Abstract
BACKGROUND:
The IL-4 receptor alpha subunit (IL-4Rα), when associated with the common gamma chain receptor, or the IL-13Rα1 subunit, transduces signals to STAT6 in response to IL-4 and IL-13 stimulations. This results in a number of cell-specific responses including Th2 differentiation, lymphocyte proliferation and IgE production. Given the prominent role of IL-4Rα in allergic disorders, several single-nucleotide polymorphisms (SNPs) have been found associated with asthma and other atopic disorders, including rs1805010 (I75V) and rs1801275 (Q576R) SNPs; however, lack of significant association have also been reported for some ethnic groups. The objective of this study was to determine whether IL-4Rα rs1805010 and rs1801275 polymorphisms are associated with asthma in patients from Saudi Arabia.
MATERIALS AND METHODS:
One hundred and ninety severe asthmatic patients (11-70 years old) and 194 healthy subjects of equivalent age range were recruited for blood donation. DNA was purified and genotyping for rs1801275 and rs1805010 polymorphisms in the IL-4Rα gene was performed by PCR amplification, followed by cycle sequencing of the purified PCR fragments using BigDye chain terminator and capillary electrophoresis.
RESULTS:
Pearson's Chi-square tests showed that the minor alleles, G, for both rs1805010 and rs1801275 SNPs, were significantly more frequent in asthmatics than in the healthy group (Yates’ P < 0.05); conversely, the major alleles, A, were significantly more frequent in healthy than in asthmatics (P < 0.05). Concerning association analysis, odds for A/G-G/G genotypes were significantly higher to be associated with asthma predisposition (rs1801275: OR = 2.12; 95% CI = 1.39-3.22; P < 0.001*; rs1805010: OR = 1.6; 95% CI = 1.01-2.53; P < 0.05*; dominant model). Analysis of gender-genotype interactions, with genders nested within A/G-G/G, indicated higher odds for females than males of significant association with asthma (rs1801275: OR = 5.19, 95% CI = 2.09-12.94*; rs1805010: OR = 3.73, 95% CI = 2.06-6.74*). Rs1805010 and rs1801275 were in linkage disequilibrium (D’ = 0.27; P < 0.0004*), with G-G haplotype being more frequent in asthmatics than in healthy subjects (OR = 2.43, 95% CI = 1.59-3.71*).
CONCLUSIONS:
The risk alleles, G, of IL-4Rα rs1805010 and rs1801275 SNPs and corresponding A/G-G/G genotypes were significantly associated with asthma predisposition in asthmatics from Saudi Arabia.
Keywords: Asthma, allergy, association, genotype, IL-4 receptor alpha, susceptibility, STAT6
Introduction
The IL-4 receptor alpha (IL-4Rα, also called CD124) gene encodes a single-pass trans-membrane subunit protein which, when associated with the common gamma chain (γc, also called Interleukin-2 Receptor Gamma subunit, IL-2Rγ) constitute the IL-4Rα/IL-2Rγ complex receptor for its ligand, IL-4 cytokine.[1,2] IL-4Rα subunit can also form another receptor complex with the trans-membrane IL-13Rα1 subunit, serving as docking site for IL-13 cytokine signaling.[3] Although ubiquitously expressed, the highest levels of IL-4Rα expression are observed in B lymphocytes; other cells expressing relatively high levels are CD4+ and CD8+ lymphocytes, as well as monocytes, dendritic cells and macrophages.[4,5,6,7] The high expression levels of IL-4Rα in CD19+ B lymphocytes is congruous with its active role in isotype class-switch recombination and IgG4 and IgE production, in response to IL-4/IL-13 or allergen stimulations.[8,9] Thus, the crucial importance of IL-4Rα in IgE production was demonstrated in mice, in which inactivation of the receptor results in complete suppression of the IgE immune response.[10] In addition, IL-4Rα and its ligand, IL-4, stimulate T lymphocyte proliferation, and promotes apoptosis resistance and differentiation of naïve CD4+ cells into Th-2 lineage cells.[4,11] Furthermore, IL-4Rα is a vital component of the allergen sensitization pathway and allergy induction, and plays an essential role in the pathogenesis of asthma through IL-13 and IL-4 cytokines signaling.[12,13,14] Not surprisingly, IL4Rα is currently considered a key therapeutic target, where its partial blockage could help relieve symptoms in atopic asthmatics.[2,12,15,16]
Considerable interest and effort have been dedicated to determine the possible contribution of single nucleotide polymorphisms (SNPs) on the susceptibility or predisposition of atopic patients to develop asthma.[17,18,19,20] In fact, genetic association studies identified a large number of genes having significant association with asthma susceptibility, including IL-4Rα.[21,22,23,24] However, the challenge remains to replicate those findings in different ethnic populations, and to understand the gene-environment interactions, which very likely contribute to the heterogeneity in the responses of patients to asthma medications.[25,26] The IL-4Rα gene is highly polymorphic and several SNPs have been associated with different immune-related disorders, including atopy and asthma, although not without discrepancies.[8,10,27,28,29] The rs1805010 polymorphism (A→G) is a type of ‘gain-of-function’ mutation, which results in substitution of isoleucine for valine (I75V) in the extracellular domain of IL-4Rα subunit; in vitro, this mutant receptor with enhanced IL-4 signaling appears to promote a sustained phosphorylation and hence, keep STAT6 transcription factor abnormally active, even upon withdrawal of IL-4 stimulation.[28,30] The rs1801275 (A→G) is another gain-of-function mutation also aberrantly enhancing signal transduction, which results in glutamine being substituted by arginine (Q576R) in the intracellular domain of IL-4Rα; it is situated beside the critically important tyrosine-2 residue, which is part of the docking site for STAT6.[31] Since its identification, the Q576R mutation was found to significantly predispose people to atopy, and was also associated with elevated CD23 levels in response to IL-4 stimulation.[31,32]
