Abstract
Objective: The Unified Medical Language System's (UMLS's) Semantic Network's (SN's) two-tree structure is restrictive because it does not allow a semantic type to be a specialization of several other semantic types. In this article, the SN is expanded into a multiple subsumption structure with a directed acyclic graph (DAG) IS-A hierarchy, allowing a semantic type to have multiple parents. New viable IS-A links are added as warranted.
Design: Two methodologies are presented to identify and add new viable IS-A links. The first methodology is based on imposing the characteristic of connectivity on a previously presented partition of the SN. Four transformations are provided to find viable IS-A links in the process of converting the partition's disconnected groups into connected ones. The second methodology identifies new IS-A links through a string matching process involving names and definitions of various semantic types in the SN. A domain expert is needed to review all the results to determine the validity of the new IS-A links.
Results: Nineteen new IS-A links are added to the SN, and four new semantic types are also created to support the multiple subsumption framework. The resulting network, called the Enriched Semantic Network (ESN), exhibits a DAG-structured hierarchy. A partition of the ESN containing 19 connected groups is also derived.
Conclusion: The ESN is an expanded abstraction of the UMLS compared with the original SN. Its multiple subsumption hierarchy can accommodate semantic types with multiple parents. Its representation thus provides direct access to a broader range of subsumption knowledge.
The Unified Medical Language System (UMLS) is a large terminology whose Metathesaurus (META) contains approximately 900,000 concepts.1,2,3 The Semantic Network (SN) has 135 (134 at the time of our study) semantic types and provides a high-level abstract view of the META.4,5 As expressed in the study by McCray and Nelson,6 “The Semantic Network encompasses and provides a unifying structure for the Metathesaurus constituent vocabularies.”
The SN contains a hierarchy consisting of two trees rooted at the semantic types Event and Entity,* respectively.7 This hierarchy is based on the IS-A (subsumption) relationship, which connects a more specialized semantic type (a child) to a more generalized semantic type (its parent). Each semantic type, except for Event and Entity, is a specialization of exactly one semantic type (its parent) and inherits semantic relationships only from this unique parent.
Many researchers have suggested that concept-oriented8,9 and logic-based10,11,12,13 approaches are beneficial for creating categorical terminological structures,14 especially when their purpose is to support, as the UMLS does, cross-thesaurus mappings.14,15 However, the SN does not provide sufficient logic-based structures to apply such methods; we must, therefore, seek alternative methods to improve the consistency and utility of the SN.
Although the SN's tree structure is easy to implement and process, it is restrictive in that it does not allow multiple parent types when warranted. There are, in fact, some semantic types that could naturally be specializations of more than one semantic type. For example, Gene or Genome could conceptually be a child of two semantic types: one is its current parent Fully Formed Anatomical Structure; the other is Molecular Sequence. Hence, Gene or Genome should inherit from Molecular Sequence the semantic relationship result_of to Mental Process. In a case such as this, the modeling of the SN omits an aspect of current medical knowledge.
In this article, we present two methodologies to structurally enrich the SN by transforming its hierarchy into a directed acyclic graph (DAG) structure that allows multiple parents. The methodologies are based on the identification of viable new IS-A relationships currently not included in the SN. These omissions may have been due to the tree-structure restriction on the SN, noted previously in this article. We add new semantic types to the SN as necessary to accommodate the new multiple subsumption framework. In the first methodology, the identification of new IS-As is guided by imposing connectivity on an existing partition of the SN.16 In the second methodology, the identification is based on partial string matching between names of semantic types and the definitions of other semantic types. These identified potential IS-A relationships are then reviewed by a domain expert to decide whether they are semantically valid. With the addition of these new IS-A relationships, we get a new DAG version of the SN that we refer to as the Enriched Semantic Network (ESN). Furthermore, we obtain a partition of the ESN consisting of groups, each of which has a tree structure. The ESN serves as an enhanced abstraction of the UMLS. The accompanying partition enables the creation of a metaschema,17 an additional abstract layer of the ESN that can help users in their orientation to the UMLS.
Methods to Enhance the SN'S IS-A Hierarchy
Imposing Connectivity on an SN Partition
Basis
McCray et al.16 presented a partition of the SN into 15 groups, with each group representing a subject area. Six principles that such a partition should satisfy were proposed. One of them, semantic validity, can be assessed by seeing if a group's semantic types together with their IS-A links form a connected subgraph of the SN. We refer to this as the “connectivity property.” Because the SN's IS-A hierarchy consists of two trees, such a connected subgraph in the current SN must form a tree with a unique root.
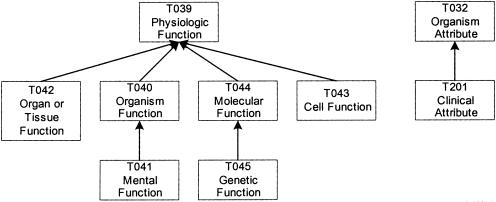
In the analysis of McCray et al.,16 it was noted that: “In some cases, it was not possible to resolve anomalies in our attempt to create a coherent and semantically valid set of groupings.” In fact, some of the partition's groups do not satisfy the connectivity property. Such groups contain a forest, comprising two or more trees, or perhaps isolated semantic types. (Some groups have both.) For example, the Physiology† group contains a forest of two trees (▶). There are no hierarchical relationships between a semantic type of one tree and a semantic type of the other tree. Therefore, the Physiology group is not connected.
Figure 1.
Physiology group.
