Abstract
Wolbachia pipientis is one of the most widely studied endosymbionts today, yet we know little about its short-term adaptation and evolution. Here, using a set of 91 inbred Drosophila melanogaster lines from five populations, we explore patterns of diversity and recent evolution in the Wolbachia strain wMel. Within the D. melanogaster lines, we identify six major mitochondrial clades, including one not yet described in the literature. Using Bayesian analysis informed with demographic estimates of colonization times, we estimate that all extant D. melanogaster mitochondrial haplotypes coalesce to a Wolbachia-infected ancestor approximately 2,200 years ago. Concordant with past studies, the Wolbachia haplotypes contain an overall low level of nucleotide diversity, yet they still display geographic structuring. Finally, we show that fly populations vary in wMel titre. This demonstration of local phenotypic divergence suggests that intra-specific host genetic variation plays a key role in shaping this model symbiotic system.
Keywords: Wolbachia, Drosophila melanogaster, endosymbiosis, mtDNA, population genetics
Introduction
Endosymbiotic relationships are increasingly recognized as key drivers of adaptation and speciation (McFall-Ngai et al. 2013). Genomic comparisons — both among endosymbionts as well as between endosymbionts and their nearest free-living relatives — have brought to light a number of key observations about the evolution of bacterial symbionts in general and intracellular symbionts in particular (Medina & Sachs 2010; Moran et al. 2008; Moya et al. 2008), but we are only beginning to understand the intraspecific variation that affects their short-term evolution (Moran et al. 2009; Richardson et al. 2012).
One of the more widely studied endosymbionts is Wolbachia pipientis, an α-Proteobacterium estimated to infect 40% of terrestrial arthropods (Zug & Hammerstein 2012) as well as some nematodes (Taylor et al. 2005). Wolbachia resides in both somatic and gonadal insect tissue and is transferred from mother to offspring through the egg cytoplasm. Despite this reliance on vertical transmission, however, Wolbachia evolution has been marked by frequent host-jumps that have been accompanied with high levels of genetic recombination (Baldo et al. 2006b; Werren et al. 1995). These large evolutionary transitions are marked by recombination and genomic rearrangements (Baldo et al. 2006a; Klasson et al. 2009), which have perhaps been enabled by key genomic characteristics — in particular, the maintenance of functional DNA repair and recombinational machinery (Wu et al. 2004). Still, it is unknown what genetic factors influence population dynamics within single Wolbachia lineages.
Wolbachia induces a range of phenotypic changes in its hosts. While acting as an obligate mutualist in some filarial nematodes, it is best known as a reproductive parasite in insects, inducing cytoplasmic incompatibility (CI), parthenogenesis, feminization, and male killing. Compared to many other Wolbachia strains, wMel, the strain that infects the fruit fly Drosophila melanogaster, causes more moderate phenotypic effects. These include fitness-enhancing phenotypes such as heightened viral resistance (Teixeira et al. 2008) and increased iron tolerance (Brownlie et al. 2009), as well as low levels of CI (Friberg et al. 2011; Reynolds & Hoffmann 2002).
Developing a deeper understanding of the persistence and ecological importance of wMel infections will rely on a more thorough description of the mutational processes and selection pressures that shape the bacterium's evolution. Regions that are known to be variable among different Wolbachia strains have shown essentially no variation within wMel. Until recently, previous analyses of global genetic diversity have been limited to a few known structural variants (Nunes et al. 2008b; Riegler et al. 2005). Importantly, these studies identified a number of divergent wMel lineages and showed that the frequencies of these haplotypes dramatically changed in the latter half of the 20th century. More recently, Richardson et al. (2012) provided a first look at genome-wide wMel diversity. Their study leveraged data from two different large-scale D. melanogaster sequencing efforts (the Drosophila Population Genomics Project and the Drosophila Genetic Reference Panel) that focused on multiple sparsely-sampled populations within Africa, one sparsely-sampled population within Europe, and one deeply-sampled population within North Carolina. This previous study provided key insights into wMel transmission, depth of coverage, and nucleotide evolution, but the different sequencing and sampling approaches used by the two sequencing efforts makes comparisons between the populations difficult. Furthermore, the African and European sequences were derived from haploid embryos, making phenotypic analyses of adults impossible.
Here we present genomic sequences of 65 wMel strains from five geographically diverse populations of D. melanogaster, providing a picture of global genome-wide nucleotide diversity in this model endosymbiont. Combined with the reconstruction of mtDNA sequences from these same fly lines, this high-resolution dataset allows us to address three aspects of recent wMel evolution that are key to advancing our understanding of the D. melanogaster-wMel symbiosis. First, we analyze patterns of molecular evolution in the wMel genome to provide a summary of its global genetic diversity and patterns of transmission. Second, we combine demographic information with a Bayesian phylogenetic reconstruction to estimate the date of the cytoplasmic Most Recent Common Ancestor (MRCA). Finally we examine the extent to which a key phenotype, the within-fly density of wMel, is determined by genotypic differences among its D. melanogaster host.
Materials and Methods
Drosophila melanogaster lines, DNA extraction, and sequencing
We used 91 inbred Drosophila melanogaster lines from 5 populations: Beijing (China), Ithaca (NY, USA), Netherlands, Tasmania, and Zimbabwe (Table 1). The lines were established from isofemale lines and then inbred for 12 generations, as described in Greenberg et al. (2010). DNA was extracted from pools of 50 adult female flies using Qiagen DNeasy Blood & Tissue kits. Samples were then sequenced to approximately 12× nuclear genomic depth at the Beijing Genomics Institute. Sequencing was performed on an Illumina HiSeq2000 using 100-bp paired-end reads with a 450-500 bp insert size (manuscript in preparation).
Table 1.
Geographic distribution of mtDNA and wMel haplotypes.
| Beijing (China) | Ithaca, NY (USA) | Netherlands | Tasmania | Zimbabwe | Total | |
|---|---|---|---|---|---|---|
| I | 1 (1) | 19 (14) | 1 (0) | 4 (1) | 1 (1) | 26 (17) |
| II | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1) | 1 (1) |
| III | 3 (2) | 0 (0) | 15 (12) | 15 (15) | 16 (14) | 49 (41) |
| IV | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| V | 0 (0) | 0 (0) | 2 (0) | 0 (0) | 0 (0) | 2 (0) |
| VI | 0 (0) | 0 (0) | 1 (0) | 0 (0) | 0 (0) | 1 (0) |
| VIII | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| VIII | 12 (6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 12 (6) |
| Total | 16 (9) | 19 (14) | 19 (12) | 19 (14) | 18 (16) | 91 (65) |
For each population, the number of sampled mtDNA haplotypes within each major clade is listed, followed by the number of lines carrying a Wolbachia infection in parentheses. Haplotypes correspond with those given in Fig 2. Clades I-VI are defined by Richardson et al. (2012); Clade VII is defined in Ilinsky (2013); clade VIII is described in this paper.
