Abstract
A total of 395 Haemophilus influenzae strains from 226 Japanese institutions participating in the Nationwide Surveillance Study Group for Bacterial Meningitis were received from 1999 to 2002. All strains were analyzed by PCR to identify the resistance genes, and their susceptibilities to β-lactam agents were determined. Of these strains, 29.1% were β-lactamase nonproducing and ampicillin (AMP) susceptible (BLNAS) and lacked all resistance genes; 15.4% were β-lactamase producing and AMP resistant and had the blaTEM-1 gene; 30.6% were β-lactamase nonproducing and AMP resistant (low-BLNAR) and had a Lys-526 or His-517 amino acid substitution in ftsI encoding PBP 3; 13.9% were β-lactamase nonproducing and AMP resistant (BLNAR) and had an additional substitution of Thr-385 in ftsI; 9.1% were amoxicillin-clavulanic acid resistant (BLPACR I) and had the blaTEM-1 gene and a Lys-526 or His-517 amino acid substitution in ftsI; and 1.8% showed resistance similar to that of the BLPACR I group (BLPACR II) but had blaTEM-1 gene and ftsI substitutions, as was the case for the BLNAR strains. All but three strains were serotype b. The prevalence of BLNAR strains has increased rapidly: 0% in 1999, 5.8% in 2000, 14.1% in 2001, and 21.3% in 2002. The MICs at which 90% of BLNAR isolates were inhibited were as follows: AMP, 16 μg/ml; cefotaxime, 1 μg/ml; ceftriaxone, 0.25 μg/ml; and meropenem, 0.5 μg/ml. All of these values were higher than those for the BLNAS counterpart strains. The relatively wide distributions of the β-lactam MICs for BLNAR strains presumably reflect variations in ftsI gene mutations. Pulsed-field gel electrophoresis suggested the rapid spread of specific H. influenzae type b strains throughout Japan. Expedited vaccination, rapid identification, and judicious antibiotic use could slow their spread.
Penicillin-intermediate Streptococcus pneumoniae and penicillin-resistant S. pneumoniae isolates from patients with respiratory tract infections (RTIs) have emerged and increased in number in Japan since 1988 (1). In contrast to the phenotypes of S. pneumoniae isolates from the United States (15), strains with an abnormal PBP 2X and strains for which penicillin G MICs at which 50% of isolates are inhibited (MIC50s) were 0.06 μg/ml and cefotaxime (CTX) MIC50s were 0.125 to 0.5 μg/ml accounted for 20% of all strains collected throughout Japan between 1998 and 2000 (22). The high prevalence of these resistant organisms is attributed to the frequent use of oral and intravenous cephem antibiotic agents in Japan (15, 22). A similar situation is now evident among Haemophilus influenzae isolates from patients with RTIs, in which a rapid increase in the number of β-lactamase-nonproducing, ampicillin (AMP)-resistant (BLNAR) H. influenzae strains has been detected (11, 23). Resistance in BLNAR strains results from mutations in the ftsI gene encoding PBP 3, which mediates septal peptidoglycan synthesis (24). Amino acid substitutions identified at three positions in the ftsI gene are very important for resistance phenotypes: (i) Asn-526→Lys, (ii) Arg-517→His, and (iii) Ser-385→Thr (24). On the basis of these characteristic substitutions, BLNAR strains are classified as low-BLNAR strains if they possess the Asn-526→Lys or Arg-517→His substitution and as BLNAR strains if they possess the Asn-526→Lys and Ser-385→Thr substitutions or the Arg-517→His and Ser-385→Thr substitutions.
The MICs of cephalosporins such as CTX and ceftriaxone (CRO) for BLNAR strains are 32 times those for β-lactamase-nonproducing, AMP-susceptible (BLNAS) strains (24). The poor activities of cephalosporins against BLNAR strains presumably reflect the higher affinities of these agents for PBP 3 than those of the penicillins.
We have constructed PCR kits using six sets of primers to identify six elements: the p6 gene, which encodes the P6 membrane protein specific to H. influenzae (17, 27); the serotype b capsule gene (26); the TEM-1-type β-lactamase (blaTEM-1) gene (21); the ROB-1-type β-lactamase (blaROB-1) gene (10); a portion of the ftsI gene present in BLNAS strains (8); and a portion of the ftsI gene with a Ser-385→Thr amino acid substitution plus an Asn-526→Lys substitution (9). Isolates from RTI patients in Japan and the United States have been compared by using the primer kit (9). The respective prevalences of each resistance class in Japan and the United States were 55.1 and 46.0% for BLNAS, 3 and 26% for the blaTEM-1 gene (28), 0 and 10% for the blaROB-1 gene (12), 26.4 and 13% for low-BLNAR, and 13.2 and 0% for BLNAR. No BLNAR strains have been detected in the United States. However, according to a nationwide surveillance study conducted in the United States in 1994 and 1995, BLNAR strains have been very uncommon (5). Recently, low-BLNAR strains with ftsI mutations were reported in France (3) and the United Kingdom (20).