In our previous work,18 we presented an alternative partition of the SN based on the sets of relationships exhibited by the semantic types. In our technique, we required that the hierarchy of each group of the partition be a tree exhibiting the connectivity property. In this way, we obtained a partition that is strictly semantically uniform. A difference between the partition of McCray et al.16 and that of Chen et al.18 is that the connectivity is only a preferred, not required, property in McCray et al.,16 whereas it is required and enforced in Chen et al.18
In our semantic technique, we use the partition of McCray et al.16 as a basis for augmenting the SN's hierarchy, and, in particular, for identifying new viable IS-A relationships. The basic idea is to bridge the gap between the two partitioning techniques by imposing the connectivity property on the partition of McCray et al.16 To convert the disconnected groups of McCray et al.16 into connected groups, we need to identify and insert additional IS-As. This will yield a first version of our desired multiple subsumption hierarchy and an accompanying partition. Analysis of the definitions of semantic types within each disconnected group will guide the introduction of the new IS-A links. In this context, we will use four kinds of transformations with respect to the groups of McCray et al.16 Another methodology using exact string matching will then be used in a following subsection.
Four transformations to identify new IS-A links
The possible transformations that can be applied to disconnected groups to make them connected are listed in the following. The choice of which transformation to use is based on reviews of the definitions of all semantic types within a group.
IS-A Addition Transformation: Identify a viable IS-A and add it to transform the group into a connected subtree.
Split Transformation: Split a group into several groups, each of which is either a rooted tree structure or can be transformed into a rooted tree structure by adding IS-A relationships.
Root-addition Transformation: Create a new semantic type that will be an ancestor of all roots in the group. Make the new semantic type the group's root by adding additional IS-A relationships from all the roots of the group's connected components (either directly or through more new semantic types, if necessary).
Root-moving Transformation: Locate a semantic type (from another group) that is a lowest common ancestor of the roots of all the disconnected group's subtrees and/or isolated semantic types. Move that lowest common ancestor into the disconnected group, making it the root of the group and thereby connecting the group. Also, move all the new root's existing descendants into the group.
The new network obtained by applying these transformations is called the Enriched Semantic Network (ESN). It has a DAG structure rather than a two-tree structure. We now demonstrate the various transformations and show their impact on different disconnected groups.
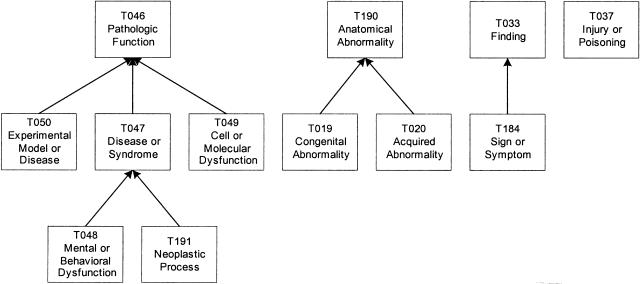
As demonstrations of the IS-A addition transformation and split transformation, we consider the group Disorders (▶). This group contains 12 semantic types, 11 of which belong to three trees rooted at Pathologic Function, Anatomical Abnormality, and Finding, respectively. Injury or Poisoning is an isolated member of the group. Clearly, Disorders does not satisfy connectivity.
Figure 2.
Disorders group.
The IS-A addition transformation is first applied to this group to connect Injury or Poisoning to the tree rooted at Pathologic Function. Actually, Injury or Poisoning should have a subsumption relationship to Disease or Syndrome and inherit its semantic relationships. Thus, we add an IS-A link to capture this.
Because in the original SN, Disease or Syndrome is a descendant of Phenomenon or Process, the original IS-A from Injury or Poisoning to Phenomenon or Process can be removed because it can be inferred transitively through the new IS-A link from Injury or Poisoning to Disease or Syndrome.
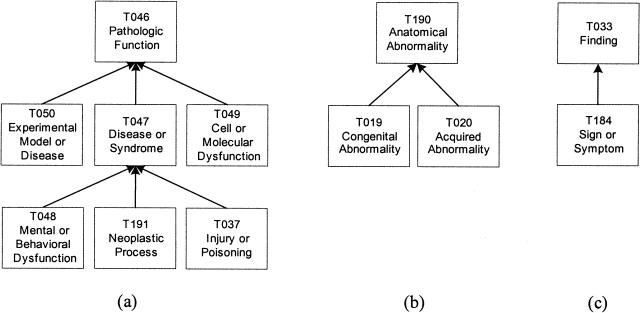
At this point, the group is still a collection of disconnected trees. To rectify this, we apply the split transformation to form three new groups. According to the definitions of the 12 semantic types, we find that Pathologic Function and its six descendant semantic types, including the new descendant Injury or Poisoning, emphasize phenomenon or process and are in the Event tree, whereas the remaining semantic types emphasize an entity or object and are in the Entity tree. Furthermore, Anatomical Abnormality and its children are descendants of Physical Object, whereas Finding and its child are conceptual entities. So, it is natural to partition this group into three smaller connected groups, each comprising a tree. These groups, Pathologic Function, Anatomical Abnormality, and Finding, are shown in ▶. Note that using a root-addition transformation for all or any two trees is not an option because this new root could not be placed anywhere in the SN due to the differences in the contents of the trees. The new groups are named after their roots.
Figure 3.
Three new groups: (a) Pathologic Function, (b) Anatomical Abnormality, and (c) Finding (via IS-A Addition transformation and Split transformation).