However, discrepancies in genetic association studies for various ethnic groups have also been reported. A number of studies found significant association of SNPs with allergy-related diseases and autoimmune disorders; for instance, both the rs1805010 (A/A genotype) and the rs1801275 (A/A and A/G genotypes) were reportedly associated with incidence of rheumatoid arthritic nodules.[36] Other studies found these SNPs significantly associated with asthma and asthma-related symptoms.[29,36,38,39,40,41] Yet some studies failed to find significant association with asthma,[10,42] or with other immune disorders such as eczema[43] and diabetes.[44] Possibly, discrepancies among the studies could result from the influence of both the environment and distinct genetic backgrounds among ethnic groups. Gene association studies in asthmatics from Saudi Arabia are very limited;[22] therefore, we sought to examine the possible association of rs1805010 and rs1801275 SNPs with asthma prevalence in asthmatic patients from Saudi Arabia.
Materials and Methods
One hundred and ninety severe asthmatic patients (age from 11-70 years old), and 194 healthy subjects of equivalent age range were recruited for blood donation at the King Khalid University Hospital (KKUH), Riyadh, Saudi Arabia. Identification for severe asthma was determined based on symptoms and physician diagnosis following the criteria described by the American Thoracic Society.[45] All adult donors and the parents/guardians of children donors (<18 years old) were requested to sign a fully informed and written consent, approved by the Ethics Review Committee of the King Khalid Hospital. This study was approved by the Ethical Committee of the KKUH. Peripheral blood was extracted and DNA was purified from 2 mL of whole blood, using a QIAamp DNA Blood Midi Kit (QIAGEN Sciences, Maryland, USA), following the manufacturer's instructions. DNA concentrations were determined using a spectrophotometer (Nano Drop 2000c, Thermo Scientific, Wilmington, USA). Genotyping for rs1801275 and rs1805010 polymorphisms in the IL-4Rα gene was performed by PCR amplification, followed by sequencing. PCR amplification of the two regions of the gene encompassing the variants was done using Taq Polymerase (New England Bio Labs, Quick-Load Taq 2X Master Mix) and specific primers; primers for rs1801275: 5’-AGG AAT GAG GTC TTG GAA AGG (right) and 5’-AGG AAG TAG AAC CCG AGA TGC (left); primers for rs1805010: 5’-AGC CCA CAG GTC CAG TGT AT (right) and 5’-GAA GAG CTT GAT GCG GTT CC (left). Cycle sequencing of the purified PCR fragments was performed using the Big Dye Terminator kit and capillary electrophoresis (Applied Biosystems, Sequencer model 3730xl), following the manufacturer's instructions. Allele and genotype frequencies were analyzed by Pearson's Chi-square tests; Yates’ P-values were also calculated using an interactive calculation tool;[46] P-values < 0.05 were considered statistically significant. Hardy-Weinberg equilibrium tests, linkage disequilibrium and analysis of interaction with covariate gender (both, crude analysis and adjusted for covariate gender) were performed using SNPStats tool.[47] Odds ratios (OR) and confidence intervals (95% CI) were calculated using an interactive calculation tool.[48] Best fit genetic model tests (co-dominant, dominant, recessive and over-dominant) were performed using the Chi-square test, and the odds ratios (OR) and 95% confidence intervals (CI) were calculated with the SNPStats,[47] or with Bland's interactive calculator.[48]
Results
IL-4Rα polymorphisms and asthma susceptibility in a Saudi Arabian population
Analysis of both allele and genotype frequencies for rs1801275 and rs1805010 polymorphisms indicated that both SNPs were distributed in Hardy-Weinberg equilibrium. Chi-square (X2) calculations tests of goodness of fit for allele frequencies and genotype frequencies showed that rs1801275 (Q576R) polymorphism was significantly associated with asthma predisposition [Table 1]. The minor allele, G, (21% of the total, including healthy and asthmatics) of rs1801275 SNP was significantly more frequent in asthmatics (7% more than expected) than in the healthy group (7% less than the expected); likewise, the major allele, A, was significantly more frequent in the healthy group (26% greater than the expected value) than in asthmatics (25.8% smaller than the expected value) (Yates’ X2 = 14.1; Yates’ P-value = 0.0002*) [Table 1]. Concerning genotype frequencies, Yates’ X2 tests demonstrated significant differences between asthmatics and healthy subjects: The most important deviation from expected values was the minor G/G genotype, which was more frequent in the asthmatics (56% greater than the expected) than in the healthy group (Yates’ X2 = 14.7; Yates’ P-value = 0.0006*) [Table 1].