We also noted that BLNAR strains have emerged among H. influenzae type b (Hib) isolates from patients with meningitis, in parallel with the increase in the number of BLNAR strains from patients with RTIs (23). Unfortunately, Hib vaccination has not yet begun in Japan; consequently, an increase in the numbers of cases of meningitis caused by Hib BLNAR strains is feared.
To carry out molecular epidemiologic surveillance, H. influenzae strains isolated from the cerebrospinal fluid of meningitis patients were collected in a cooperative multi-institutional program, known as the Nationwide Surveillance for Bacterial Meningitis (NSBM) study, that we initiated throughout Japan between January 1999 and December 2002.
In this report we describe the prevalence and antibiotic susceptibilities of BLNAS strains and β-lactamase-producing, AMP-resistant (BLPAR), low-BLNAR, BLNAR, and β-lactamase-producing and amoxicillin-clavulanic acid (AMC)-resistant (BLPACR I and BLPACR II) strains identified by PCR, as well as substitutions in the ftsI gene in BLNAR and BLPACR II strains. We also describe the results of pulsed-field gel electrophoresis (PFGE) for the resistant strains.
MATERIALS AND METHODS
Strains.
Clinical bacteriological laboratories at 226 Japanese medical institutions participating in the NSBM study submitted 395 H. influenzae strains between January 1999 and December 2002. These institutions were recruited on the basis of their comparable proximities to major metropolitan areas. After arrival, the isolates were immediately analyzed by PCR as described below, and the results were sent by facsimile or e-mail to each laboratory that had submitted H. influenzae strains. After PCR the tested strains were stored at −80°C for subsequent testing.
PCR.
PCR was carried out for H. influenzae isolates by using five sets of primers reported previously (9): P6 primers to amplify the p6 gene, which encodes the P6 membrane protein specific for H. influenzae (17); TEM-1 primers to amplify a part of the blaTEM-1 gene (21); PBP3-S primers to identify an Asn-526→Lys amino acid substitution in the ftsI gene (9); PBP3-BLN primers to identify Asn-526→Lys and Ser-385→Thr amino acid substitutions in the ftsI gene (9); and serotype b primers to amplify a portion of the gene encoding the serotype b capsule (26). PCR cycling conditions were 35 cycles at 94°C for 15 s, 53°C for 15 s, and 72°C for 15 s.
Because useful primers for the detection of all isolates suspected to be low-BLNAR strains with the Arg-517→His substitution, according to disk testing of susceptibilities, could not be designed, these strains were subjected to direct sequencing to detect this substitution (24).
On the basis of the PCR results, all H. influenzae strains tested could be placed in one of six classes: BLNAS strains, which lack all resistance genes; BLPAR strains; low-BLNAR strains, which show low-level resistance associated with a substitution of Lys-526 or His-517 in ftsI; BLNAR strains; BLPACR I strains, which have the blaTEM-1 gene and ftsI with the same substitution as the low-BLNAR strains; and BLPACR II strains, which have the blaTEM-1 gene and ftsI with the same substitution as the BLNAR strains.
Antibiotic susceptibilities.
The susceptibilities of the H. influenzae isolates to intravenous β-lactam antibiotics were determined by a previously described agar dilution method (25). Mueller-Hinton agar (Becton Dickinson and Company, Sparks, Md.) with 0.5% yeast extract, 2% horse blood lysate instead of hematin (16), and β-NAD (15 μg/ml) was used. To prepare the horse blood lysate, defibrinated horse blood was treated at 65°C for 2 h, centrifuged at 100,000 × g for 1 h, and filtered through a 0.45-μm-pore-size Millipore filter. Strains from vials of 10% skim milk stored at −80°C were inoculated onto chocolate II agar plates (Nippon Becton Dickinson Co., Ltd., Tokyo, Japan) and were incubated for 18 h. On the next day, the strains were suspended in Mueller-Hinton broth, adjusted to a McFarland turbidity of 0.5, and diluted 100-fold; and 5 μl of each diluted strain was inoculated onto a susceptibility testing plate with a Steers replicator. The MIC was defined as the lowest concentration of antibiotic that inhibited visible growth of the inoculum in comparison with the growth on antibiotic-free plates. The plates were examined after 20 h of incubation in 5% CO2 at 35°C. H. influenzae strains ATCC 49247 and ATCC 49766 were used as quality controls.