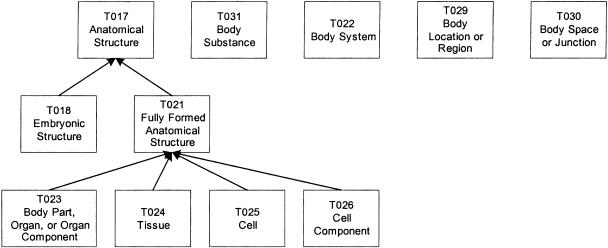
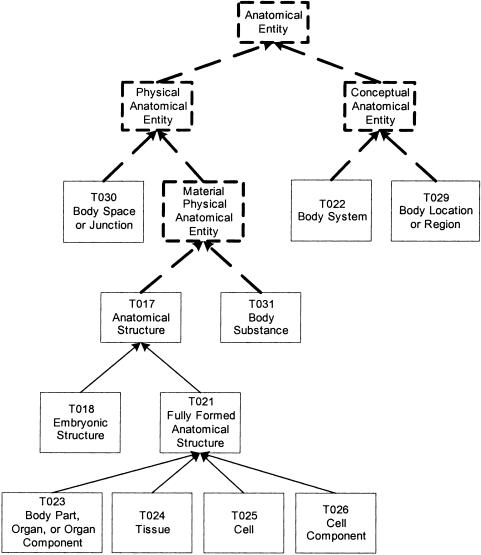
In the next example, the Anatomy group undergoes a root-addition transformation; that is, we add new semantic types to make the group connected. The group contains a tree of seven semantic types rooted at Anatomical Structure and four isolated semantic types: Body Substance, Body System, Body Location or Region, and Body Space or Junction (▶). In carrying out this transformation, we follow the analysis of Michael et al.19 for definitions of anatomical concepts. For example, the new semantic type Material Physical Anatomical Entity is defined as “IS-A Physical Anatomical Entity which has a mass.”19 Body Substance is not an Anatomical Structure because it does not have a three-dimensional shape, but it is a Material Physical Anatomical Entity because it has mass. Thus, both Body Substance and Anatomical Structure are made children of the new semantic type Material Physical Anatomical Entity.
Figure 4.
Anatomy group.
Furthermore, Body Space or Junction is not a Material Physical Anatomical Entity, but it is a Physical Anatomical Entity (defined in Michael et al.19 to have spatial dimensions). Hence, both Body Space or Junction and Material Physical Anatomical Entity are made children of the newly introduced Physical Anatomical Entity, which in turn IS-A Physical Object. The original IS-A from Anatomical Structure to Physical Object is cut because it can be inferred from the new IS-A from Anatomical Structure to Physical Anatomical Entity.
On the other hand, Body Location or Region and Body System do not have either mass or spatial dimension and thus cannot be descendants of Physical Anatomical Entity. Nevertheless, both obviously should belong to the Anatomy group. Following Michael et al.,19 we introduce the new semantic type Conceptual Anatomical Entity, which in turn IS-A Conceptual Entity, to complement Physical Anatomical Entity and serve as the parent of Body Location or Region and Body System.
Finally, the new semantic type Anatomical Entity is added as the parent of both Physical Anatomical Entity and Conceptual Anatomical Entity. In turn, Anatomical Entity IS-A Entity. In this way, the whole Anatomy group is transformed into the new group Anatomical Entity (▶). The dashed rectangles in the figure represent the newly added semantic types, and the dashed arrows represent the newly added IS-A links. We note that each of the four new semantic types should have at least the corresponding concepts suggested in Michael et al.19 assigned to it. (These concepts have been submitted to the NLM for inclusion in the next UMLS release [Rosse C, personal communication, 2002].)
Figure 5.
Anatomical Entity group.
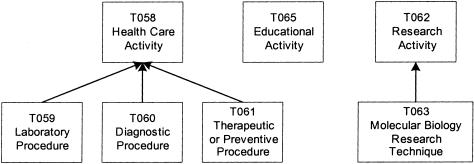
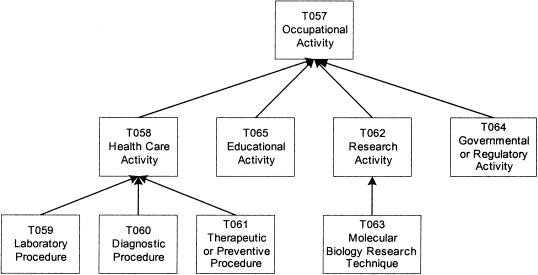
In the next example, the root-moving transformation is applied to the disconnected Procedures group to make it connected. The group contains seven semantic types, with two trees rooted at Health Care Activity and Research Activity, respectively, and the isolated Educational Activity (▶). These three are children of Occupational Activity, which has another child, Governmental or Regulatory Activity. Both of these semantic types, in turn, belong to the Activities and Behaviors group. In the context of the UMLS, these five semantic types refer to health care-related issues. They describe activities of health care professionals. Thus, Occupational Activity, the lowest ancestor of the seven semantic types in the group, and its child Governmental or Regulatory Activity are moved to this group. By doing this, the group is transformed into the new Occupational Activity connected group (▶).
Figure 6.
Procedures group.
Figure 7.
Occupational Activity group.
String Matching
Additional IS-A links can be found by using string matching involving names and definitions of various semantic types in the SN. To be more formal, we define a string match as follows:
Definition (string match)
A string match from a semantic type T1 to another semantic type T2 is a triple (T1; T2; S) such that S is a string appearing both in the definition of T1 and in the name of T2. S is called the common string and must contain one or more (not necessarily consecutive) complete words (ignoring case).
For example, the definition of Plant contains the word “organism,” which happens to be the name of a semantic type. Hence, a string match (Plant, Organism, “organism”) exists.
The motivation for using this kind of string matching to find viable new IS-A links is based on the evaluation of string matches among the 132 pairs of semantic types that currently have IS-A relationships between them in the SN. By analyzing the definitions of the children in the pairs, we found that there are string matches from 88 children to their respective parents. The string match (Plant, Organism, “organism”) is one of them. Thus, the sensitivity of this approach with known IS-A links is 67%. This finding leads us to the following observation.