Read alignment and genomic variant detection
Raw reads from each D. melanogaster line were aligned to the D. melanogaster mitochondrial genome (RefSeq NC001709.1, r5.44) and to the Wolbachia pipientis strain wMel genome (GenBank AE017196) using Mosaik v2.1.33 (http://bioinformatics.bc.edu/marthlab/Mosaik). Duplicate reads were marked using Picard v1.56 (http://picard.sourceforge.net). The resulting alignments were then fed through a standard Picard-GATK pipeline to call nucleotides at each site and to identify indels under five bp in length (DePristo et al. 2011; McKenna et al. 2010). Briefly, BAM files were merged and indexed with Picard, then realigned and genotyped with GATK. As the genomes were small, we used hard filtering instead of GATK's variant quality score recalibration pipeline. For the Wolbachia data set, we based our filters on GATK's best practices v.3. Because of the extremely high coverage of the mitochondrial genomes, we modified the filters for these sequences (for indels: QD < 2.0, ReadPosRankSum < −20.0, and FS > 400; for SNPs: QD<3.0, MQ<35.0, HaplotypeScore > 13.0, MQRankSum < −45, and ReadPosRankSum < −8.0). To obtain a representative Canton-S mitochondrial genome, we aligned Illumina reads from Canton-S ovaries (Sequence Read Archive, SRR353680; Soshnev et al. 2012) to the mitochondrial reference genome using filters recommended in GATK's best practices v. 3, except we set the mapping quality cutoff to 17.
A site was masked if a base call was made for fewer than 50% of the Drosophila lines or if it overlapped a GATK-called indel. Alternate allele calls were marked as missing in individual lines if the read depth in the line at that position was less than three. For the purpose of our analyses, we disregarded heterozygous calls made by GATK, calling the site based on the most frequent nucleotide at that position in that line's alignment. Essentially, this means we sampled a single strain of Wolbachia from each fly line. (Similar to how inbred fly lines sample a single chromosome from the original, wild-caught fly.) The decision to follow this procedure was three-fold. First, as discussed in the Results, we determined that no fly lines carried multiple haplotypes representative of different clades. Second, due to inbreeding, relaxed selection, and potential within-fly drift, it was unclear how heteroplasmy within inbred lab lines would inform our study of diversity in the wild. And finally, calculating diversity statistics would require development and fitting of a complex statistical/population genetic model that would account for these “heterozygous” sites which arise in the context of a Wolbachia infection not from diploidy but rather under a wide range of potential allele frequencies segregating within a single fly. Note that the impact of this approach was likely minimal: among the calls in the final wMel data set at variable sites, GATK had made 9,419 homozygous calls and only 19 heterozygous calls. In the mtDNA dataset, these numbers were 14,965 and 19, respectively.
Pindel v.0.2.4 (Ye et al. 2009) was used to identify inversions, tandem duplications, insertions between 5 and 80 bp, and deletions of 5 bp or greater. This program uses mapping information from paired-end reads to infer the presence of structural variants. Initial filtering removed calls with only single strand support. We subsequently removed weakly supported calls that had high strand bias and low read count. Post-hoc, as a means of determining whether these filters were overly stringent, we noted that all the removed calls were incongruous with our SNP-constructed phylogenetic trees. Riegler et al. (2005) used two variable number tandem repeat loci, a large inversion, and two IS5 transposon insertion sites to distinguish among five different Wolbachia genotypes in D. melanogaster. To determine which of these haplotypes were present in our dataset, we compared the locations of the large inversion and IS5 insertion sites to the break points identified by Pindel. These two structural variants are sufficient to distinguish among all haplotypes except wMelCS and wMelCS2.
For each genome from each line, mean read depth (mean number of reads mapped at each nucleotide position) was calculated using GATK's DepthofCoverage analysis. To create standardized estimates of Wolbachia and mtDNA density within each fly line, we calculated the ratio of aligned wMel or mtDNA reads to aligned D. melanogaster nuclear genome reads (Supplementary Information, Table S1; data not shown). Nucleotide diversity (π) was calculated using custom Perl scripts.
Testing for lateral genetic transfer
To test whether portions of the wMel genome have been transferred to the nuclear genome of its host, we looked for evidence of paired reads where one mate read aligned to the wMel genome while the other aligned to the D. melanogaster genome. Such read pairs could suggest that a piece of the wMel genome relocated to a D. melanogaster chromosome. Using SAMtools v0.1.18 (Li et al. 2009), we separated out all read pairs where one read mapped to wMel while the other was unmapped. The unmapped reads were then aligned to the complete D. melanogaster reference genome (r5.46) using the Mosaik protocol outlined above.
Phylogenetic analyses
The ancestral states of wMel SNPs were determined using W. pipientis strain wRi (RefSeq NC_012416.1). Homologous gene regions were identified using the Ensembl database. As the mutation rate of mitochondrial DNA is high and recurrent mutation is possible, mitochondrial SNPs were polarized using 15 complete D. simulans genomes (Ballard 2000; AF200833.1- AF200842.1, AF200844.1- AF200846.1, AF200848.1- AF200849.1). Sequences were aligned with Muscle v3.8.31 (Edgar 2004) and the ancestral state at each SNP was determined by eye. After initial tree building, we used parsimony to infer the ancestral states of wMel SNPs that lacked clear wRi homologs and mtDNA SNPs where the ancestral state was ambiguous due to segregating variants in D. simulans (Supporting Information, Tables S2 and S4).
We constructed phylogenetic trees using both maximum likelihood and Bayesian methods. Maximum likelihood trees were constructed with RAxML v7.2.8 (Stamatakis 2006) using the GTRCAT method, 300 multiple inferences, and the default hill-climbing algorithm. The best-scoring maximum likelihood tree was chosen and support for the tree was calculated with 10,000 bootstrap replicates. Bayesian trees were constructed with MrBayes v3.2 (Ronquist et al. 2012) using reversible jump MCMC to estimate the number of independent substitution rate parameters (nst=mixed). We ran the MCMC analysis for 5 million generations and discarded a 25% burn-in fraction prior to analysis. Results were checked by eye in Tracer v1.5 (http://beast.bio.ed.ac.uk/Tracer) to ensure convergence. Bayesian trees were also constructed with BEAST (Drummond & Rambaut 2007) during the course of the Most Recent Common Ancestor analysis discussed below. For both methods, we included either wRi or the D. simulans sequences described above in order to infer the root of the tree. In addition to these trees, we also constructed phylogenetic networks using the Neighbor-Net method in SplitsTree4 v4.12.3 (Huson & Bryant 2006). A tanglegram combining the wMel and mtDNA trees was constructed with Dendroscope v3.2.3 (Huson & Scornavacca 2012). For the Neighbor-Net analysis, ambiguous sites were inferred with parsimony where possible. We removed any remaining sites with missing data prior to analysis. Unless otherwise stated, the Wolbachia trees were constructed with a concatenated sequence composed of all identified variable sites. Mitochondrial analyses were performed using the first 14,916 nucleotides of the genome. After tree construction, we identified major clades and named them based on the system established in Richardson et al. (2012).
Trees with additional D. melanogaster mitochondrial haplotypes were constructed with MrBayes using the settings outlined above. In addition to the Canton-S sequence assembled from reads in SRA (see above), we downloaded the following mitochondrial sequences from GenBank: Alstonvl (FJ190106.1), Barcelona (JX266575.1), BER1 (JQ686694.1), Brownsvl (FJ190107.1), CO3 (JQ686695.1), Dahomey (FJ190108.1), Hawaii (JX266576.1), Israel (JX266577.1), Japan (FJ190109.1), Madang (JX266578.1), Mysore (FJ190110.1), Oregon R (AF200828.1), Oregon R-C (JQ686698.1), Puerto Montt (JX266579.1), QI2 (JQ686696.1), Reids1 (JQ686697.1), Sweden (JX266580.1), tko25t (JQ686693.1), w1118iso (FJ190105.1), and Zimbabwe 53 (AF200829.1). We aligned all sequences (approximately 12,300 bp) with Muscle v3.8.31 (Edgar 2004) prior to tree construction.