The antibiotics used in this study were AMP (Meiji Seika Kaisya, Tokyo, Japan), CTX (Aventis Pharma Ltd., Tokyo, Japan), CRO (Nippon Roch KK, Tokyo, Japan), cefotiam and cefozopran (CTM and CZP, respectively; Takeda Chemical Industries, Ltd., Osaka, Japan), meropenem (MEM; Sumitomo Pharmaceuticals, Osaka, Japan), and panipenem (PAM; Sankyo, Tokyo, Japan).
Sequencing.
The 1.0-kb DNA fragment encoding the PBP 3 transpeptidase domain was amplified from the chromosomal DNA of H. influenzae by PCR (24). One colony of H. influenzae grown on a chocolate II agar plate was picked up and placed in 30 μl of lysis solution. This suspension was incubated at 60°C for 20 min and then at 94°C for 5 min. Subsequently, 2 μl of the lysate was added to 50 μl of PCR solution for PCR; the solution and conditions were reported previously (24).
The sequencing reaction was conducted with a DYEnamic ET Terminator Cycle Sequencing kit (Amersham Pharmacia Biotech, Piscataway, N.J.). The reaction mixtures were placed in a thermal cycler and denatured at 95°C for 10 s and were then subjected to 30 PCR cycles, each of which consisted of 95°C for 20 s, 50°C for 15 s, and 60°C for 60 s. The nucleotide sequences were determined with an ABI PRISM 377 DNA sequencer.
PFGE analysis.
PFGE analysis was carried out by a modification of the method of Yano et al. (29). H. influenzae grown on a chocolate II agar plate was suspended in 2 ml of saline-EDTA solution (0.15 M NaCl, 10 mM EDTA [pH 8.0]), and the bacterial density was adjusted to a McFarland turbidity of 1.0. The bacterial suspension was harvested by centrifugation at 3,000 × g and 4°C for 5 min, followed by two washes with saline-EDTA solution. After the cells were resuspended in Pet IV solution (1 M NaCl, 10 mM EDTA), 1.6 times this volume of melted 2.0% low-melting-point agarose was added (InCert agarose; FMC Bioproducts, Rockland, Maine). The mixture was poured into an insert mold and chilled at 4°C for 20 min. Plugs were removed from this mold and treated at 37°C with 0.05 mg of lysozyme (Sigma Chemical, St. Louis, Mo.) per ml in lysis solution (1 M NaCl, 0.1 M EDTA [pH 8.0], 10 mM Tris-HCl [pH 8.0], 0.5% Brij 58, 0.2% sodium deoxycholate, 0.5% Sarkosyl). After 1 h, the lysis solution was decanted and replaced with 0.2 mg of proteinase K (Sigma) per ml in ES solution (0.25 M EDTA [pH 8.0], 1% Sarkosyl) at 50°C for 17 h. The ES solution was decanted, and the plugs were placed in TE buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA [pH 8.0] containing 1 mM phenylmethylsulfonyl fluoride) at room temperature for 3 h. Next, the plugs were washed three times in TE buffer for 15 min at room temperature and placed in TE buffer (10 mM Tris-HCl [pH 8.0], 0.1 mM EDTA [pH 8.0]). For restriction endonuclease digestion, the plugs were incubated in restriction enzyme buffer for 30 min at room temperature to remove the EDTA and were then incubated in restriction enzyme buffer with 30 U of SmaI at 30°C for 16 h. Electrophoresis was performed with a CHEF Mapper (Bio-Rad Laboratories, Hercules, Calif.). Separation of fragments was carried out at 6 V/cm at 14°C for 20 h.
RESULTS
Susceptibilities of H. influenzae strains to agents typically given intravenously.
Table 1 shows the MIC ranges, the MIC50s, and the MIC90s of β-lactam agents for intravenous use for the 395 H. influenzae strains classified into six groups according to the PCR results and ftsI sequences. The antibiotics used in this study were AMP, four cephalosporins (CTX, CRO, CTM, and CZP), and two carbapenems (MEM and PAM). The prevalence of each class among the 395 H. influenzae strains was as follows: 29.1% (n = 115) for BLNAS, 15.4% (n = 61) for BLPAR, 30.6% (n = 121) for low-BLNAR, 13.9% (n = 55) for BLNAR, 9.1% (n = 36) for BLPACR I, and 1.8% (n = 7) for BLPACR II.
TABLE 1.