Observation
If T1 IS-A T2, then there is a high likelihood of a string match from T1 to T2.
This leads us to formulate the inverse hypothesis.
Hypothesis
If there is a string match from one semantic type to another, then it is likely to imply a viable subsumption relationship between them.
Based on this hypothesis, we developed the string matching method to identify additional viable IS-A relationships not already appearing in the SN. Our methodology is a human–computer interactive methodology and contains three steps:
Preprocess names and definitions of semantic types to obtain the input file;
Apply the “AllMatches” algorithm to the input file to get all string matches; and
Manually review all resulting string matches and determine which constitute additional viable IS-A links between semantic types.
In step 1, we use some common techniques from the data mining and information retrieval fields.20
Stop-words: All stop-words such as “a,” “the,” “of,” “for,” “with,” and so on are removed from names and definitions.
Verb variant processing: All verbs and verb variants are removed from definitions of semantic types. In the string matching, we do not consider verbs and verb variants. The reason is that most semantic types' names consist only of nouns, adjectives, and adverbs.
Lexical normalization: The Specialist Lexicon (coupled with highly efficient “lexical variant generator” code)21 is applied to stem-word variations. All adjectives and adverbs are converted to nouns, and all plurals are converted to singular forms. Also, uppercase letters are changed to corresponding lowercase.
In step 2, the following AllMatches algorithm is used to find string matches between any two semantic types not currently connected by a single IS-A link or a path of such links. The input file to the algorithm contains the names and definitions of semantic types after the preprocessing step.
In the description of the AllMatches algorithm, we assume that T1, T2, …, Tm are all semantic types in the SN. (In the 2002 version, m = 134.) We use the notation DEF(Ti) to represent the definition of the semantic type Ti, 1≤i≤134, after preprocessing. NAME(Ti) is used to represent the name of Ti, in the form of a string, after preprocessing. For example, suppose Ti = Anatomical Structure, which is defined as: “a normal or pathological part of the anatomy or structural organization of an organism.” After preprocessing, NAME(Ti) = “anatomy structure” and DEF(Ti) = “normal pathology part anatomy structure organization organism.”
In the following AllMatches algorithm, we use a list L to hold all common strings. We also use the following functions defined for lists:
Length(): Return the number of elements in the list
Retrieve(k): Retrieve the kth element of the list
AllMatches algorithm: Find all string matches in the SN.
For (i = 1 to m)
For (all Tj, 1≤j≤m & j ≠ i)
If (Tj is not the parent or an ancestor of Ti)
{ L = FindCommonStrings(DEF(Ti),NAME(Tj));
//write string matches to the output file
For (k = 1 to L.Length())
{ S = L.Retrieve(k); // get kth element of the list
write (Ti; Tj; S) to output file;
} }
The function FindCommonStrings(R1, R2) is used to find all common strings involving a given pair of strings R1 and R2. During a call, R1 is the definition of a semantic type Ti in a string format, and R2 is the name of a semantic type Tj as a string. For each pair (Ti, Tj) that has no direct IS-A relationship or directed path of IS-A relationships between its components, we call FindCommonStrings(DEF(Ti), NAME(Tj)) to get all possible common strings between DEF(Ti) and NAME(Tj). Each such common string is inserted into L. We say that a match M is redundant if its constituent common string S is a substring of another match's common string (again, ignoring case). FindCommonStrings(DEF(Ti), NAME(Tj)) identifies the redundant matches and does not return them. So, L contains no redundant common strings. Finally, all string matches (Ti; Tj; S) are written to the output file. After AllMatches has been executed, we have a file containing all string matches between pairs of semantic types not connected by IS-A relationships in the SN.
As an example, consider Enzyme whose definition is “a complex chemical, usually a protein, that is produced by living cells and which catalyzes specific biochemical reactions.” The AllMatches algorithm finds three string matches:
(Enzyme, Cell, “cell”)
(Enzyme, Cell Component, “cell”)
(Enzyme; Amino Acid, Peptide, or Protein; “protein”)
In step 3, an expert is called on to review all resulting string matches to find new IS-A links not currently appearing in the SN. These newly discovered IS-A links can then be added to the ESN. As it happens, in the case of the three string matches involving Enzyme, the third match implies the existence of a new IS-A link, because any enzyme must be a kind of protein. Hence, Enzyme IS-A Amino Acid, Peptide, or Protein.
As noted previously in this article, the sensitivity of the string matching approach, when applied to known IS-A links, is 67%. To determine the sensitivity of our method for detecting unknown IS-A links, we established a gold standard by performing a manual review of randomly generated relationship pairs.
Results
Results of Imposing Connectivity on the Partition
Besides the three disconnected groups described in Methods, the original partition of McCray et al.16 contains six other disconnected groups. ▶ presents the six groups and the six transformations applied to them.
Table 1.