Node age, root age, and substitution rate estimates
To estimate the date of the most recent cytoplasmic coalescence, we calculated the divergence times of our mitochondrial haplotypes with BEAST v1.7.2 (Drummond & Rambaut 2007). To most closely approximate unconstrained, neutrally evolving sequences, we created a concatenated dataset of all third codon sites. BEAST analyses were then run with a strict molecular clock, the Hasegawa-Kishino-Yano (HKY) substitution model, no site heterogeneity, and constant population size. The MCMC chain was run for 100 million generations and a burn-in fraction of 10% was discarded prior to analysis. We examined log files in Tracer to ensure we acquired an adequate Effective Sample Size for each parameter. First, we estimated the age of the root by using a strict clock based on the mitochondrial mutation rate estimated by Haag-Liautard et al. (2008) (6.2 × 10−8 mutations per site per fly generation) and assuming 10 fly generations per year.
Second, to incorporate demographic estimates into our analysis, we placed age priors at nodes C and D (Fig 2; normal distribution with a mean of 200 years and a standard deviation of 50 years). The clock rate prior was set at 6.2 × 10−7 mutations per site per year with one of three standard deviations (1 × 10−7, 1 × 10−6, or 1 × 10−5). All other assumptions were the same as above. For each model, we estimated the marginal maximum likelihood using both path sampling and stepping-stone analyses (Baele et al. 2012; Baele et al. 2013). Bayes Factors were calculated to choose among models.
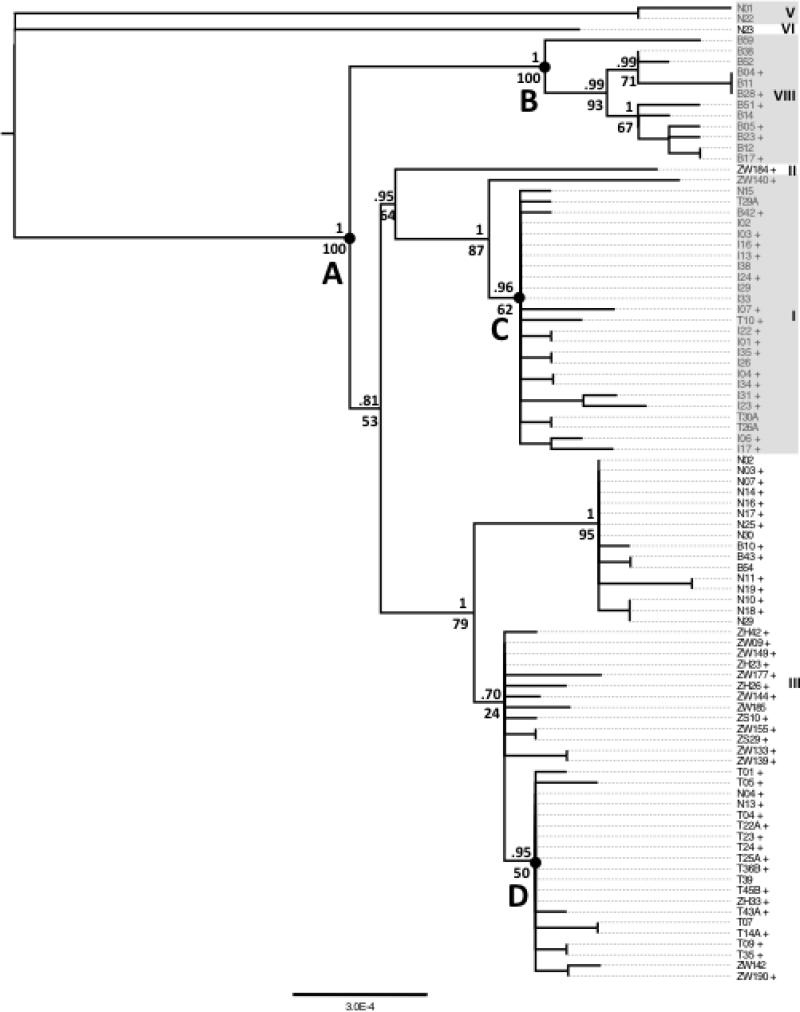
Fig. 2.
Mitochondrial phylogenetic tree calculated with RAxML. Major clades are marked on the right. Values above nodes are Bayesian probabilities calculated with MrBayes. Those below the nodes are bootstrap values calculated with RAxML. Lines followed by a + carry a Wolbachia infection. The ages of the marked nodes were calculated with a Bayesian Skyline analysis in BEAST using internal calibration points at nodes C and D. 95% HPD intervals are noted in parentheses. Root, 2239 ya (1100 – 3592); A, 957 ya (462 – 1556 ya); B, 425 ya (137 – 767 ya); C, 202 ya (91 – 311 ya); D, 192 ya (108 – 279 ya).
The wMel substitution rate was calculated relative to the mtDNA rate by running in parallel the same BEAST analyses with the wMel sequence data partitioned into codon positions and intergenic regions. The root age and clock rates for each genomic region were then scaled by the mtDNA results.
Molecular evolutionary analyses
SNPs were functionally annotated using Ensembl's Variant Effect Predictor v2.3 (McLaren et al. 2010). Genome-wide Ka and Ks values were calculated with KaKs Calculator (Zhang et al. 2006) using the Goldman-Yang (GY) maximum likelihood method on concatenated codon-aligned coding sequences. To determine mutational bias, the total number of each nucleotide within the wMel genome was counted from the wMel reference genome. Similarly, mutational bias calculations were based on changes to the reference strand. Codon usage was calculated from both confirmed and predicted protein-coding sequences as annotated in the Ensembl Bacteria database, release 15 (McLaren et al. 2010). All calculations were based only on the sites covered in our alignments. Statistical analyses were conducted in R (R Development Core Team 2011).
Quantification of wMel density
We reared flies from 61 of the 65 Wolbachia-infected lines at room temperature in vials of standard glucose-yeast media. At the larval stage, we chose two replicate vials from each line, ensuring a comparable, moderate larval density across all lines. Pools of twenty mated females, aged 6-8 days, were chosen from each vial. Flies were ground and DNA extracted with a Qiagen DNeasy Blood & Tissue Kit. DNA concentration was determined on a Nanodrop ND-1000 spectrophotometer.
To measure relative Wolbachia load, we performed two quantitative PCR (qPCR) assays. The first targeted Dfd, a single-copy nuclear gene in D. melanogaster (Dfd For 5’ GTAGCGAAGAAACCCACCAA 3’; Dfd Rev 5′ ACGCTCCACTCACCTCATTC 3’). The second used the primers wspFQALL and wspRQALL to target the wsp gene of Wolbachia (Osborne et al. 2009). Each 10 μl reaction contained 10 mM Tris 8.0, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.25 mM SYBR green, 5% DMSO, 0.25 μM of each primer, Taq Polymerase, and 25 ng of DNA. Reactions were run in triplicate on a Roche LightClycler 480 with the following conditions: one cycle of 95 C for 5 min, followed by 45 cycles of 95 C for 15 sec, 60 C for 30 and 72 for 10 sec. For each fly line, we tested two pools of 20 flies that were sampled from separate vials. A known Wolbachia-free fly was used as a negative control. For each line, we calculated relative Wolbachia density as 2(CPDfd-CPwsp). To test whether there was population-level variability in endosymbiont density, we ran an ANOVA on a Phylogenetic Generalized Least Squares (PGLS) model that tested for the effect of population while controlling for the phylogenetic relationships between wMel lineages. Phylogenetic correlations among wMel strains were derived from the MrBayes analysis described above. We performed PGLS analyses using both Brownian Motion and Ornstein-Uhlenbeck Motion models. Analyses were conducted using the R packages ape v3.0-3 (Paradis et al. 2004) and nlme v3.1-102 (Pinheiro et al. 2012).