MIC distributions and resistance genes identified by PCR in H. influenzae strains isolated in Japana
| Antimicrobial agent and resistance class | MIC (μg/ml)
|
||
|---|---|---|---|
| 50% | 90% | Range | |
| AMP | |||
| BLNAS (n = 115) | 0.25 | 0.5 | 0.031-0.5 |
| BLPAR (TEM-1) (n = 61) | 8 | 16 | 2-64 |
| Low-BLNAR (n = 121) | 1 | 2 | 0.5-2 |
| BLNAR (n = 55) | 2 | 16 | 1-16 |
| BLPACR I (n = 36) | 32 | 64 | 2-64 |
| BLPACR II (n = 7) | 32 | 64 | 16-64 |
| CTX | |||
| BLNAS | 0.016 | 0.031 | 0.004-0.063 |
| BLPAR (TEM-1) | 0.016 | 0.031 | 0.008-0.063 |
| Low-BLNAR | 0.063 | 0.125 | 0.031-0.25 |
| BLNAR | 0.5 | 1 | 0.125-2 |
| BLPACR I | 0.063 | 0.125 | 0.016-0.25 |
| BLPACR II | 0.5 | 1 | 0.25-1 |
| CRO | |||
| BLNAS | 0.004 | 0.008 | 0.002-0.016 |
| BLPAR (TEM-1) | 0.004 | 0.008 | 0.002-0.008 |
| Low-BLNAR | 0.016 | 0.031 | 0.004-0.063 |
| BLNAR | 0.125 | 0.25 | 0.031-0.5 |
| BLPACR I | 0.016 | 0.031 | 0.004-0.031 |
| BLPACR II | 0.125 | 0.25 | 0.063-0.25 |
| CTM | |||
| BLNAS | 1 | 2 | 0.063-8 |
| BLPAR (TEM-1) | 1 | 2 | 0.5-2 |
| Low-BLNAR | 8 | 16 | 2-32 |
| BLNAR | 32 | 64 | 4-64 |
| BLPACR I | 4 | 16 | 1-32 |
| BLPACR II | 64 | 64 | 8-64 |
| CZP | |||
| BLNAS | 0.25 | 0.25 | 0.016-1 |
| BLPAR (TEM-1) | 0.125 | 0.25 | 0.063-0.5 |
| Low-BLNAR | 1 | 2 | 0.5-4 |
| BLNAR | 8 | 16 | 2-64 |
| BLPACR I | 1 | 2 | 0.25-4 |
| BLPACR II | 16 | 32 | 8-32 |
| MEM | |||
| BLNAS | 0.063 | 0.063 | 0.008-0.125 |
| BLPAR (TEM-1) | 0.063 | 0.063 | 0.031-0.063 |
| Low-BLNAR | 0.125 | 0.25 | 0.031-0.5 |
| BLNAR | 0.25 | 0.5 | 0.031-0.5 |
| BLPACR I | 0.25 | 0.25 | 0.063-0.5 |
| BLPACR II | 0.25 | 0.25 | 0.125-0.25 |
| PAM | |||
| BLNAS | 0.25 | 1 | 0.031-1 |
| BLPAR (TEM-1) | 0.125 | 1 | 0.125-1 |
| Low-BLNAR | 1 | 2 | 0.25-8 |
| BLNAR | 1 | 2 | 0.25-4 |
| BLPACR I | 1 | 2 | 0.125-4 |
| BLPACR II | 1 | 2 | 0.5-2 |
A total of 395 isolates were tested.
The MIC50s of the intravenous agents for the low-BLNAR isolates were two to eight times higher than those for the BLNAS isolates. The MIC50s of all cephalosporin agents for BLNAR isolates were 32 times higher than those for BLNAS isolates. However, the MIC50s of AMP and MEM for the BLNAR isolates were only four to eight times higher than those for the BLNAS isolates.
Of the 395 H. influenzae strains, 392 were identified as serotype b by PCR, while the remaining 3 were nontypeable.
Sequencing of ftsI gene.
Table 2 shows the deduced amino acid substitutions in the ftsI genes in the BLNAR and BLPACR II strains. These strains were classified into six subgroups on the basis of different substitution patterns.
TABLE 2.
Amino acid substitutions identified in ftsI genes from BLNAR and BLPACR II H. influenzae strainsa
| Subgroup | Strain or no. of strains | Amino acid substitution
|
MIC50 (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Surrounding SSN motif
|
Near KTG motif
|
|||||||||
| Met-377 | Ser-385 | Leu-389 | Arg-517 | Asn-526 | AMP | CTX | CRO | MEM | ||
| Control | MT196 | 0.125 | 0.016 | 0.004 | 0.031 | |||||
| BLNAR | ||||||||||
| I | 9 | Ile | Thr | His | 1 | 0.25 | 0.063 | 0.125 | ||
| II | 2 | Ile | Thr | Phe | His | 8 | 1 | 0.25 | 0.125 | |
| III | 7 | Thr | Lys | 2 | 0.25 | 0.063 | 0.25 | |||
| IV | 1 | Ile | Thr | Lys | 1 | 0.125 | 0.031 | 0.25 | ||
| V | 1 | Thr | Phe | Lys | 2 | 0.125 | 0.063 | 0.125 | ||
| VI | 35 | Ile | Thr | Phe | Lys | 4 | 0.5 | 0.125 | 0.25 | |
| BLPACR II | ||||||||||
| VII | 1 | Thr | His | 32 | 1 | 0.25 | 0.125 | |||
| VIII | 6 | Ile | Thr | Phe | Lys | 64 | 0.5 | 0.125 | 0.25 | |
A total of 55 BLNAR strains and 7 BLPACR II strains were tested.