Table 1.
Allele and genotype frequencies for IL-4Ralpha SNPs
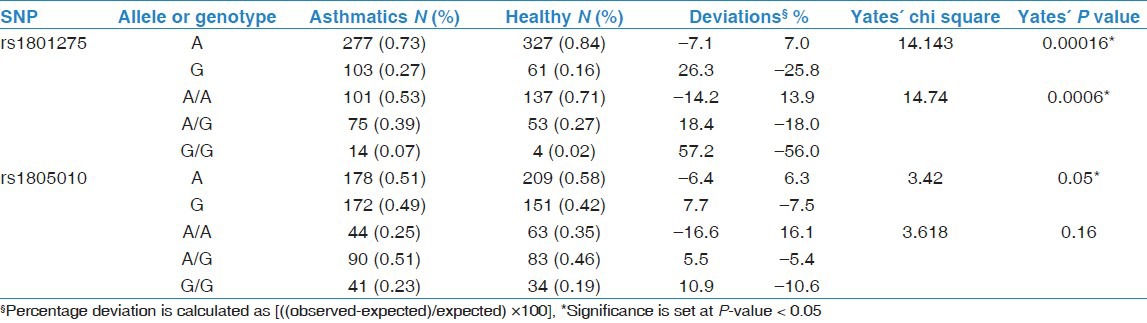
Association of rs1805010 (I75V) allele frequency with asthma was marginal (P = 0.05*). Concerning genotypes, A/A was more frequent in the healthy subjects (16% greater than expected) whereas G/G was more frequent in the asthmatics (11% greater than expected); however, these differences were not significant (Yates’ X2 = 3.61; Yates’ P-value = 0.16) [Table 1].
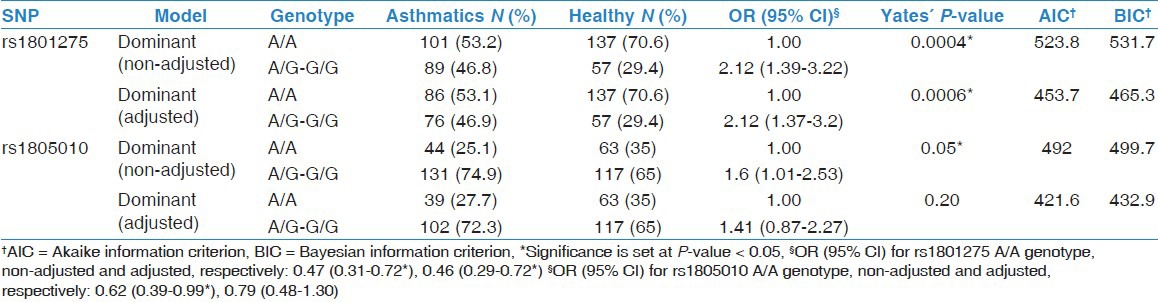
A non-adjusted association analysis of genotypes in which several genetic models were tested indicated that both IL-4Rα SNPs were significantly associated with asthma prevalence (P < 0.05) [Table 2]. In this analysis, the best fit genetic model was the Dominant model for both IL-4Rα SNPs, suggesting that a single copy of the minor allele, G, is enough to increase the risk of asthma (e.g., A/G and G/G genotypes would have same effect) (rs1801275: OR = 2.12; 95% CI = 1.39-3.22; P < 0.001*; rs1805010: OR = 1.6; 95% CI = 1.01-2.53; P < 0.05*). To rule out a possible bias due to incomplete matching by gender on recruitment, an analysis adjusted by gender was performed: Only the association of rs1801275 (Q576R) with asthma remained significant [Table 2].