The Six Transformations Applied to Six Disconnected Groups of the Partition of McCray et al.16
| Old Group Name | Isolated Semantic Types and/or Trees in a Disconnected Group | Transformation Type | Transformations Applied | New Group Name |
|---|---|---|---|---|
| Chemicals and Drugs | Clinical Drug | Split transformation | Split into two connected groups; the group Clinical Drug contains just one semantic type | Two groups: Chemical; Clinical Drug |
| Devices | Research Device; Medical Device | Root-moving transformation | Move Manufactured Object from the Objects group and make it the new root of the Devices group | Manufactured Object |
| Genes and Molecular Sequences | Gene or Genome | IS-A addition transformation | Add (Gene or Genome IS-A Molecular Sequence) link | Molecular Sequence |
| Living Beings | {Organism; Fungus; Alga; Virus; Human; Plant; Archaeon; Reptile; Rickettsia or Chlamydia; Amphibian; Mammal; Fish}; {Group; Family Group; Age Group; Population Group; Professional or Occupational Group; Patient or Disabled Group} | Split transformation | Split into two smaller connected groups | Two groups: Organism; Group |
| Phenomena | Laboratory or Test Result | IS-A addition transformation | Add (Laboratory or Test Result IS-A Phenomenon or Process) link | Phenomenon or Process |
| Physiology | {Organism Attribute; Clinical Attribute} | IS-A addition transformation | Add (Organism Attribute IS-A Physiologic Function) link | Physiologic Function |
For each such group, ▶ shows the isolated semantic types or trees that existed in the group and the transformations used. In the second column of ▶, each tree in the group is denoted by placing its constituent semantic types in braces “{}.” In the fourth column, we use the notation (A IS-A B) to denote a single IS-A link that was added to the group, where A and B are semantic types. The new groups are named after their respective roots.
Overall, using the four kinds of transformations, we converted all disconnected groups into new connected groups, each with an internal tree structure. During this process, a total of ten transformations were applied: the IS-A addition transformation was used four times; the split transformation was used three times; the root-addition transformation was used once (on the Anatomy group); and the root-moving transformation was used twice. Note that multiple transformations might have been applied to a single group. For example, see the Disorders group. The application of the four transformations yielded the preliminary ESN with 15 new IS-A links. Its DAG structure allows semantic types to have multiple parents.
A total of 19 disjoint groups, which together constitute a partition of the ESN, was also obtained. See ▶, where we use an asterisk (*) to denote a group different from that originally appearing in McCray et al.16 Each group is a connected subgraph of the ESN. Hence, the partition satisfies the connectivity property preferred for semantic validity.
Table 2.
Partition of the ESN into 19 Connected Groups
| Group | No. of STs | Group | No. of STs |
|---|---|---|---|
| Anatomical Abnormality* | 3 | Anatomical Entity* | 15 |
| Chemical* | 25 | Clinical Drug* | 1 |
| Conceptual Entity | 12 | Entity* | 4 |
| Event* | 7 | Finding* | 2 |
| Geographic Area | 1 | Group* | 6 |
| Manufactured Object* | 3 | Molecular Sequence | 5 |
| Occupation or Discipline | 2 | Occupational Activity* | 9 |
| Organism* | 17 | Organization | 4 |
| Pathologic Function* | 7 | Phenomenon or Process | 6 |
| Physiologic Function | 9 |
STs = Semantic Types; ESN = Enriched Semantic Network.
Results of String Matching
For our manual review, we randomly selected 550 (3%) of the 17,396 possible pairs of semantic types for which no ancestor/descendant relationship currently exists. Neither of the two reviewers judged any of the 550 pairs to represent a true parent–child relationship. This corresponds to a prevalence of unknown pairs of 0%, with a 95% confidence interval of 0–0.54%.
A total of 665 string matches were found by our algorithm. Only five of these were judged to represent true parent–child relationships, for a precision of 0.75%. However, these five positive results suggest a prevalence of 0.029% (five of 17,396), which is within the 95% confidence interval of our gold standard analysis.
Our semantic method resulted in the addition of 15 new IS-A links. However, 11 of these links involved the addition of new semantic types, leaving four previously undiscovered IS-A links. One of these, Gene or Genome IS-A Molecular Sequence, was also detected by the string matching method. Thus, a total of eight new parent–child relationships were discovered (prevalence eight of 17,396 = 0.046%, still within the range found by the gold standard). The string matching method detected five of the eight true parent–child relationships discovered by both methods, yielding a sensitivity (or recall) of 62.5%. At the maximum prevalence suggested by the 95% confidence interval (0.54%), the sensitivity could be as low as 5.3%.
Let us review the four additional IS-A links. One is the new IS-A link from Enzyme to Amino Acid, Peptide, or Protein, which was demonstrated above.
Another example relates to Receptor, for which there were five string matches:
(Receptor, Cell Component, “cell”)
(Receptor, Cell, “cell”)
(Receptor, Anatomical Structure, “structure”)
(Receptor; Amino Acid, Peptide, or Protein; “protein”)
(Receptor, Hormone, “hormone”)
In accordance with the review of the domain expert, an IS-A link from Receptor to Cell Component was added. The other string matches did not imply IS-A links.
The third valid IS-A link involves Vitamin, which had four matches:
(Vitamin, Pharmacologic Substance, “substance”)
(Vitamin, Organic Chemical, “organic chemical”)
(Vitamin, Body Substance, “substance”)
(Vitamin, Animal, “animal”)
Based on the domain expert's review, two IS-A links were added: one IS-A from Vitamin to Pharmacologic Substance, and another IS-A from Vitamin to Organic Chemical.
Summary of Results of Two Methodologies
After adding the new IS-A links derived by our two methodologies, we get the new ESN. Compared with the original SN, the ESN has four new semantic types and 19 new IS-A links. Two IS-A links appearing in the SN were not included in the ESN. Hence, the ESN has 149 IS-A links and 138 semantic types, among which 12 semantic types (approximately 8%) have multiple parents, giving the ESN a DAG-structured IS-A (subsumption) hierarchy. See ▶ for these 12 semantic types and their parents.
Table 3.