In subgroups I (n = 9) and II (n = 2), both Arg-517→His and Ser-385→Thr substitutions were present, and additionally, Met-377→Ile and Leu-389→Phe substitutions were identified in subgroup II. Strains with the Asn-526→Lys substitution were classified into subgroups III to VI. These strains had the Ser-385→Thr substitution in common and, in addition, frequently had the Met-377→Ile and Leu-389→Phe substitutions in common. Subgroups VI, I, and III predominated, in that order. Strains with three substitutions surrounding the Ser-Ser-Asn (SSN) motif showed slightly higher levels of resistance to the β-lactam agents than the strains in the other subgroups.
Subgroups VII and VIII were both recognized among the BLPACR II strains.
PFGE.
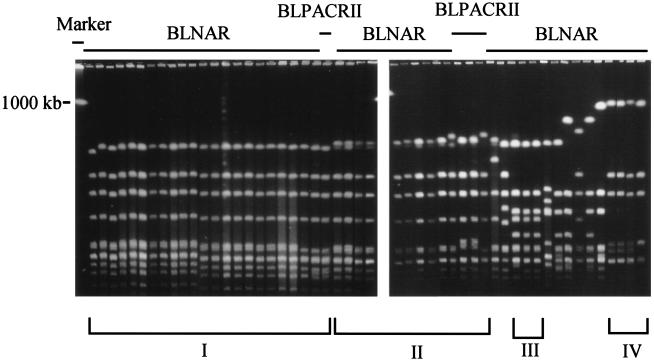
All BLNAR and BLPACR II strains (n = 62) were examined by PFGE after treatment with the endonuclease SmaI. Figure 1 shows the PFGE profiles of 51 strains selected at random from among the 62 strains. The profiles showed pattern I and pattern II in 23 and 13 strains, respectively; and the strains with these two profiles showed similarities. Strains with PFGE patterns I and II were widely disseminated. The remaining strains had different profiles, such as patterns III (n = 3) and IV (n = 4). Eight strains did not fit any pattern. The BLNAR strains isolated from pneumonia patients during the study period showed a variety of PFGE profiles (data not shown).
FIG. 1.
Results of PFGE showing four DNA patterns among BLNAR and BLPACR II isolates (n = 51) after treatment with the SmaI endonuclease.
Changes in resistance among strains by year.
Table 3 shows the yearly changes in resistance among the strains between 1999 and 2002. Both the numbers of meningitis cases and the prevalence of BLNAR strains among those cases have rapidly increased since 2000: 0% (n = 0) in 1999, 5.8% (n = 4) in 2000, 14.1% (n = 19) in 2001, and 21.3% (n = 32) in 2002. The prevalences of BLPACR I and II strains also increased every year. In contrast, the incidence of BLPAR strains gradually decreased.
TABLE 3.
Changes in resistance among H. influenzae strains from meningitis patients, by year, as identified by PCR
| Resistance class | No. (%) of strains in the following yr:
|
||||
|---|---|---|---|---|---|
| 1999 | 2000 | 2001 | 2002 | Total | |
| BLNAS | 14 (34.1) | 24 (34.8) | 45 (33.3) | 31 (21.3) | 115 (29.1) |
| BLPAR | 13 (31.7) | 14 (20.3) | 17 (12.6) | 17 (11.3) | 61 (15.4) |
| Low-BLNAR | 12 (29.2) | 24 (34.8) | 40 (29.6) | 45 (30.0) | 121 (30.6) |
| BLNAR | 0 (0.0) | 4 (5.8) | 19 (14.1) | 32 (21.3) | 55 (13.9) |
| BLPACR I | 1 (2.4) | 3 (4.3) | 12 (8.9) | 19 (13.3) | 36 (9.1) |
| BLPACR II | 1 (2.4) | 0 (0.0) | 2 (1.5) | 4 (2.7) | 7 (1.8) |
| Total | 41 | 69 | 135 | 150 | 395 (100.0) |
Age distribution of meningitis patients.