Table 2.
Association of IL-4R-alpha SNPs with asthma (non-adjusted and adjusted by gender)
Interaction of genotype frequencies with gender and susceptibility to asthma
We next analyzed whether the interaction of SNP with gender could alter the risk of asthma; this analysis involved testing several genetic models of inheritance [Table 3]. In the best fit Dominant model, with genotypes nested within genders, the major A/A genotype of rs1801275 (Q576R) had significantly lower odds to be associated with asthma in both genders (males: OR = 0.50, 95% CI = 0.30-0.84*; females: OR = 0.34, 95% CI = 0.12-0.90*) [Table 3]. Likewise, the A/G-G/G genotypes combined had significantly higher odds to be associated with asthma, in both genders, suggesting that allele G increases the risk of asthma (males: OR = 2.0, 95% CI = 1.19-3.39*; females: OR = 2.92, 95% CI = 1.11-7.71*). In the analysis done with genders nested within A/A genotype, females but not males, had higher odds of significant association with asthma. Similarly, when genders were nested within A/G-G/G genotype, females had higher odds of significant association with asthma, whereas males had lower odds of being associated with asthma [Table 3]. These results indicate that this SNP is more prominent in asthmatic females than males.
Table 3.
Interaction analysis of single IL-4Rα SNP with covariate gender; with genotypes nested within genders (left) and genders nested within genotypes (right); Dominant model tested (see Table 2)
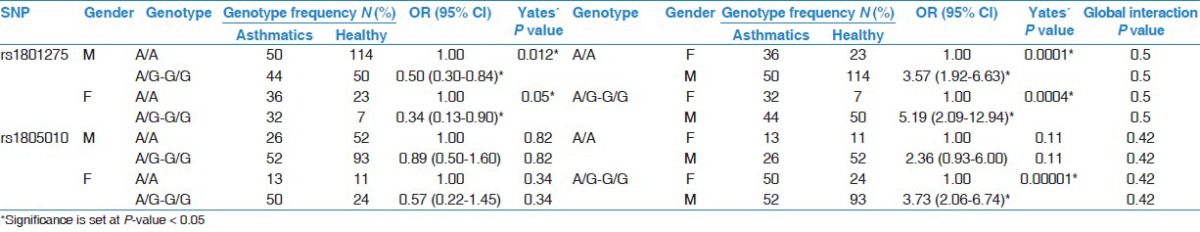
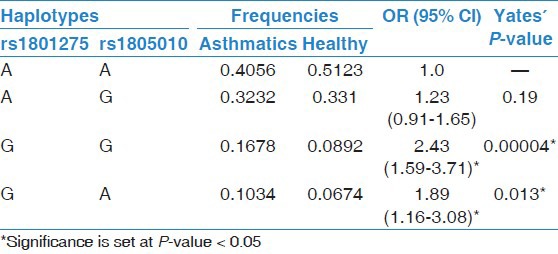
In the case of rs1805010 (I75V) SNP, data analysis with genders nested within A/G-G/G genotype, indicated that females had significantly higher odds of association with asthma; no other significant interactions were observed [Table 3]. Furthermore, both SNPs were found in linkage disequilibrium (D’ = 0.27; P = 0.003*). In addition, the G-G haplotype was significantly more frequent in the asthmatics than in healthy group (OR = 2.43, 95% CI = 1.59-3.71; Yates’ P < 0.001*), suggesting that when G alleles from both SNPs are inherited together, they tend to increase the susceptibility or risk of asthma [Table 4].
Table 4.