Semantic Types (STs) with Multiple Parents in the ESN
| Child ST | Old Parent ST | New Parent ST | New Parent ST |
|---|---|---|---|
| Body Location or Region | Spatial Concept | Conceptual Anatomical Entity | — |
| Body Space or Junction | Conceptual Entity | Physical Anatomical Entity | — |
| Body Substance | Substance | Material Physical Anatomical Entity | — |
| Body System | Functional Concept | Conceptual Anatomical Entity | — |
| Conceptual Anatomical Entity | — | Conceptual Entity | Anatomical Entity |
| Enzyme | Biologically Active Substance | Amino Acid, Peptide, or Protein | — |
| Gene or Genome | Fully Formed Anatomical Structure | Molecular Sequence | — |
| Laboratory or Test Result | Finding | Phenomenon or Process | — |
| Organism Attribute | Conceptual Entity | Physiologic Function | — |
| Physical Anatomical Entity | — | Physical Object | Anatomical Entity |
| Receptor | Biologically Active Substance | Cell Component | — |
| Vitamin | Biologically Active Substance | Organic Chemical | Pharmacologic Substance |
ESN = Enriched Semantic Network.
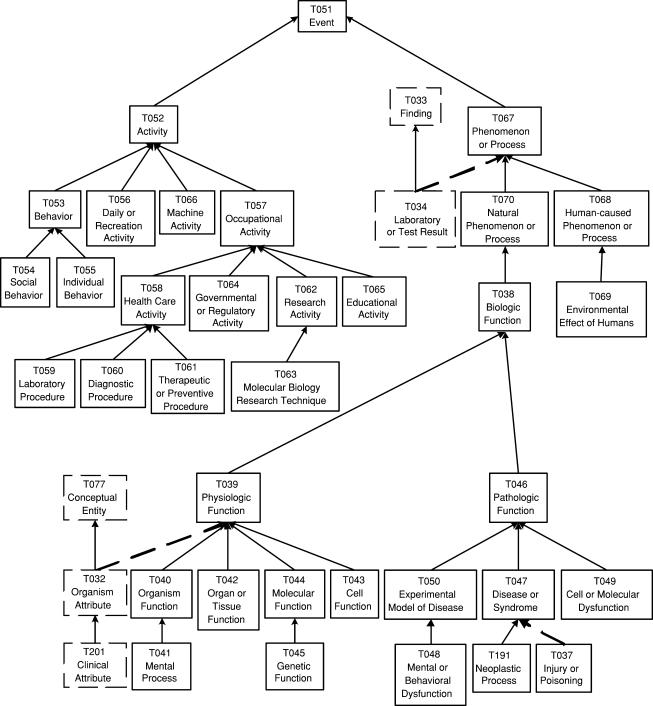
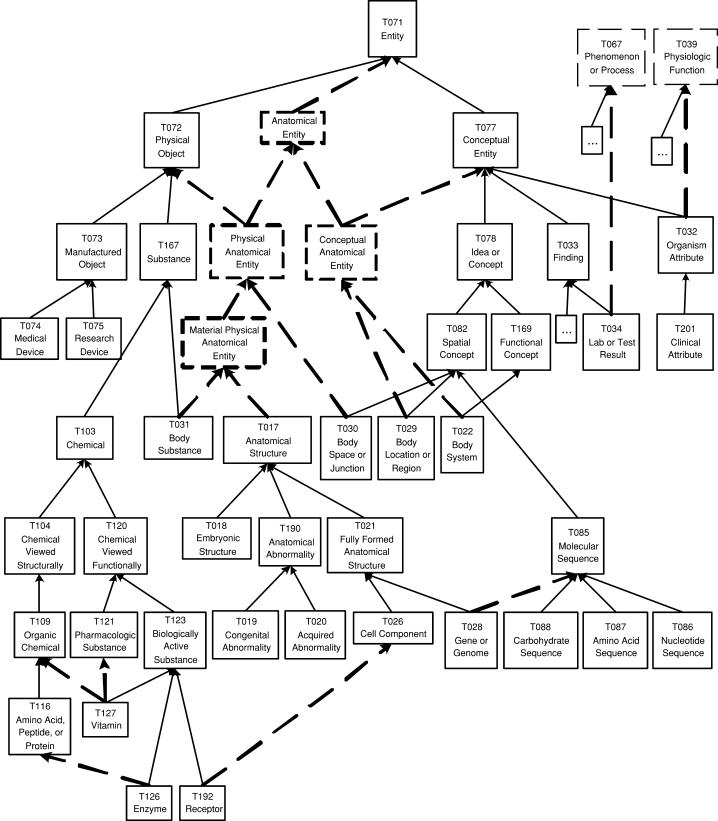
▶ shows the portion of the ESN's hierarchy rooted at Event, and ▶ shows part of the portion rooted at Entity. To emphasize the changes from the original SN, we use dashed arrows to denote the new IS-A links and thick dashed rectangles to denote new semantic types. Thin dashed rectangles denote semantic types that originally resided in the other tree of the SN. Ellipses in a rectangle indicate that the names of one or several semantic types are not shown due to lack of space.
Figure 8.
Event Portion of the Enriched Semantic Network.
Figure 9.
Part of the Entity portion of the Enriched Semantic Network.
Discussion
Advantages of the Enriched Semantic Network
The ESN has 12 semantic types with multiple parents. As it happens, most such semantic types are leaves or parents of leaves. As such, the changes are local, not influencing other semantic types. An exception is the modeling of the four new semantic types, Anatomical Entity, its two children Conceptual Anatomical Entity and Physical Anatomical Entity, and the child of the latter, Material Physical Anatomical Entity. This is the most visible difference from the original SN's two-tree structure, because it happens close to the root Entity rather than at the bottom levels of the SN. As such, it is not a local change.