The relationship between the resistance status of 395 causative strains and the ages of the meningitis patients is considered in Table 4. The prevalence of H. influenzae isolates was highest in patients 1 year of age (n = 90; 22.8%) and was next highest in patients between the ages of 7 and 11 months (n = 85; 21.5%). Patients 1 to 6 months old showed a relatively high prevalence (14.9%). H. influenzae gradually became less prevalent after the age of 2 years (n = 56; 14.2%) and was rare by age 5 years. Low-BLNAR and BLNAR strains were prominent among the causative organisms.
TABLE 4.
Relationship between resistant isolates and age of patients with meningitis
| Resistance class | No. (%) of isolates from patients of the following age:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-6 mo | 7-11 mo | 1 yr | 2 yr | 3 yr | 4 yr | 5 yr | 6 yr | 7 yr | Unknown | Total | |
| BLNAS | 17 | 24 | 22 | 11 | 11 | 10 | 4 | 1 | 4 | 11 | 115 (29.1) |
| BLPAR | 12 | 9 | 15 | 12 | 7 | 4 | 0 | 0 | 0 | 2 | 61 (15.4) |
| Low-BLNAR | 16 | 28 | 37 | 11 | 15 | 11 | 1 | 1 | 0 | 1 | 121 (30.6) |
| BLNAR | 4 | 18 | 9 | 14 | 6 | 2 | 0 | 0 | 1 | 1 | 55 (13.9) |
| BLPACR I | 9 | 5 | 5 | 6 | 5 | 4 | 1 | 0 | 0 | 1 | 36 (9.1) |
| BLPACR II | 1 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 7 (1.8) |
| Total | 59 (14.9) | 85 (21.5) | 90 (22.8) | 56 (14.2) | 45 (11.4) | 31 (7.8) | 6 (1.5) | 2 (0.5) | 5 (1.3) | 16 (4.1) | 395 (100.0) |
DISCUSSION
Worldwide, resistant strains of S. pneumoniae and H. influenzae pose significant clinical problems for those treating patients with RTIs. In the United States, however, where penicillins are frequently prescribed for outpatients, penicillin-resistant S. pneumoniae strains with abnormal penicillin-binding proteins (PBPs) have been noted since the 1980s. H. influenzae strains with PBPs showing decreased affinities for β-lactam agents were also described in the United States in the 1980s (13, 14, 18), but they were not considered important clinical problems. Instead, TEM-1- and ROB-1-type BLPAR strains have been prevalent in the United States; accordingly, AMC has been recommended as first-line oral antibiotic therapy (4-6, 19). BLNAR isolates have rarely been reported in the United States (5, 9).
In Japan, on the other hand, TEM-1-type BLPAR appeared in the early 1970s and now has a relative prevalence of ≤10%. Low-BLNAR strains, for which the MICs of cephalosporin agents are slightly increased, have been isolated since 1985 (unpublished data), recently amounting to 20 to 25% of isolates in our retrospective studies. BLNAR strains, for which the increases in the MICs of cephalosporin agents are greater, appeared in 1997 and increased rapidly among isolates from pediatric outpatients with RTIs or acute otitis media. The increase in the number of BLNAR strains is apparently related to the clinical use of low doses of oral cephalosporin agents, mainly by pediatric patients. A main factor contributing to the increase is that the MICs of most oral agents for BLNAR strains typically exceed the concentrations in serum obtained clinically by use of the doses approved by the Japanese Ministry of Health Labor and Welfare (7). In Japan, the dosages of oral AMP and AMC commonly used are generally half to a third of those used in the United States. Additionally, the bactericidal activities of cephalosporin agents against BLNAR strains apparently decreased more than their activities against BLNAS strains. Lysis was rarely induced, even by the use of concentrations that exceed the MIC of each agent for BLNAR strains. Instead, morphological alterations such as filamentous forms and/or spheroplast cells were observed. Such forms readily reverted to typical bacillary forms in vitro after the agent was removed.
As mentioned in the Results section, the NSBM study noted that over 3 years cases of meningitis caused by Hib BLNAR strains have increased in parallel with increases in the numbers of BLNAR isolates from patients with RTIs and acute otitis media (22). In particular, the increase in the number of cases of meningitis caused by Hib BLNAR strains can be related to vaccination not having begun yet for children against Hib in Japan. Adverse effects after inoculation of the measles-mumps-rubella (MMR) vaccine became a public health problem in 1989, and since then, a consensus to initiate a new vaccination program has been difficult to attain.
Although our surveillance study does not cover all hospitals throughout Japan, we were able to calculate a Hib attack rate of approximately 10/100,000 children younger than age 5 years.