Haplotype frequencies for IL-4Rα
Discussion
The IL-4 receptor alpha is a vital element through which extracellular IL-4 and IL-13 cytokines signaling are transduced into alternative cell-specific responses, affecting many processes including the immune response.[1,2,4] This receptor mediates in B lymphocytes the class-switch recombination mechanism and generation of immunoglobulins IgE and IgG, which happens in response to IL4/IL-13 and allergens/antigens.[4,14,16] The crucial role of IL-4Rα in the pathogenesis of allergic disorders has driven multiple studies aiming toward the development of therapeutic compounds to modulate its function.[2,12,15,16]
In this study, we found that two SNPs of the IL-4Rα gene, rs1801275 and rs1805010, were significantly linked with increased risk of asthma in a Saudi Arabian population. Thus, our findings are in agreement with previous reports on different ethnic backgrounds.[9,28,31,35,37,38,39,40,41] In this study, we confirmed that the minor allele, G, was found more frequently in asthmatics than in healthy subjects; likewise, the major allele, A, was found more frequently in the healthy subjects than in asthmatics. Genotype frequencies with allele G, namely A/G and G/G, were significantly more frequent in asthmatics, thus supporting a possible link of the mutant allele, G, with asthma prevalence. The dominant effect of the mutant G alleles predicted from our study was previously reported in in vitro studies.[33,35] Importantly, linkage disequilibrium analysis confirmed a significant association of V75/R576 with asthma, suggesting that both risk mutations inherited together enhance the predisposition to asthma in the Saudi population; it was reported previously in atopic asthmatics from USA that both V75/R576 SNPs inherited together can result in greater association with atopic asthma than either allele alone.[35] A theoretical explanation for the apparent dominant effect of risk allele, G, may result from the direct interaction of IL-4Rα subunit with STAT6, which is then activated by phosphorylation.[28,30] On the other hand, IL-4Rα polymorphisms alone may not always influence the susceptibility to disease;[29,42,49] however, the combined effect of IL-4Rα Q576R (rs1801275) SNP with mutant alleles in other genes could contribute significantly to disease susceptibility, as determined by in vitro functional studies,[10,33] albeit not without discrepancies.[34] The consequence of these findings is that common variants alone cannot account for a given disease; asthma is a complex disease where many genes are involved.[19,20,23] Given the complexity of asthma, it could be speculated that in an individual, multiple SNPs in several genes could act in concert or synergy to produce a significant effect.[17]
A great deal of research has been dedicated to the functional relevance of this receptor in many inflammatory disorders, including asthma.[8,21,36,43,44] Although we have a fairly good understanding of the molecular mechanisms by which the IL-4Rα and its ligands IL-4 and IL-13 induce a stimulatory cell signaling, we still have yet to determine the impact or relevance of the many mutation variants of this gene in asthma and other diseases. The IL-4Rα is highly polymorphic, but only a few of these mutations have been functionally studied. Nevertheless, many polymorphisms occurring within the coding regions of genes do not always produce a significant phenotype change (on health or in development).[50] Both rs1801275 and rs1805010 variants are considered ‘gain-of-function’ mutations, where they abnormally enhance or promote STAT6 signaling: In vitro experimental evidence suggests that these mutations contribute to maintain STAT6 in a phosphorylated state, hence active even after the ligand has been removed, which is observed as increased CD23 expression.[30,33,35] However, in vivo evidence supporting that these particular mutations contribute to enhanced STAT6 activity is missing: One study showed that the rs1801275 mutant was not associated with serum IgE levels in Chinese asthmatic children.[49] Moreover, functional studies testing the effect of a single SNP not always have demonstrated an association with response.[34,35] Another source of uncertainty is that genetic association analyses do not consistently agree on whether a given variant (SNP) contributes significantly to asthma predisposition, and the question remains unanswered of whether this is influenced by the ethnic background and/or environment.[17,25] In this respect, the Saudi Arabian population has a high frequency of asthma, reaching 11%.[22,51] Consistently, allele variants of IL-17A, IL-17F and ADAM33 genes were found associated with asthma in this population;[22,52] however, more studies are needed to further identify critical genes influencing asthma susceptibility in the Saudi population.
Conclusion
Although much remains to be done, our data provide important information supporting the association of two IL-4Rα variants with asthma in this ethnic group. This could be useful in future studies to assess the risk of individuals to asthma and allergy-related diseases, with the aim to implement a personalized, management medical program for individuals with high genetic risk.
Acknowledgments
We would like to thank Kristine Alba-Conception, Mary A. Esperas and Amer Jamhawi for technical help. This study was supported by grants from the National Plan for Sciences and Technology, King Saud University, Riyadh, Saudi Arabia (grant number 10-MED1224-02) and Merck Frost (grant number 38259).
Footnotes
Source of Support: National Plan for Sciences and Technology, King Saud University, Riyadh, Saudi Arabia (grant number 10-MED1224-02) and Merck Frost (grant number 38259)
Conflict of Interest: None declared.