Results
Genome alignments and variant discovery
Wolbachia genome
Based on alignment to the W. pipientis wMel reference genome, 65 of the 91 D. melanogaster lines showed strong evidence of Wolbachia infection (Table 1). For each of these lines, Mosaik mapped more than 90,000 reads to the wMel reference genome, giving an average read depth of 7 or greater for each line (Supporting Information, Table S1). Conversely, 25 lines had fewer than 1,500 reads that mapped to the wMel reference and so were considered Wolbachia-free. One line (B59) was intermediate to these two groups with 12,500 mapped reads. This could indicate an unusually low level of Wolbachia infection or a small amount of contamination. Because of the low genome coverage (about 1×), genotype calls could not be made with reasonable accuracy, and we excluded line B59 from the subsequent Wolbachia analyses. Across the 65 Wolbachia-infected lines used in the subsequent analyses, GATK made base calls at 1,134,595 positions within the 1,267,782 bp genome (89.5%) and called single nucleotide polymorphisms (SNPs) at 174 positions. With additional filtering, we removed 22 sites where only heterozygous calls were made or where more than 10 lines were called as heterozygous or missing. Because of the repetitive nature of the wMel genome, there was a high probability that these calls resulted from misalignments, a hypothesis supported by the observation that 55% of these discarded calls were within 20 bp of a second low-confidence site. After this additional filtering, the final dataset contained 145 SNPs, of which 51 were detected in only a single line (Supporting Information, Table S2). Assuming each fly line carried a single wMel copy (as discussed in the Methods), average genome-wide nucleotide diversity (π) across all populations was 1.8 × 10−5 (Table 2).
Table 2.
Wolbachia and mitochondrial genomic diversity.
| N | π | S | D | ||
|---|---|---|---|---|---|
| wMel | |||||
| All | 65 | 1.8 × 10−5 | 145 | −1.1 | |
| Beijing | 9 | 2.8 × 10−5 | 75 | 0.7 | |
| Ithaca, NY | 14 | 4.0 × 10−6 | 24 | −1.7 | |
| Netherlands | 12 | 3.1 × 10−6 | 12 | −0.5 | |
| Tasmania | 14 | 3.7 × 10−6 | 29 | −2.3 | |
| Zimbabwe | 16 | 7.9 × 10−6 | 50 | −1.7 | |
| mtDNA | |||||
| All | 91 | 1.02 × 10−3 | 147 | −1.63 | |
| Beijing | 16 | 1.51 × 10−3 | 112 | −1.48 | |
| Ithaca, NY | 19 | 2.76 × 10−4 | 33 | −2.28 | |
| Netherlands | 19 | 1.05 × 10−3 | 73 | −1.07 | |
| Tasmania | 19 | 4.34 × 10−4 | 26 | −0.56 | |
| Zimbabwe | 18 | 3.84 × 10−4 | 39 | −2.05 |
N, number of lines; π, average pairwise nucleotide diversity; S, number of segregating sites; D, Tajima's D
Using our GATK pipeline and subsequent filtering, we identified 22 single bp indels within the wMel genome. Pindel analysis identified six deletions and two small insertions (Supporting Information, Table S3). We compared all Pindel-identified breakpoints to those described in Riegler et al. (2005), and found that all of our lines matched the wMel genotype. No inversions, tandem duplications, or additional IS-element insertions were identified.
Mitochondrial genome
After MOSAIK alignment, the average mitochondrial read depth was 303.7 (Supporting Information, Table S1). For our analyses, we considered the GATK nucleotide calls for the first 14,916 bp of the chromosome. This includes all the coding regions, but excludes the repetitive AT-rich region where short-read alignments were unreliable. Within this region, calls were made at 14,661 positions (98.3%). GATK identified 166 SNPs, eight of which failed to pass our additional filters (Supporting Information, Table S4). Of the 158 SNPs in our final dataset, 11 were fixed within our sample (representing differences with the reference only) and 55 were singletons. In relation to the reference, Pindel analysis identified one 6 bp deletion present in all of our lines and one 5 bp insertion present in a subset of the lines (Supporting Information, Table S5). We found no inversions or tandem duplications. Average genome-wide nucleotide diversity (π) across all populations was 1.02 × 10−3 (Table 2).
Heteroplasmy
To determine whether any of our inbred fly lines were heteroplasmic, we examined closely all sites where multiple alleles were called within a single fly line. To be as rigorous as possible, we also analyzed 29 heterozygous wMel sites that were filtered from our final dataset (making 40 sites in total for wMel and seven for mtDNA). When haplotypes within a fly differed by only one or two SNPs, we could not determine whether heteroplasmy was caused by horizontal transfer of a closely related haplotype or by a mutation within the maternal lineage. We therefore disregarded extremely low levels of heteroplasmy and instead determined whether flies carried two divergent mtDNA or wMel haplotypes (for instance, haplotypes from both clades I and III, which are known to segregate in the same geographic areas). Upon examination, no fly lines had more than two heterozygous sites in their mtDNA (17 had one and a single line had two). While more diverse, the wMel data yielded similar results. Examining only the sites in our final dataset, 12 lines had a single heterozygous wMel site while two lines had two. Only a single fly line contained three heterozygous sites, but the alleles at these sites were not consistently shared with a one single known wMel clade. The heterozygous calls in the low-confidence wMel sites formed no informative pattern. In the wild, upwards of 14% of D. melanogaster may carry multiple mtDNA haplotypes (Nunes et al. 2013), however, our failure to find segregating divergent cytotypes here is not unexpected: these lines have been maintained in the lab beyond the 100 generations that Nunes et al. (2013) estimates is needed for complete sorting of mtDNA haplotypes. We conclude that the mtDNA heterozygosity seen is not due to paternal leakage of disparate haplotypes that would cause true heteroplasmy, but instead is some combination of somatic mutations and sequencing error. For this reason we used a single haplotype sampled from each line in subsequent analysis.
Read depth comparison
We standardized mitochondrial read depth (as described in Methods) then compared standardized read depth in infected and uninfected lines and found no significant difference between the two groups (ANOVA, P = 0.1233). Similarly we tested whether Wolbachia read depth correlated with mitochondrial read depth and found no significant correlation (Pearson's product-moment correlation, P = 0.7966). These results are concordant with earlier findings in parasitic wasps (Mouton et al. 2009).
Lateral genetic transfer
Previously, transfer of Wolbachia genetic material into a host nuclear genome has been observed (Hotopp et al. 2007; Kondo et al. 2002; Nikoh et al. 2008). To test whether any portion of the wMel genome has been transferred into the nucleus of any of our D. melanogaster genomes, we examined pairs of reads where one read mapped to the wMel genome while the paired read mapped to the D. melanogaster genome. For such paired reads, the only regions of the D. melanogaster genome mapped with an aligned read depth greater than two were highly repetitive and noninformative, providing no evidence of lateral genetic transfer in our sample.
