), and employees' fringe benefits that support tuition for part-time training experiences. Emphasis in evaluating training programs should fall on the quality of the experiences the programs provide, not the source of funding. The ACMI Task Force believes that the features of good informatics training provided in this report will assist in the assessment of training program quality and may also provide a guide for improvement. We believe that these features apply in equal measure to all formal training in biomedical informatics, including programs funded under NLM's existing T15 program, those supported under the BISTI initiative (whether directly funded by NLM or another NIH agency), and those supported by other sources.
The Task Force's vision for education in biomedical informatics is consistent with the vision of “cross-disciplinary” education expressed in the BISTI report, but we take a stronger position. The BISTI report advocated for curricula that combine formal training in computer science with formal training in biology. A curriculum that consists of courses in computer science (absent specific discussion of biology as an application domain) and courses in biology (absent specific discussion of computational aspects) would meet the BISTI criterion for effective training. This curriculum would not, however, meet our criterion, because it would lack the key element of integration described above. The training model advanced in the BISTI report is one of “combined training in biology and computer science.” It lacks the elements that explicitly connect the two fields. As such, it is not training in informatics.
The 1999 BISTI report asserted that there were few programs providing such combined training. In making this assertion, we believe that the BISTI panel failed to recognize that many of the existing NLM-supported T15 programs were doing this. Perhaps because these T15 programs were not exclusively devoted to biological science as an application domain, they were unknown to, or discounted by, the BISTI panel. Perhaps the integrated nature of the training in many of these programs concealed them. Regardless of why they went unnoticed, many T15s are doing what the BISTI report proposed, and more. Also, as stated earlier, we believe that the capability of supporting training in several application domains (for example, bioinformatics and clinical informatics), by intermingling faculty and trainees with these diverse interests around a common theme, is a factor that strengthens rather than weakens these programs.
In this report, the ACMI Task Force takes no formal position on optimal length of training or the relative value of different kinds of academic degrees that training programs might award. Training programs come in many shapes and sizes; trainees have a wide range of needs. The full range of options currently available—from nondegree certificate programs to PhD-granting programs—is almost certainly necessary to meet these diverse needs. At the same time, we would emphasize that serious training in biomedical informatics, irrespective of application domain, typically requires focused and extended study. It is not possible to become an informatician, in any sense of the word, through attendance of a lecture series or participation in a series of workshops.
Many of the T15 training programs currently supported by the NLM have a long history of successful training. They have invented means to implement the desiderata for excellent training in biomedical informatics, as described above—and they continue to evolve and refine these means. Many of the NIH/NLM-supported T15 programs are, for these and other reasons, very strong and should continue. At the same time, the Task Force can express concern about the T15 programs that mirrors the concern expressed about the training model advocated in the BISTI report. The T15 programs can weaken themselves by becoming or remaining dominantly focused on clinical informatics in the face of an emergent and diversifying set of application areas. Programs can also weaken themselves by producing graduates whose training experiences are narrowly defined by or “overfitted” to specific scientific problems and who, upon completion of training, cannot generalize from their experience to address new problems with novel and unanticipated features.
The ACMI Task Force encourages the directors of the T15 programs, and the directors of all biomedical informatics training activities, to diversify their curricula so as to address the BISTI training challenges. We also encourage individuals who are creating new training programs in biomedical informatics and related fields, whether under the BISTI initiative or in response to other opportunities, to consider these principles when designing their curricula and training environments. Those seeking to respond to the BISTI training challenge may wish to explore existing training programs in their own institutions to determine whether creating a new emphasis within an existing program might be a more efficacious pathway than establishing an entirely new program. All other things remaining equal, it is usually easier to diversify an existing curriculum to include new application domains than it is to create an entirely new one.
Supported in part by the National Library of Medicine, through purchase order 467-MZ-200555.
References
- 1.BISTI. The Biomedical Information Science and Technology Initiative. Bethesda, MD: National Institutes of Health. Available at: http://www.nih.gov/about/director/060399.htm. June 1999. Accessed: Feb 25, 2004.
- 2.Friedman CP, Ozbolt JG, Masys DR. American College of Medical Informatics. Toward a new culture for biomedical informatics: report of the 2001 ACMI symposium. J Am Med Inform Assoc. 2001;8:519–26. [DOI] [PMC free article] [PubMed] [Google Scholar]