References
- 1.Zhang JL, Buehner M, Sebald W. Functional epitope of common gamma chain for interleukin-4 binding. Eur J Biochem. 2002;269:1490–9. doi: 10.1046/j.1432-1033.2002.02796.x. [DOI] [PubMed] [Google Scholar]
- 2.Andrews AL, Holloway JW, Holgate ST, Davies DE. IL-4 receptor alpha is an important modulator of IL-4 and IL-13 receptor binding: Implications for the development of therapeutic targets. J Immunol. 2006;176:7456–61. doi: 10.4049/jimmunol.176.12.7456. [DOI] [PubMed] [Google Scholar]
- 3.Tabata Y, Khurana Hershey GK. IL-13 receptor isoforms: Breaking through the complexity. Curr Allergy Asthma Rep. 2007;7:338–45. doi: 10.1007/s11882-007-0051-x. [DOI] [PubMed] [Google Scholar]
- 4.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: Signaling mechanisms and biologic function. Annu Rev Immunol. 1999;17:701–38. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 5.Pangault C, Amé-Thomas P, Ruminy P, Rossille D, Caron G, Baia M, et al. Follicular lymphoma cell niche: Identification of a preeminent IL-4-dependent T(FH)-B cell axis. Leukemia. 2010;24:2080–9. doi: 10.1038/leu.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan C, Taylor AA, Coburn MZ, Marino JH, Van De Wiele CJ, Teague TK. Ten-color flow cytometry reveals distinct patterns of expression of CD124 and CD126 by developing thymocytes. BMC Immunol. 2011;12:36. doi: 10.1186/1471-2172-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford AQ, Dasgupta P, Mikhailenko I, Smith EM, Noben-Trauth N, Keegan AD. Adoptive transfer of IL-4Rα + macrophages is sufficient to enhance eosinophilic inflammation in a mouse model of allergic lung inflammation. BMC Immunol. 2012;13:6. doi: 10.1186/1471-2172-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Di Rienzo A, Ober C. A population genetics study of single nucleotide polymorphisms in the interleukin 4 receptor alpha (IL4RA) gene. Genes Immun. 2001;2:128–34. doi: 10.1038/sj.gene.6363746. [DOI] [PubMed] [Google Scholar]
- 9.Howard TD, Koppelman GH, Xu J, Zheng SL, Postma DS, Meyers DA, et al. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet. 2002;70:230–6. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franjkovic I, Gessner A, König I, Kissel K, Bohnert A, Hartung A, et al. Effects of common atopy-associated amino acid substitutions in the IL-4 receptor alpha chain on IL-4 induced phenotypes. Immunogenetics. 2005;56:808–17. doi: 10.1007/s00251-004-0763-1. [DOI] [PubMed] [Google Scholar]
- 11.Chomarat P, Banchereau J. An update on interleukin-4 and its receptor. Eur Cytokine Netw. 1997;8:333–44. [PubMed] [Google Scholar]
- 12.Zavorotinskaya T, Tomkinson A, Murphy JE. Treatment of experimental asthma by long-term gene therapy directed against IL-4 and IL-13. Mol Ther. 2003;7:155–62. doi: 10.1016/s1525-0016(02)00050-3. [DOI] [PubMed] [Google Scholar]
- 13.Nishikubo K, Murata Y, Tamaki S, Sugama K, Imanaka-Yoshida K, Yuda N, et al. A single administration of interleukin-4 antagonistic mutant DNA inhibits allergic airway inflammation in a mouse model of asthma. Gene Ther. 2003;10:2119–25. doi: 10.1038/sj.gt.3302131. [DOI] [PubMed] [Google Scholar]
- 14.Nieuwenhuizen N, Herbert DR, Lopata AL, Brombacher F. CD4+ T cell-specific deletion of IL-4 receptor alpha prevents ovalbumin-induced anaphylaxis by an IFN-gamma-dependent mechanism. J Immunol. 2007;179:2758–65. doi: 10.4049/jimmunol.179.5.2758. [DOI] [PubMed] [Google Scholar]
- 15.Corren J, Busse W, Meltzer EO, Mansfield L, Bensch G, Fahrenholz J, et al. A randomized, controlled, phase 2 study of AMG 317, an IL-4Ralpha antagonist, in patients with asthma. Am J Respir Crit Care Med. 2010;181:788–96. doi: 10.1164/rccm.200909-1448OC. [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Lu J. Retrovirus-mediated delivery of an IL-4 receptor antagonist inhibits allergic responses in a murine model of asthma. Sci China Life Sci. 2010;53:1215–20. doi: 10.1007/s11427-010-4067-4. [DOI] [PubMed] [Google Scholar]
- 17.Barnes KC. Genomewide association studies in allergy and the influence of ethnicity. Curr Opin Allergy Clin Immunol. 2010;10:427–33. doi: 10.1097/ACI.0b013e32833de6ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poon A, Litonjua AA, Laprise C. Relevance and implication of genetic determinants to asthma pathophysiology. Curr Opin Allergy Clin Immunol. 2011;11:407–13. doi: 10.1097/ACI.0b013e32834a9540. [DOI] [PubMed] [Google Scholar]