According to a report of the Centers for Disease Control and Prevention, in the United States, meningitis cases caused by Hib have decreased significantly as a result of vaccination with the Hib conjugate vaccine since 1990, and the severe infection rate in the United States has been reported to be <1 case per 100,000 children <5 years old (2). This underscores the importance of initiating vaccination with the Hib conjugate vaccine in Japan as soon as possible.
A variety of substitutions in ftsI were identified and allowed the isolates to be classified into six subgroups. The somewhat wide distribution of the β-lactam MICs for BLNAR strains is believed to reflect the differences among strains with these mutations. Furthermore, highly resistant BLNAR strains with additional substitutions will likely emerge as long as the use of oral β-lactam agents, which are used at present, continues in Japan. Instead, effective doses are required for the treatment of infections caused by resistant strains, with the particular antibiotics used being chosen on the basis of PCR results.
Even so, some meningitis patients infected with BLNAR strains have failed chemotherapy with CTX or CRO at the doses recommended in the United States (data not shown). Combination therapy, such as CTX with MEM, which shows a different affinity than CTX monotherapy, might be more effective than monotherapy. Resolution of these therapeutic issues requires concerted efforts, and these issues are being evaluated in our centers.
According to the results of PFGE analysis presented at the end of the Results section, isolates with similar profiles were identified; and it was found that certain subtypes are rapidly spreading throughout Japan.
Important steps that can be taken to prevent further increases in the numbers of severe infections caused by Hib are vaccination, rapid identification of the causative pathogens, proper use of antibiotics on the basis of pharmacokinetics and pharmacodynamics, and epidemiologic surveillance based on molecular analysis.
Acknowledgments
This work was supported in part by two grants, one from the Japanese Ministry of Health Labor, and Welfare (The Research Project for Emerging and Re-emerging Infectious Diseases, April 2000 to March 2003) and one from the Japanese Ministry of Education, Culture, Sports, Science and Technology (the 21st Century COE Program).
REFERENCES
- 1.Asahi, Y., and K. Ubukata. 1998. Association of a Thr-371 substitution in a conserved amino acid motif of penicillin-binding protein 1A with penicillin resistance of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2267-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2002. Haemophilus influenzae type b. http://www.cdc.gov/nip/publications/pink/hib.pdf.
- 3.Dabernat, H., C. Delmas, M. Seguy, R. Pelissier, G. Faucon, S. Bennamani, and C. Pasquier. 2002. Diversity of β-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob. Agents Chemother. 46:2208-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David, N. G., C. M. Robert, and A. S. Merle. 2000. Guide to antimicrobial therapy, p. 7. Antimicrobial therapy, Inc., Hyde Park, Vt.
- 5.Doern, G. V., A. B. Brueggemann, G. Pierce, H. P. Horry, Jr., and A. Rauch. 1997. Antibiotic resistance among clinical isolates of Haemophilus influenzae in the United States in 1994 and 1995 and detection of β-lactamase-positive strains resistant to amoxicillin-clavulanate: results of a national multicenter surveillance study. Antimicrob. Agents Chemother. 41:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doern, G. V., J. H. Jorgensen, C. Thornsberry, D. A. Proston, and the Haemophilus influenzae Surveillance Group. 1986. Prevalence of antimicrobial resistance among clinical isolates of Haemophilus influenzae: a collaborative study. Diagn. Microbial. Infect. Dis. 4:95-107. [DOI] [PubMed] [Google Scholar]
- 7.Drusano, G. L., and W. A. Craig. 1997. Relevance of pharmacokinetics and pharmacodynamics in the selection of antibiotics for respiratory tract infections. J. Chemother. 9(Suppl.):38-44. [PubMed] [Google Scholar]
- 8.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. G. Sutton, W. FitzHugh, C. A. Fields, J. D. Gocayne, J. D. Scott, R. Shirley, L. I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 9.Hasegawa, K., K. Yamamoto, N. Chiba, R. Kobayashi, K. Nagai, M. R. Jacobs, P. C. Appelbaum, K. Sunakawa, and K. Ubukata. 2003. Diversity of ampicillin-resistance genes in Haemophilus influenzae in Japan and the United States. Microb. Drug Resist. 9:39-46. [DOI] [PubMed] [Google Scholar]