The ESN has a number of advantages over the original SN. The multiple subsumption hierarchy enables better modeling of IS-A relationships for those semantic types having multiple parents. In the ESN, some semantic types will have more semantic relationships than they had in the SN. Specifically, semantic types with multiple parents will inherit relationships independently from each of those parents. Thus, such a semantic type will have a larger relationship set than before. For example, Organism Attribute and its child Clinical Attribute will now have a relationship result_of to Anatomical Abnormality. This relationship is inherited by Organism Attribute from its new parent Physiologic Function, and further inherited by its child Clinical Attribute.
One might consider the introduction of multiple inheritance as a potential problem in that inconsistent information from different parents might be inherited. However, when the placement of a concept into two classes is semantically correct, then the inheritance of definitional attributes from multiple parents is, by definition, also correct. Multiple inheritance will allow the identification of inconsistencies that were already there implicitly; it will not introduce new ones.
The addition of the IS-A links helps to expose missing classifications of concepts of the META to semantic types. Let us demonstrate this with regard to the concepts assigned to the semantic type Vitamin.
We checked all 1,204 concepts from the META assigned to Vitamin and found that 957 are also assigned to Pharmacologic Substance. One of them is also assigned to Antibiotic, which is a child of Pharmacologic Substance. The other 246 concepts are not assigned to Pharmacologic Substance. For example, the concept Folate‡ is not assigned to Pharmacologic Substance. However, we know that some drugs, for example, vitamins given to pregnant women, contain folate to prevent possible congenital deficiencies of the baby. Hence, Folate should indeed be assigned to Pharmacologic Substance. As a matter of fact, all the remaining 246 concepts should also have been assigned to Pharmacologic Substance because all vitamins can be ingredients of drugs.
Similarly, all concepts in Vitamin should also be assigned to Organic Chemical or one of its descendants. Among the 1,204 concepts assigned to Vitamin, 735 are also assigned to Organic Chemical or children of Organic Chemical. However, there are 469 concepts assigned to Vitamin that are not assigned to Organic Chemical or to any of its children. An example is 24,25-Dihydroxyvitamin D, which is a kind of vitamin D that is helpful for the absorption of calcium and is certainly an organic chemical. In fact, all these 469 concepts should have been assigned to Organic Chemical.
Another advantage of the multiple-parent hierarchy is that it can simplify the assignment of META's concepts to semantic types. An important rule promoted by the SN's designers states that a concept should be explicitly assigned to the most specialized possible semantic type in the SN's IS-A hierarchy.6 Suppose a concept was assigned to two semantic types, T1 and T2, that originally had no IS-A path between them in the SN. If in the ESN there is a direct IS-A link or path from T1 to T2 (i.e., T2 is now a parent or ancestor of T1), then the assignment of the concept to T2 is considered redundant22,23 and should be removed because it can be inferred from the assignment to T1.
As an example, consider the new IS-A link from Vitamin to Pharmacologic Substance. After adding this IS-A link, the 957 assignments of Vitamin's concepts to Pharmacologic Substance should be removed because Vitamin is now more specialized than Pharmacologic Substance in the network. The 957 concepts should only be assigned to Vitamin, because implicit assignments to Pharmacologic Substance can be inferred via the new IS-A. After the addition of Vitamin IS-A Organic Chemical, the 735 assignments to Organic Chemical should also be removed for the same reason.
Let us note that in the preceding discussion, we proposed both the assignment of additional concepts to Organic Chemical and then their subsequent removal. However, the proposed additions were strictly in the context of the current SN hierarchy, in which simultaneous assignment to Vitamin and Organic Chemical is not redundant and is in fact warranted. In the ESN, with Vitamin now being a child of Organic Chemical, such assignments become redundant and therefore unnecessary. This further supports the validity of the new IS-A link and demonstrates that the ESN hierarchy requires fewer explicit assignments of META's concepts to the semantic types.
The partition of the ESN can enable the design of a metaschema,24 a higher-level abstraction network that can aid user orientation. Among other things, a metaschema allows a user to focus on a subject area of interest without losing sight of the overall ESN layout. Previously, we developed metaschemas for the current SN. The connected partition of the ESN allows us to do the same for the ESN.17
Regarding limitations, our first methodology was applied only to the partition presented in McCray et al.,16 and decisions were made with respect to the current definitions of semantic types. Of course, there are many possible partitions of the SN. If we consider other partitions, we might decide on different IS-A links.
The string matching methodology is dependent on the definitions of the semantic types. We realize that the current definitions are not necessarily the only ones possible for the given semantic types. Another team of designers might come up with slightly different definitions. Because we used the exact wording of the definitions, our results may very well be altered by alternative definitions. Furthermore, the average time complexity of the algorithm is approximately O(n2), and this limits its scalability. It is thus applicable only to a compact upper-level abstraction ontology (like the SN), not a full-scale ontology. For example, the algorithm would be very time-consuming if it were applied to find string matches for the META's concepts. Word-level synonymy (or phrase synonymy) was not considered in our algorithm. If used, it could increase the string match cases and maybe the number of new viable IS-A links found. However, this would likely erode the algorithm's efficiency, which is already low, and might increase the number of false-positives, which is already high.
Evaluation
Without an exhaustive examination of all 17,396 pairs of unrelated semantic types, it is impossible to know the exact prevalence of undiscovered parent–child relationships. However, all of the methods we used (semantic modeling, manual review, and string matching) suggest that the number of such relationships is very low. In the absence of a precise figure for prevalence, estimating the sensitivity of our automated methods is impossible. However, the semantic modeling revealed 15 links and the string matching revealed five links; either of these counts represents a significant contribution to the number of links in the SN; taken together as 19 (because one was repeated), they increase the number of links by 14.3%.