Cytoplasmic haplotypes show geographic structuring
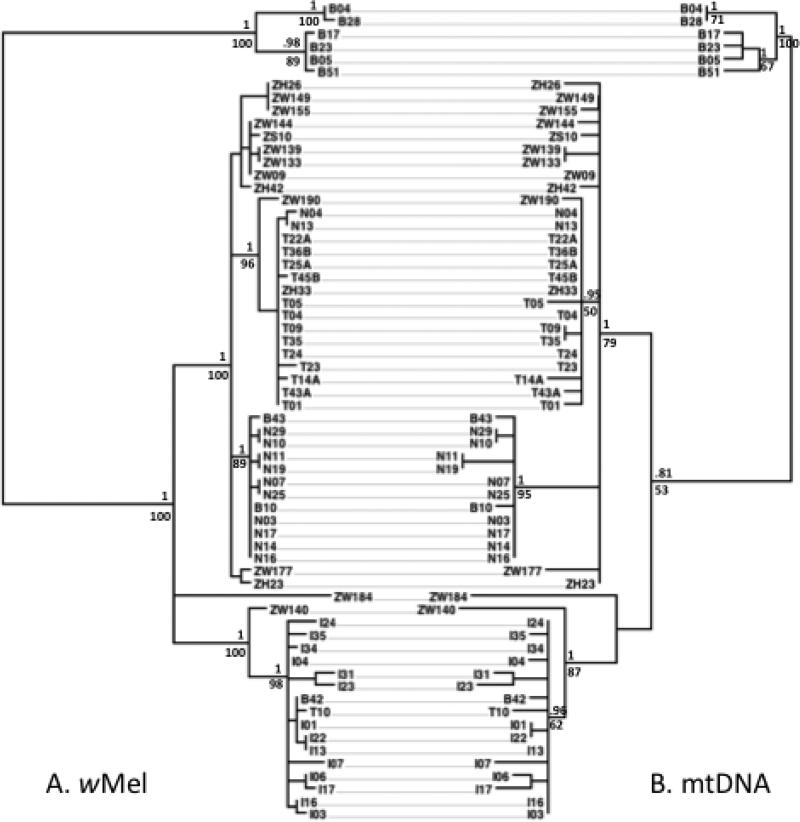
For both the wMel and mtDNA data, we constructed phylogenetic trees using three methods: maximum likelihood using RAxML v7.2.8 (Stamatakis 2006), Bayesian inference using MrBayes v3.2 (Ronquist et al. 2012), and Bayesian inference using a strict molecular clock in BEAST v1.7.2 (Drummond & Rambaut 2007). The wRi reference sequence or the D. simulans mitochondrial sequences described in the Methods were included to infer the root. Ignoring branching patterns with little support, all methods yielded identical topologies for the Wolbachia data (Fig 1A). For the mitochondria data, all methods resulted in identical branching patterns (again ignoring branches with little support), but these methods differed in the placement of the root. Because of this difficulty in resolving the root of the phylogeny, we constructed a phylogenetic network using the Neighbor-Net method in SplitsTree4 (Huson & Bryant 2006). The results showed the presence of conflicting phylogenetic signals in the mitochondria data (Supporting Information, Fig S1A). Because the haplotype divergence was so low, we did not have the power to test whether this pattern could have resulted from recombination. The high mutation rate of D. melanogaster mtDNA, however, makes it likely that this pattern is the result of recurrent mutation in divergent lineages. We noted that one SNP and two indels from our dataset arose independently in a set of mutation accumulation lines (Haag-Liautard et al. 2008). In addition, 23 polymorphic sites within our lines are also known SNP locations in D. simulans mtDNA (Ballard 2000). No conflicting signals were detected in the Wolbachia data (Supporting Information, Fig S1B). The final rooting of the mtDNA tree is based on mid-point rooting and BEAST analyses, and is concordant with the rooting of the Wolbachia tree (Fig 1B).
Fig. 1.
Tanglegram showing concordance of the A) wMel and B) mitochondrial phylogenetic trees. Only infected lines are included in the mitochondrial tree. Trees are midpoint rooted. The first letter of the line name represents the geographic population of origin: B, Beijing, China; I, Ithaca, NY; N, Netherlands; T, Tasmania; Z, Zimbabwe.
Our sample contained six of the eight previously defined mtDNA clades (Fig 2; Ilinsky 2013; Richardson et al. 2012). In addition, we describe here an additional clade (VIII), which segregates at a high frequency in the Beijing population. To further determine the extent to which our samples captured the full breadth of global D. melanogaster mitochondrial diversity, we also constructed mitochondrial trees that included both our samples and the 19 partial D. melanogaster mitochondrial genomes currently available in GenBank. In addition, we aligned short-read mtDNA sequences from whole-genome sequencing of a Canton-S line. These additional sequences clustered within or near already identified haplotypes showing that our samples capture a wide range of extant global genetic diversity (Supporting Information, Fig S2). As in Richardson et al. (2012) and Ilinsky (2013), our cytoplasmic genomes show strong geographic structuring. All sampled populations contained one high frequency cytotype and with the exception of the Ithaca, NY population, at least one lower frequency cytotype (Fig 1 and 2; Table 1; Supporting Information, Fig S4). Despite this structuring, however, most cytotypes are not geographically isolated. The exceptions include clade VII, which was found only in the Beijing population, and clades V and VI, which were only detected in the Netherlands.
Evidence for strictly vertical wMel transmission with occasional loss events
If wMel is transmitted exclusively maternally, it will show a tight evolutionary correlation with D. melanogaster mtDNA. Alternatively, if horizontal transmission has played a role in shaping wMel evolution, we expect to see at least one of three possible patterns. First, we could directly find multiple wMel haplotypes within a single inbred fly line. Second, we could infer past co-infection by detecting recombination between divergent wMel haplotypes. Or third, we could infer horizontal transmission or paternal leakage by finding the same wMel haplotype associated with different mitochondrial backgrounds. As noted above, we found no support for the first two patterns: we did not detect multiple divergent haplotypes segregating within a single fly line, and the Neighbor-Net analysis provided no evidence of potential recombination. To test for the final pattern, we compared the branching patterns in the Wolbachia and mitochondrial phylogenies. With the exception of the Ithaca, NY population, all the sampled fly populations contained multiple segregating mitochondrial haplotypes, showing that opportunities exist for Wolbachia to contact new cytoplasmic backgrounds. However, like Richardson et al. (2012) we found the mtDNA and wMel trees completely congruent, suggesting that horizontal transmission has not played a major role in recent wMel evolution or ecology (Fig 1).
A second observation that comes from looking at infection patterns in the mtDNA phylogeny is that Wolbachia infections have been repeatedly lost since the most recent cytoplasmic coalescence (Fig 2; Richardson et al. 2012). All common mtDNA haplotypes contained a mix of infected and uninfected cytoplasmic backgrounds, suggesting recent losses within these lineages. Within our sample, the two rare mitochondrial clades (V and VI) show no Wolbachia association, however, evidence from other studies suggest these lineages likely lost an ancestral infection. In Richardson et al. (2012), clade VI was shown to associate with Wolbachia. As for clade V, we noted that it clustered with the COI haplotype 10 whose members were infected with either wMel or an undetermined strain of Wolbachia (Supporting Information, Fig S3; Nunes et al. 2008a). This provides evidence of past infections in both these lineages, and suggests that the coalescence of our mtDNA tree would also represent the MRCA of the extant global wMel population.
Most Recent Common Cytoplasmic Ancestor
Previous analyses have shown recent global shifts in the prevalence of particular wMel haplotypes, as defined by the five previously identified structural variants (Ilinsky & Zakharov 2007; Nunes et al. 2008b; Riegler et al. 2005). Specifically, since the 1960s the global frequency of wMel-like haplotypes (to which all our Wolbachia samples belong) has risen sharply whereas the frequency of the wMelCS haplotype has rapidly declined. Our results support the hypothesis that this sweep was acting on standing variation (Richardson et al. 2012), as the wMel haplotype and its corresponding mitochondrial background do not represent recent mutation events. To more precisely date the age of the major haplotypes and to determine the Most Recent Common Ancestor (MRCA) of all our cytoplasmic samples, we estimated divergence times and node ages of the mtDNA haplotypes with BEAST.