- 10.Juteau, J. M., and R. C. Levesque. 1990. Sequence analysis and evolutionary perspectives of ROB-1 β-lactamase. Antimicrob. Agents Chemother. 34:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markowitz, S. M. 1980. Isolation of an ampicillin-resistant, non-β-lactamase-producing strain of Haemophilus influenzae. Antimicrob. Agents Chemother. 17:80-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medeiros, A. A., R. Levesque, and G. A. Jacoby. 1986. An animal source for the ROB-1 β-lactamase of Haemophilus influenzae type b. Antimicrob. Agents Chemother. 29:212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendelman, P. M., D. O. Chaffin, J. M. Musser, R. DeGroot, D. A. Serfass, and R. K. Selander. 1987. Genetic and phenotypic diversity among ampicillin-resistant, non-β-lactamase-producing, nontypeable Haemophilus influenzae isolates. Infect. Immun. 55:2585-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendelman, P. M., D. O. Chaffin, T. L. Stull, C. E. Rubens, K. D. Mack, and A. L. Smith. 1984. Characterization of non-β-lactamase-mediated ampicillin resistance in Haemophilus influenzae. Antimicrob. Agents Chemother. 26:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagai, K., Y. Shibasaki, K. Hasegawa, T. A. Davies, M. R. Jacobs, K. Ubukata, and P. C. Appelbaum. 2001. Evaluations of the primers for PCR to screen Streptococcus pneumoniae isolates, β-lactam resistance and to detect common macrolide resistance determinants. J. Antimicrob. Chemother. 48:915-918. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial susceptibility testing. Fifth informational supplement M100-S10. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Nelson, M. B., M. A. Apicella, T. F. Murphy, H. Vankeulen, L. D. Spotila, and D. Rekosh. 1988. Cloning and sequencing of Haemophilus influenzae outer membrane protein P6. Infect. Immun. 56:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parr, T. R., Jr., and L. E. Bryan. 1984. Mechanism of resistance of an ampicillin-resistant, β-lactamase-negative clinical isolate of Haemophilus influenzae type b to β-lactam antibiotics. Antimicrob. Agents Chemother. 25:747-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter, S. S., A. B. Brueggemann, H. K. Huynh, P. R. Rhomberg, E. M. Wingert, R. Flamm, and G. V. Doern. 1999. A 1997-1998 national surveillance study: Moraxella catarrhalis and Haemophilus influenzae antimicrobial resistance in 34 US institutions. Int. J. Antimicrob. Agents 13:99-107. [DOI] [PubMed] [Google Scholar]
- 20.Straker, K., M. Wootton, A. M. Simm, P. M. Bennett, A. P. MacGowan, and T. R. Walsh. 2003. Cefuroxime resistance in non-β-lactamase Haemophilus influenzae is linked to mutations in ftsI. J. Antimicrob. Chemother. 51:523-530. [DOI] [PubMed] [Google Scholar]
- 21.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ubukata, K. 2001. Three years of surveillance of invasive Streptococcus pneumoniae and Haemophilus influenzae by the Study Group for Community-Acquired Infections. Jpn. J. Antibiot. 54:72-79. (In Japanese.) [Google Scholar]
- 23.Ubukata, K. 2003. Problems associated with high prevalence of multi-drug resistant bacteria in patients with community-acquired infections. J. Infect. Chemother. 9:285-291. [DOI] [PubMed] [Google Scholar]
- 24.Ubukata, K., Y. Shibasaki, K. Yamamoto, N. Chiba, K. Hasegawa, Y. Takeuchi, K. Sunakawa, and M. Inoue. 2001. Association of amino acid substitutions in penicillin-binding protein 3 with β-lactam resistance in β-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 45:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ubukata, K., N. Chiba, K. Hasegawa, Y. Shibasaki, K. Sunakawa, M. Nonoyama, S. Iwata, and M. Konno. 2002. Differentiation of β-lactamase-negative ampicillin-resistant Haemophilus influenzae from other H. influenzae strains by a disc method. J. Infect. Chemother. 8:50-58. [DOI] [PubMed] [Google Scholar]
- 26.Van Eldere, J., L. Brophy, B. Loynds, P. Celis, I. Hancock, S. Carman, J. S. Kroll, and E. R. Moxon. 1995. Region II of the Haemophilus influenzae type b capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol. Microbiol. 15:107-118. [DOI] [PubMed] [Google Scholar]
- 27.van Ketel, R. J., B. de Wever, and L. van Alphen. 1990. Detection of Haemophilus influenzae in cerebrospinal fluids by polymerase chain reaction DNA amplification. J. Med. Microbiol. 33:271-276. [DOI] [PubMed] [Google Scholar]
- 28.Vega, R., H. L. Sadoff, and M. J. Patterso. 1976. Mechanism of ampicillin resistant in Haemophilus influenzae type b. Antimicrob. Agents Chemother. 9:164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yano, H., M. Suetake, A. Kuga, K. Irinoda, R. Okamoto, T. Kobayashi, and M. Inoue. 1999. Pulsed-field gel electrophoresis analysis of nasopharyngeal flora in children attending a day care center. J. Clin. Microbiol. 38:625-629. [DOI] [PMC free article] [PubMed] [Google Scholar]