Although at first glance, the precision of the string matching method (0.75%) appears poor, applying it to the SN reduces the number of semantic-type pairs that must be manually reviewed by 94%. We note that there is no inherent reason why a string match from the definition of a semantic type to the name of another semantic type would necessarily indicate an IS-A link. The match could indicate another kind of connection such as a semantic relationship. Our hypothesis was that if there is a shared string, then the likelihood of a parent–child relationship is substantially higher. The results support our hypothesis.
Conclusion
We have enhanced the UMLS's SN hierarchy by adding new IS-A links and new semantic types to accommodate multiple parents. We obtained a new semantic network that has a DAG structure instead of a two-tree structure. This new semantic network, containing 138 semantic types and 149 IS-A relationships, is referred to as the ESN. The ESN can express cases of multiple subsumption for some semantic types. Furthermore, a partition of the ESN comprising 19 groups was derived; each group in the partition exhibits connectivity and semantic uniformity. This new partition can enable the design of a metaschema17 to help further improve user orientation to the ESN.
This research was partially supported by contract #N01-1-3543 from the National Library of Medicine (NLM), and by the New Jersey Commission for Science and Technology.
Semantic types appear in boldface type.
Group names appear in italic type.
Concept names appear in small caps.
References
- 1.Campbell KE, Oliver DE, Shortliffe EH. The Unified Medical Language System: toward a collaborative approach for solving terminologic problems. J Am Med Inform Assoc. 1998;5:12–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Humphreys BL, Lindberg DAB, Schoolman HM, Barnett GO. The Unified Medical Language System: an informatics research collaboration. J Am Med Inform Assoc. 1998;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.US Department of Health and Human Services, National Institutes of Health, National Library of Medicine. Washington, DC: Unified Medical Language System (UMLS), 2003.
- 4.McCray AT, Hole WT. The scope and structure of the first version of the UMLS Semantic Network. Proc Annu Symp Comput Appl Med Care. 1990:126–30.
- 5.McCray AT. Representing biomedical knowledge in the UMLS Semantic Network. In: Broering NC, (ed). High-Performance Medical Libraries: Advances in Information Management for the Virtual Era. Westport, CT: Mekler, 1993, pp. 45–55.
- 6.McCray AT, Nelson SJ. The representation of meaning in the UMLS. Methods Inf Med. 1995;34:193–201. [PubMed] [Google Scholar]
- 7.McCray AT. UMLS Semantic Network. Proc Annu Symp Comput Appl Med Care. 1989:503–7.
- 8.Chute CG. The Copernican era of healthcare terminology: a re-centering of health information systems. Proc AMIA Annu Symp. 1998:68–73. [PMC free article] [PubMed]
- 9.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37:394–403. [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell KE, Das AK, Musen MA. A logical foundation for representation of clinical data. J Am Med Inform Assoc. 1994;1:218–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell KE, Cohn SP, Chute CG, Shortliffe EH, Rennels G. Scalable methodologies for distributed development of logic-based convergent medical terminology. J Am Med Inform Assoc. 1998;37:426–39. [PubMed] [Google Scholar]
- 12.Rector AL, Rogers J, Roberts A, Wroe C. Scale and context: issues in ontologies to link health and bio-informatics. Proc AMIA Annu Symp. 2002:642–4. [PMC free article] [PubMed]
- 13.Rector AL. Thesauri and formal classifications: terminologies for people and machines. Methods Inf Med. 1998;37:501–9. [PubMed] [Google Scholar]
- 14.Rossi Mori A, Consorti F, Galeazzi E. Standards to support development of terminological systems for healthcare telematics. Methods Inf Med. 1998;37:551–63. [PubMed] [Google Scholar]
- 15.Dessena S, Rossi Mori A, Galeazzi E. Building cross-thesauri with the support of UMLS. In: Cesnik B, (ed). Medinfo 98. Amsterdam, The Netherlands: IOS Press, 1998, pp. 654–9. [PubMed]
- 16.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Medinfo 2001. Amsterdam: IOS Press, 2001, pp 171–5. [PMC free article] [PubMed]
- 17.Zhang L, Perl Y, Halper M, Geller J. Designing metaschemas for the UMLS Enriched Semantic Network. J Biomed Inform. 2003;36:433–49. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Perl Y, Halper M, Geller J, Gu H. Partitioning the UMLS Semantic Network. IEEE Trans Inf Technol Biomed. 2002;6:102–8. [DOI] [PubMed] [Google Scholar]
- 19.Michael J, Mejino J, Rosse C. The role of definitions in biomedical concept representation. In: Bakken S (ed). Proc AMIA Annu Symp. 2001:463–7. [PMC free article] [PubMed]
- 20.Han J, Kamber M. Data Mining: Concepts and Techniques. San Francisco, CA: Morgan Kaufmann Publishers, 2000.
- 21.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Applic Med Care. 1994:235–9. [PMC free article] [PubMed]
- 22.Gu H, Perl Y, Geller J, Halper M, Liu L, Cimino JJ. Representing the UMLS as an OODB: modeling issues and advantages. J Am Med Inform Assoc. 2000;7:66–80. Selected for reprint in Haux R, Kulikowski C (eds). Yearbook of Medical Informatics. Rotterdam: International Medical Informatics Association, 2001, pp 271–285.
- 23.Peng Y, Halper M, Perl Y, Geller J. Auditing the UMLS for redundant classifications. Proc AMIA Annu Symp. 2002:612–6. [PMC free article] [PubMed]
- 24.Perl Y, Chen Z, Halper M, Geller J, Zhang L, Peng Y. The cohesive metaschema: a higher-level abstraction of the UMLS Semantic Network. J Biomed Inform. 2003;35:194–212. [DOI] [PubMed] [Google Scholar]