We conducted the analyses under two different sets of assumptions. The first model assumed a strict molecular clock, a constant population size, 10 fly generations per year (as in Richardson et al. 2012), and neutral evolution of third codon sites at the Haag-Liautard et al. (2008) estimate of the mitochondrial mutation rate. This gave a root age of 5,958 ya (95% Highest Posterior Density (HPD): 4,216 – 7,886 ya) and internal node ages (defined in Fig 2) of: A, 2,578 ya (1,644 – 3,575 ya); B, 1,132 ya (482 – 1843 ya); C, 62 ya (279 – 1017 ya); and D, 568 ya (199 – 1027 ya).
Our second set of models similarly assumed a strict molecular clock and neutral evolution at third codon positions. However, rather than calibrate the tree with the mitochondrial mutation rate, we placed age priors at nodes C and D (Fig 2), representing the estimated colonization of North America and Australia, respectively (approximately 200 years ago; David & Capy 1988; Keller 2007). Three separate models were run with different standard deviations placed on the clock-rate prior (6.2 × 10−7 mutations per site per year; standard deviations of 1 × 10−7, 1 × 10−6, and 1 × 10−5). The node-calibrated models had higher marginal maximum likelihoods than the uncalibrated analysis, and the model with the greater support contained a clock standard deviation of 1 × 10−6 (Bayes Factor = 3.86, compared to the strict clock model). It dated the root of the tree to 2,239 years ago (95% HPD: 1100 – 3592 ya) and estimated the third codon position clock rate to be 1.75 × 10−6 substitutions/site/year (95% HPD: 9.5 × 10−7 – 2.6 × 10−6 substitutions/site/year). Internal node ages are given in Fig 2.
Molecular evolution of wMel
Because of its small effective population size, Wolbachia is expected to show reduced efficacy of selection. To test for this, we calculated the ratio of non-synonymous to synonymous amino acid substitutions (Ka/Ks) and compared SNP density as well as the estimated substitution rate within coding and noncoding regions. Calculated with the GY method in KaKs calculator (Zhang et al. 2006), average genome-wide Ka and Ks values were 1.58 × 10−5 and 1.89 × 10−5 respectively, giving a Ka/Ks ratio of 0.875 which is not statistically different from the expectation of neutral evolution with no constraint (P = 0.508), a finding also reached by Richardson et al. (2012). Additionally, we examined whether mutational patterns varied along the branches of the phylogenetic trees. By comparing mutations at the tips (singletons) to mutations on deeper branches, we found no significant differences in the ratio of non-synonymous to synonymous mutations.
To test whether SNPs were evenly distributed between coding and noncoding regions, we calculated the proportion of sites in both regions that were polymorphic. SNP density in intergenic regions (0.248 SNPs/kb) was higher than in protein-coding regions (0.112 SNPs/kb; Fisher's exact test (FET); P = 2.34 × 10−4; Table 3). Our BEAST models, however, did not find any significant difference in the substitution rate of coding versus intergenic regions (relative clock rates; Protein-coding: 0.928 (95% HPD: 0.075 – 2.35), Intergenic: 1.824 (95% HPD: 0.144 – 4.60)).
Table 3.
Distribution of variant and invariant nucleotide sites in the wMel genomes
| Ancestral State | Location | ||||||
|---|---|---|---|---|---|---|---|
| A | C | G | T | Protein Coding | Intergenic | Total | |
| Invariant | 366,331 | 199,828 | 198,234 | 370,074 | 961,702 | 133,078 | 1,134,450 |
| Variant | 30 | 36 | 37 | 25 | 108 | 33 | 145 |
Bacteria generally show a GC to AT mutational bias and maintain constant GC levels only through selection or after equilibrium nucleotide levels are reached (Hershberg & Petrov 2010). wMel has an AT-rich genome (35% GC content), but it is unknown whether this is stable or whether the genome is evolving toward a still higher AT content. Consistent with a GC to AT mutational bias, the results show a higher relative number of polymorphisms at ancestral C and G nucleotides compared to A and T nucleotides (FET, P = 5.87 × 10−7; Table 3). The majority of these mutations were transitions (Ti=102 and Tv=26), leading to an overall increase in AT content (70 GC-to-AT mutations versus 54 AT-to-GC mutations; FET, P = 1.727 × 10−6).
We did not find any evidence for codon selection. The wMel genome shows strong codon usage bias that correlates with codon AT-content (Wu et al. 2004). In our dataset, the strength of codon bias did not correlate with the direction of synonymous codon mutations (Pearson's product-moment correlation, P = 0.5291), suggesting these observed patterns of variation are due to mutation, not selection.
BEAST analyses showed that the substitution rate in intergenic regions of the wMel genome is 91 times slower than the substitution rate at third codon positions in the D. melanogaster mitochondrial genome. Assuming the Haag-Liautard et al. (2008) mitochondrial mutation rate, this yields a wMel mutation rate of 6.8 × 10−10 substitutions/site/fly generation (95% HPD: 5.0 X 10−11 – 1.7 × 10−9). This is almost identical to the estimate of 6.87 × 10−10 substitutions/site/generation calculated by Richardson et al. (2012).
Host effects on wMel within-fly density
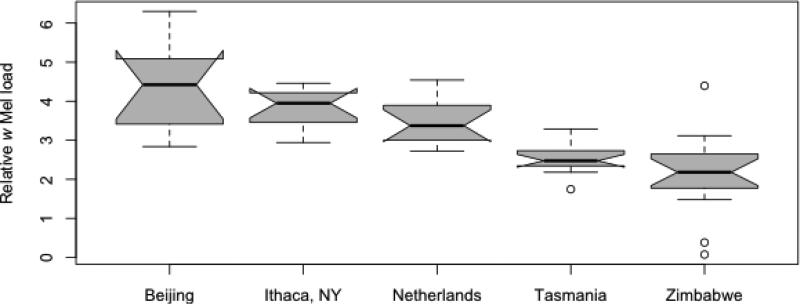
Fitness effects conferred by wMel on its fly host may vary with bacterial density (Osborne et al. 2009). We were therefore interested in examining whether populations varied in their wMel load. Across our samples, depth of coverage of the wMel genome was highly variable (Supporting Information, Table S1), an observation we validated with a qPCR analysis of additional replicates from each line. Despite the different methods, the qPCR and Illumina measurements had a Spearman's Rank Correlation of 0.787. The qPCR results confirmed that Wolbachia titre varies across lines and populations (Fig 3). Since our wMel haplotypes are highly geographically structured, we constructed a Phylogenetic Generalized Least Squares (PGLS) model that tested the effect of fly population while controlling for the phylogenetic relationships between wMel lineages. This model showed that fly populations significantly differ in their Wolbachia levels, a result that was robust under both a Brownian motion model (ANOVA, P = 0.016) and an Ornstein-Uhlenbeck model (ANOVA, P = 0.0005) of trait evolution.
Fig. 3.
Average relative Wolbachia load within each population as determined by qPCR. In a PGLS model assuming Brownian Motion trait evolution based on the wMel phylogeny, the difference among populations is still significant (ANOVA, P = 0.0156).
Discussion
Because of the diversity of hosts it inhabits and the wide range of phenotypes it induces, the endosymbiont Wolbachia is a particularly intriguing study system. Until recently, phenotypic studies performed with wMel have necessarily assumed genetic and phenotypic uniformity among infecting bacteria. Evidence over the last decade has shown that there is indeed genetic variation within this “clonal” infection (Ilinsky 2013; Ilinsky & Zakharov 2007; Nunes et al. 2008b; Richardson et al. 2012; Riegler et al. 2005); however, many aspects of this variation, including its genomic extent, global distribution, and phenotypic effects, remain under-studied. Here, using a set of globally distributed populations, we combine an in-depth molecular genetic analysis with evidence of phenotypic variation among wMel-infected host populations.