- 19.Koppelman GH, Sayers I. Evidence of a genetic contribution to lung function decline in asthma. J Allergy Clin Immunol. 2011;128:479–84. doi: 10.1016/j.jaci.2011.05.036. [DOI] [PubMed] [Google Scholar]
- 20.Cookson WO, Moffatt MF. Genetics of complex airway disease. Proc Am Thorac Soc. 2011;8:149–53. doi: 10.1513/pats.201101-003MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Postma DS, Kerkhof M, Boezen HM, Koppelman GH. Asthma and chronic obstructive pulmonary disease: Common genes, common environments? Am J Respir Crit Care Med. 2011;183:1588–94. doi: 10.1164/rccm.201011-1796PP. [DOI] [PubMed] [Google Scholar]
- 22.Al-Khayyat AI, Al-Anazi M, Warsy A, Vazquez-Tello A, Alamri AM, Halwani R, et al. T1 and T2 ADAM33 single nucleotide polymorphisms and the risk of childhood asthma in a Saudi Arabian population: A pilot study. Ann Saudi Med. 2012;32:479–86. doi: 10.5144/0256-4947.2012.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol. 2008;8:169–82. doi: 10.1038/nri2257. [DOI] [PubMed] [Google Scholar]
- 24.Mathias RA, Grant AV, Rafaels N, Hand T, Gao L, Vergara C, et al. A genome-wide association study on African-ancestry populations for asthma. J Allergy Clin Immunol. 2010;125:336–46.e4. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G, Goldblatt J, LeSouëf PN. Does the relationship between IgE and the CD14 gene depend on ethnicity? Allergy. 2008;63:1411–7. doi: 10.1111/j.1398-9995.2008.01804.x. [DOI] [PubMed] [Google Scholar]
- 26.Portelli M, Sayers I. Genetic basis for personalized medicine in asthma. Expert Rev Respir Med. 2012;6:223–36. doi: 10.1586/ers.12.9. [DOI] [PubMed] [Google Scholar]
- 27.Ober C, Leavitt SA, Tsalenko A, Howard TD, Hoki DM, Daniel R, et al. Variation in the interleukin 4-receptor alpha gene confers susceptibility to asthma and atopy in ethnically diverse populations. Am J Hum Genet. 2000;66:517–26. doi: 10.1086/302781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wenzel SE, Balzar S, Ampleford E, Hawkins GA, Busse WW, Calhoun WJ, et al. IL4R alpha mutations are associated with asthma exacerbations and mast cell/IgE expression. Am J Respir Crit Care Med. 2007;175:570–6. doi: 10.1164/rccm.200607-909OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murk W, Walsh K, Hsu LI, Zhao L, Bracken MB, Dewan AT. Attempted replication of 50 reported asthma risk genes identifies a SNP in RAD50 as associated with childhood atopic asthma. Hum Hered. 2011;71:97–105. doi: 10.1159/000319536. [DOI] [PubMed] [Google Scholar]
- 30.Ford AQ, Heller NM, Stephenson L, Boothby MR, Keegan AD. An atopy-associated polymorphism in the ectodomain of the IL-4R(alpha) chain (V50) regulates the persistence of STAT6 phosphorylation. J Immunol. 2009;183:1607–16. doi: 10.4049/jimmunol.0803266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruse S, Japha T, Tedner M, Sparholt SH, Forster J, Kuehr J, et al. The polymorphisms S503P and Q576R in the interleukin-4 receptor alpha gene are associated with atopy and influence the signal transduction. Immunology. 1999;96:365–71. doi: 10.1046/j.1365-2567.1999.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med. 1997;337:1720–5. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- 33.Mitsuyasu H, Yanagihara Y, Mao XQ, Gao PS, Arinobu Y, Ihara K, et al. Cutting edge: Dominant effect of Ile50Val variant of the human IL-4 receptor alpha-chain in IgE synthesis. J Immunol. 1999;162:1227–31. [PubMed] [Google Scholar]
- 34.Wang HY, Shelburne CP, Zamorano J, Kelly AE, Ryan JJ, Keegan AD. Cutting edge: Effects of an allergy-associated mutation in the human IL-4R alpha (Q576R) on human IL-4-induced signal transduction. J Immunol. 1999;162:4385–9. [PubMed] [Google Scholar]
- 35.Risma KA, Wang N, Andrews RP, Cunningham CM, Ericksen MB, Bernstein JA, et al. V75R576 IL-4 receptor alpha is associated with allergic asthma and enhanced IL-4 receptor function. J Immunol. 2002;169:1604–10. doi: 10.4049/jimmunol.169.3.1604. [DOI] [PubMed] [Google Scholar]