Global picture of wMel genomic diversity
Overall, our lines contain five of the seven previously described D. melanogaster cytotypes (Ilinsky 2013; Richardson et al. 2012). In addition, we name an additional mitochondrial and wMel clade (VIII), which we found only in the Beijing population. The two known haplotypes that we do not recover currently have a low global frequency (clade IV; Richardson et al. 2012) or a limited geographic distribution (clade VII; Ilinsky 2013). While evidence suggests that all our cytoplasmic backgrounds are derived from a Wolbachia-infected ancestor, only four of our six cytoplasmic groups currently include Wolbachia-infected individuals and all of these represent wMel-like infections. As in a previous study (Richardson et al. 2012), we find that genome-wide nucleotide diversity (π) among wMel isolates is low (Table 2).
Concordant with past studies (Ilinsky 2013; Nunes et al. 2008a; Richardson et al. 2012), cytotypes display strong geographic structuring, with each population containing one major haplotype. A dominant haplotype could have arisen in each population due to drift, but it is likely that selection and local adaptation have played a role as gene flow is present among these populations. Phylogenetic analysis shows that the cytoplasmic associations between mitochondrial and Wolbachia haplotypes are stable and long-lived (Fig. 1; Richardson et al. 2012), so at this time, we can only hypothesize whether any selection has primarily acted on mitochondria or Wolbachia. Wolbachia is, however, a likely target for selection. Recent studies have uncovered several fitness benefits that wMel confers to its host including iron provisioning (Brownlie et al. 2009) and viral resistance (Teixeira et al. 2008). Indeed, some combination of selection and CI must allow for the maintenance of Wolbachia infections in natural populations. Otherwise, even the rare loss events seen in the wild (due to incomplete transmission from mother to offspring) would have resulted in a lower infection prevalence than what we, and others, have observed (Hoffmann et al. 1998; Ilinsky & Zakharov 2007; Richardson et al. 2012; Verspoor & Haddrill 2011).
Date of cytoplasmic coalescence
Linking known demographic information to our phylogenetic analysis, we date the cytoplasmic coalescence in D. melanogatser to approximately 2,239 ya (95% HPD: 1100 – 3592 ya). While overlapping with their 95% confidence intervals, this estimate differs from that recently proposed by Richardson et al. (2012; 8,008 ya, 95% BCI: 3,263-13,998 ya). Our decision to use a node-calibrated analysis arose from the observation that the major haplotypes in the two most recently founded populations (Ithaca, NY and Tasmania) displayed star-like topologies. As Richardson et al. (2012) proposed for a separate North American population, these haplotypes may have been repeatedly reintroduced to Tasmania and New York. A more parsimonious explanation for these star-like topologies, however, evokes a single founding event followed by a subsequent radiation within the population. Under this scenario, the ages of these clades should be no older than the colonization of the areas in question (approximately 200 years; David & Capy 1988; Keller 2007). Our uncalibrated coalescent analysis with a strict clock rate dates both of these nodes to approximately 600 ya, while the calibrated analysis provides node estimates that are in line with demographic age estimates without deviating too far from our initial assumptions (Fig 2; Ithaca node: 202 ya, Tasmania node: 193 ya, third codon position clock rate: 1.75 × 10−6 substitutions/nucleotide/year).
Genetic variation in determinants of wMel density
Currently, little is known about the genetic interplay between Wolbachia and its hosts (Ikeya et al. 2009; Serbus et al. 2008; Yamada et al. 2011). Untangling these interactions will be key to understanding the evolution of these diverse symbioses. Here, we focus on one foundational phenotype that likely drives other phenotypic effects: Wolbachia titre. Wolbachia within-fly density has important implications for both partners as it correlates with levels of cytoplasmic incompatibility (Perrot-Minnot & Werren 1999; Poinsot et al. 1998; Unckless et al. 2009; Veneti et al. 2003) and potentially affects fitness benefits conferred to the fly (Osborne et al. 2012). Yet, despite its importance, we are only beginning to understand how bacterial density is regulated (Bordenstein et al. 2006; Serbus et al. 2011). Past studies have demonstrated a general effect of host genotypic variation on Wolbachia titer, but these studies have largely involved the transfer of Wolbachia infections among different host species (e.g., Bordenstein et al. 2003; McGraw et al. 2001).
We present evidence that Wolbachia titre varies among fly populations in a way that is independent of wMel phylogeny. A recent analysis of D. simulans lines (Correa & Ballard 2012) showed that while Wolbachia ovarian density is highly variable in wild-caught females, this variability rapidly declines with laboratory rearing (within 19 generations). This observation suggests that the variation we see is not caused by the lingering effects of the environment, but is rather the result of intra-specific nuclear genetic variation among these different populations. While we cannot conclude that the population-level variation reflects local adaptation and not the effects of drift, these results nevertheless point to the key role that host genotype plays in the regulation of wMel density. Future studies could leverage the natural variation we describe here as a way of exploring further phenotypes and the specific genetic factors that mediate the interactions within this model symbiont-host system.
Supplementary Material
Acknowledgements
We wish to thank Margarida Cardoso Moreira, Rayna Bell and Robert Unckless for analytical advice and discussions, as well as Nancy Chen, Jae Young Choi, Vanessa Bauer DuMont, and Jennifer Grenier for helpful comments on this manuscript. This study was supported by NIH grant R01 AI064950.
Footnotes
Author Contributions
AME and AGC designed the research. AME performed the research, analyzed the data and wrote the paper with contributions from AGC.
Data Accessibility
Tables S2-S5 (Supporting Information) provide information on the identified SNPs and indels in all mtDNA and Wolbachia genomes. Illumina DNA sequences will be submitted to Sequence Read Archive.
References
- Baele G, Lemey P, Bedford T, et al. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Molecular Biology and Evolution. 2012;29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Li WL, Drummond AJ, Suchard MA, Lemey P. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Molecular Biology and Evolution. 2013;30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Molecular Biology and Evolution. 2006a;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- Baldo L, Dunning Hotopp JC, Jolley KA, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006b;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JWO. Comparative genomics of mitochondrial DNA in Drosophila simulans. Journal of Molecular Evolution. 2000;51:64–75. doi: 10.1007/s002390010067. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, Marshall ML, Fry AJ, Kim U, Wernegreen JJ. The tripartite associations between bacteriophage, Wolbachia, and arthropods. Plos Pathogens. 2006;2:384393. doi: 10.1371/journal.ppat.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Uy JJ, Werren JH. Host genotype determines cytoplasmic incompatibility type in the haplodiploid genus nasonia. Genetics. 2003;164:223–233. doi: 10.1093/genetics/164.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie JC, Cass BN, Riegler M, et al. Evidence for Metabolic Provisioning by a Common Invertebrate Endosymbiont, Wolbachia pipientis, during Periods of Nutritional Stress. Plos Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa CC, Ballard JW. Wolbachia gonadal density in female and male Drosophila vary with laboratory adaptation and respond differently to physiological and environmental challenges. Journal of Invertebrate Pathology. 2012;111:197–204. doi: 10.1016/j.jip.2012.08.003. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. Genetic-Variation of Drosophila-Melanogaster Natural-Populations. Trends in Genetics. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friberg U, Miller PM, Stewart AD, Rice WR. Mechanisms Promoting the Long-Term Persistence of a Wolbachia Infection in a Laboratory-Adapted Population of Drosophila melanogaster. Plos One. 2011;6 doi: 10.1371/journal.pone.0016448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg AJ, Hackett SR, Harshman LG, Clark AG. A Hierarchical Bayesian Model for a Novel Sparse Partial Diallel Crossing Design. Genetics. 2010;185:361–U551. doi: 10.1534/genetics.110.115055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C, Coffey N, Houle D, et al. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. Plos Biology. 2008;6:1706–1714. doi: 10.1371/journal.pbio.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Evidence That Mutation Is Universally Biased towards AT in Bacteria. Plos Genetics. 2010;6 doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Hercus M, Dagher H. Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics. 1998;148:221–231. doi: 10.1093/genetics/148.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotopp JCD, Clark ME, Oliveira DCSG, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Broughton S, Alic N, Grandison R, Partridge L. The endosymbiont Wolbachia increases insulin/IGF-like signalling in Drosophila. Proceedings of the Royal Society B-BiologicalSciences. 2009;276:3799–3807. doi: 10.1098/rspb.2009.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilinsky Y. Coevolution of Drosophila melanogaster mtDNA and Wolbachia Genotypes. Plos One. 2013;8 doi: 10.1371/journal.pone.0054373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilinsky YY, Zakharov IK. The endosymbiont Wolbachia in Eurasian populations of Drosophila melanogaster. Russian Journal of Genetics. 2007;43:748–756. [PubMed] [Google Scholar]
- Keller A. Drosophila melanogaster's history as a human commensal. Current Biology. 2007;17:R77–R81. doi: 10.1016/j.cub.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Klasson L, Westberg J, Sapountzis P, et al. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Nikoh N, Ijichi N, Shimada M, Fukatsu T. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14280–14285. doi: 10.1073/pnas.222228199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci US A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw EA, Merritt DJ, Droller JN, O'Neill SL. Wolbachia-mediated sperm modification is dependent on the host genotype in Drosophila. Proceedings of the Royal Society B-Biological Sciences. 2001;268:2565–2570. doi: 10.1098/rspb.2001.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M, Sachs JL. Symbiont genomics, our new tangled bank. Genomics. 2010;95:129–137. doi: 10.1016/j.ygeno.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and Evolution of Heritable Bacterial Symbionts. Annual Review of Genetics. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran NA, McLaughlin HJ, Sorek R. The Dynamics and Time Scale of Ongoing Genomic Erosion in Symbiotic Bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- Mouton L, Henri H, Fleury F. Interactions between Coexisting Intracellular Genomes: Mitochondrial Density and Wolbachia Infection. Applied and Environmental Microbiology. 2009;75:1916–1921. doi: 10.1128/AEM.02677-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Pereto J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nature Reviews Genetics. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- Nikoh N, Tanaka K, Shibata F, et al. Wolbachia genome integrated in an insect chromosome: Evolution and fate of laterally transferred endosymbiont genes. Genome Research. 2008;18:272–280. doi: 10.1101/gr.7144908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MDS, Dolezal M, Schlotterer C. Extensive paternal mtDNA leakage in natural populations of Drosophila melanogaster. Molecular Ecology. 2013;22:2106–2117. doi: 10.1111/mec.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MDS, Neumeier H, Schlotterer C. Contrasting patterns of natural variation in global Drosophila melanogaster populations. Molecular Ecology. 2008a;17:44704479. doi: 10.1111/j.1365-294X.2008.03944.x. [DOI] [PubMed] [Google Scholar]
- Nunes MDS, Nolte V, Schlotterer C. Nonrandom Wolbachia Infection Status of Drosophila melanogaster Strains with Different mtDNA Haplotypes. Molecular Biology and Evolution. 2008b;25:2493–2498. doi: 10.1093/molbev/msn199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne SE, Iturbe-Ormaetxe I, Brownlie JC, O'Neill SL, Johnson KN. Antiviral Protection and the Importance of Wolbachia Density and Tissue Tropism in Drosophila simulans. Applied and Environmental Microbiology. 2012;78:6922–6929. doi: 10.1128/AEM.01727-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne SE, San Leong Y, O'Neill SL, Johnson KN. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. Plos Pathogens. 2009;5:e1000656. doi: 10.1371/journal.ppat.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Perrot-Minnot MJ, Werren JH. Wolbachia infection and incompatibility dynamics in experimental selection lines. Journal of Evolutionary Biology. 1999;12:272–282. [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, the R Development Core Team nlme: Linear and Nonlinear Mixed Effects Models. 2012 [Google Scholar]
- Poinsot D, Bourtzis K, Markakis G, Savakis C, Mercot H. Wolbachia transfer from Drosophila melanogaster into D-simulans: Host effect and cytoplasmic incompatibility relationships. Genetics. 1998;150:227–237. doi: 10.1093/genetics/150.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2011. [Google Scholar]
- Reynolds KT, Hoffmann AA. Male age, host effects and the weak expression or nonexpression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genetical Research. 2002;80:79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- Richardson MF, Weinert LA, Welch JJ, et al. Population Genomics of the Wolbachia Endosymbiont in Drosophila melanogaster. Plos Genetics. 2012;8:e1003129. doi: 10.1371/journal.pgen.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler M, Sidhu M, Miller WJ, O'Neill SL. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Current Biology. 2005;15:1428–1433. doi: 10.1016/j.cub.2005.06.069. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus LR, Casper-Lindley C, Landmann F, Sullivan W. The Genetics and Cell Biology of Wolbachia-Host Interactions. Annual Review of Genetics. 2008;42:683–707. doi: 10.1146/annurev.genet.41.110306.130354. [DOI] [PubMed] [Google Scholar]
- Serbus LR, Ferreccio A, Zhukova M, et al. A feedback loop between Wolbachia and the Drosophila gurken mRNP complex influences Wolbachia titer. J Cell Sci. 2011;124:4299–4308. doi: 10.1242/jcs.092510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev AA, He B, Baxley RM, et al. Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 2012;40:54155431. doi: 10.1093/nar/gks225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Ferreira A, Ashburner M. The Bacterial Symbiont Wolbachia Induces Resistance to RNA Viral Infections in Drosophila melanogaster. Plos Biology. 2008;6:27532763. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unckless RL, Boelio LM, Herren JK, Jaenike J. Wolbachia as populations within individual insects: causes and consequences of density variation in natural populations. Proceedings of the Royal Society B-Biological Sciences. 2009;276:2805–2811. doi: 10.1098/rspb.2009.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneti Z, Clark ME, Zabalou S, et al. Cytoplasmic incompatibility and sperm cyst infection in different Drosophila-Wolbachia associations. Genetics. 2003;164:545–552. doi: 10.1093/genetics/164.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspoor RL, Haddrill PR. Genetic Diversity, Population Structure and Wolbachia Infection Status in a Worldwide Sample of Drosophila melanogaster and D. simulans Populations. Plos One. 2011;6 doi: 10.1371/journal.pone.0026318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Zhang W, Guo LR. Evolution and Phylogeny of Wolbachia -Reproductive Parasites of Arthropods. Proceedings of the Royal Society B-Biological Sciences. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. Plos Biology. 2004;2:327–341. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Iturbe-Ormaetxe I, Brownlie JC, O'Neill SL. Functional test of the influence of Wolbachia genes on cytoplasmic incompatibility expression in Drosophila melanogaster. Insect Molecular Biology. 2011;20:75–85. doi: 10.1111/j.1365-2583.2010.01042.x. [DOI] [PubMed] [Google Scholar]
- Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Zhao XQ, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zug R, Hammerstein P. Still a Host of Hosts for Wolbachia: Analysis of Recent Data Suggests That 40% of Terrestrial Arthropod Species Are Infected. Plos One. 2012;7 doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.