- 36.Burgos PI, Causey ZL, Tamhane A, Kelley JM, Brown EE, Hughes LB, et al. Association of IL4R single-nucleotide polymorphisms with rheumatoid nodules in African Americans with rheumatoid arthritis. Arthritis Res Ther. 2010;12:R75. doi: 10.1186/ar2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandford AJ, Chagani T, Zhu S, Weir TD, Bai TR, Spinelli JJ, et al. Polymorphisms in the IL4, IL4RA, and FCERIB genes and asthma severity. J Allergy Clin Immunol. 2000;106:135–40. doi: 10.1067/mai.2000.107926. [DOI] [PubMed] [Google Scholar]
- 38.Hesselmar B, Enelund AC, Eriksson B, Padyukov L, Hanson LÅ, Aberg N. The heterogeneity of asthma phenotypes in children and young adults. J Allergy (Cairo) 2012. 2012 doi: 10.1155/2012/163089. 163089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi WA, Kang MJ, Kim YJ, Seo JH, Kim HY, Kwon JW, et al. Gene-gene interactions between candidate gene polymorphisms are associated with total IgE levels in Korean children with asthma. J Asthma. 2012;49:243–52. doi: 10.3109/02770903.2012.660294. [DOI] [PubMed] [Google Scholar]
- 40.Battle NC, Choudhry S, Tsai HJ, Eng C, Kumar G, Beckman KB, et al. Ethnicity-specific gene-gene interaction between IL-13 and IL-4Ralpha among African Americans with asthma. Am J Respir Crit Care Med. 2007;175:881–7. doi: 10.1164/rccm.200607-992OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beghé B, Hall IP, Parker SG, Moffatt MF, Wardlaw A, Connolly MJ, et al. Polymorphisms in IL13 pathway genes in asthma and chronic obstructive pulmonary disease. Allergy. 2010;65:474–81. doi: 10.1111/j.1398-9995.2009.02167.x. [DOI] [PubMed] [Google Scholar]
- 42.Melén E, Umerkajeff S, Nyberg F, Zucchelli M, Lindstedt A, Gullstén H, et al. Interaction between variants in the interleukin-4 receptor alpha and interleukin-9 receptor genes in childhood wheezing: Evidence from a birth cohort study. Clin Exp Allergy. 2006;36:1391–8. doi: 10.1111/j.1365-2222.2006.02577.x. [DOI] [PubMed] [Google Scholar]
- 43.Miyake Y, Tanaka K, Arakawa M. Case-control study of eczema in relation to IL4Rα genetic polymorphisms in Japanese women: The Kyushu Okinawa Maternal and Child Health Study. Scand J Immunol. 2013;77:413–8. doi: 10.1111/sji.12043. [DOI] [PubMed] [Google Scholar]
- 44.Erlich HA, Lohman K, Mack SJ, Valdes AM, Julier C, Mirel D, et al. Association analysis of SNPs in the IL4R locus with type I diabetes. Genes Immun. 2009;10(Suppl 1):S33–41. doi: 10.1038/gene.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.American Thoracic Society. Proceedings of the ATS workshop on refractory asthma: Current understanding, recommendations, and unanswered questions. Am J Respir Crit Care Med. 2000;162:2341–5. doi: 10.1164/ajrccm.162.6.ats9-00. [DOI] [PubMed] [Google Scholar]
- 46.Preacher KJ. Calculation for the chi-square test: An interactive calculation tool for chi-square tests of goodness of fit and independence. 2001, Computer software. [Last accessed on 2014 Jan 6]. Available from: http://quantpsy.org .
- 47.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: A web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–9. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 48.Bland JM, Altman DG. Statistics notes. The odds ratio. BMJ. 2000;320:1468. doi: 10.1136/bmj.320.7247.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang AM, Li HL, Hao P, Chen YH, Li JH, Mo YX, et al. Association of Q576R polymorphism in the interleukin-4 receptor gene with serum IgE levels in children with asthma. Zhongguo Dang Dai Er Ke Za Zhi. 2006;8:109–12. [PubMed] [Google Scholar]
- 50.Prans E, Kingo K, Traks T, Silm H, Vasar E, Kõks S. Copy number variations in IL22 gene are associated with Psoriasis vulgaris. Hum Immunol. 2013;74:792–5. doi: 10.1016/j.humimm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Al-Moamary MS, Al-Hajjaj MS, Idrees MM, Zeitouni MO, Alanezi MO, Al-Jahdali HH, et al. The Saudi Initiative for asthma. 2009;4:216–23. doi: 10.4103/1817-1737.56001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bazzi MD, Sultan MA, Al Tassan N, Alanazi M, Al-Amri A, Al-Hajjaj MS, et al. Interleukin 17A and F and asthma in Saudi Arabia: Gene polymorphisms and protein levels. J Investig Allergol Clin Immunol. 2011;21:551–5. [PubMed] [Google Scholar]


