Abstract
The glomerulus contains unique cellular and extracellular matrix (ECM) components, which are required for intact barrier function. Studies of the cellular components have helped to build understanding of glomerular disease; however, the full composition and regulation of glomerular ECM remains poorly understood. We used mass spectrometry-based proteomics of enriched ECM extracts for a global analysis of human glomerular ECM in vivo and identified a tissue-specific proteome of 144 structural and regulatory ECM proteins. This catalog includes all previously identified glomerular components plus many new and abundant components. Relative protein quantification showed a dominance of collagen IV, collagen I, and laminin isoforms in the glomerular ECM together with abundant collagen VI and TINAGL1. Protein network analysis enabled the creation of a glomerular ECM interactome, which revealed a core of highly connected structural components. More than one half of the glomerular ECM proteome was validated using colocalization studies and data from the Human Protein Atlas. This study yields the greatest number of ECM proteins relative to previous investigations of whole glomerular extracts, highlighting the importance of sample enrichment. It also shows that the composition of glomerular ECM is far more complex than previously appreciated and suggests that many more ECM components may contribute to glomerular development and disease processes. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD000456.
The glomerulus is a sophisticated organelle comprising unique cellular and extracellular matrix (ECM) components. Fenestrated capillary endothelial cells and overlying podocytes are separated by a specialized glomerular basement membrane (GBM), and these three components together form the filtration barrier. Mesangial cells and their associated ECM, the mesangial matrix, exist between adjacent capillary loops and maintain the three-dimensional organization of the capillary bundle. In turn, the parietal epithelial cells and ECM of Bowman’s capsule enclose this network of capillaries. Cells adhere to ECM proteins by adhesion receptors, and these interactions are required to maintain intact barrier function of the glomerulus.1,2
In addition to operating as a signaling platform, ECM provides a structural scaffold for adjacent cells and has a tissue-specific molecular composition.3,4 Candidate-based investigations of glomerular ECM have focused on the GBM and shown that it resembles the typical basal lamina found in multicellular organisms, containing a core of glycoproteins (collagen IV, laminins, and nidogens) and heparan sulfate proteoglycans (agrin, perlecan, and collagen XVIII).5 Mesangial and parietal cell ECMs have been less well investigated; nonetheless, they are also thought to contain similar core components in addition to other glycoproteins, including fibronectin.6,7 Thus, the glomerulus consists of a combination of condensed ECM within the GBM and Bowman’s capsule and loose ECM supporting the mesangial cells.
The ECM compartments in the glomerulus are thought to be distinct and exhibit different functional roles. The GBM is integral to the capillary wall and therefore, functionally linked to glomerular filtration.5 Mutations of tissue-restricted isoforms of collagen IV (COL4A3, COL4A4, and COL4A5) and laminin (LAMB2), which are found in the GBM, cause significant barrier dysfunction and ultimately, renal failure.8,9 Less is understood about the functions of mesangial and parietal cell ECMs, although expansion of the mesangial compartment is a histologic pattern seen across the spectrum of glomerular disease.10
Compositional investigation of the distinct glomerular ECM compartments is limited by the technical difficulties of separation. Early investigations of GBM constituents used the relative insolubility of ECM proteins to facilitate separation from cellular proteins in the glomerulus but did not separate the GBM from mesangial and parietal cells ECMs.11,12 Recently, studies incorporating laser microdissection of glomerular sections have been coupled with proteomic analyses.13,14 These studies report both cellular and ECM components and typically require pooled material from glomerular sections to improve protein identification. The ability of laser microdissection to separate glomerular ECM compartments has not yet been tested; however, this approach will be limited by the amount of protein that is possible to retrieve. To achieve good coverage of ECM proteins within a tissue, proteomic studies need to combine a reduction in sample complexity with maximal protein quantity. Currently, the inability to separate glomerular ECM compartments in sufficient quantity is a limitation that prohibits proteomic studies of these structures; however, for other tissues, proteomic analysis of ECM has been achieved by enrichment of ECM combined with sample fractionation.15
Although the composition of the ECM in other tissues has been addressed using proteomic approaches,15 studies of glomerular ECM to date have used candidate-based technologies. These studies have identified key molecular changes during development and disease and highlighted the compositional and organizational dynamics of glomerular ECM. Nonetheless, the extracellular environment within the glomerulus is the setting for a complex series of interactions between both structural ECM proteins and ECM-associated proteins, such as growth factors16–18 and proteases,19 which together provide a specialized niche to support glomerular cell function. Therefore, to interrogate this complexity effectively, a systems-level understanding of glomerular ECM is required. To address the need for a global analysis of the extracellular environment within the glomerulus, we used mass spectrometry (MS)-based proteomics of glomerular ECM fractions to define the human glomerular ECM proteome.
Results
Isolation of Glomerular ECM
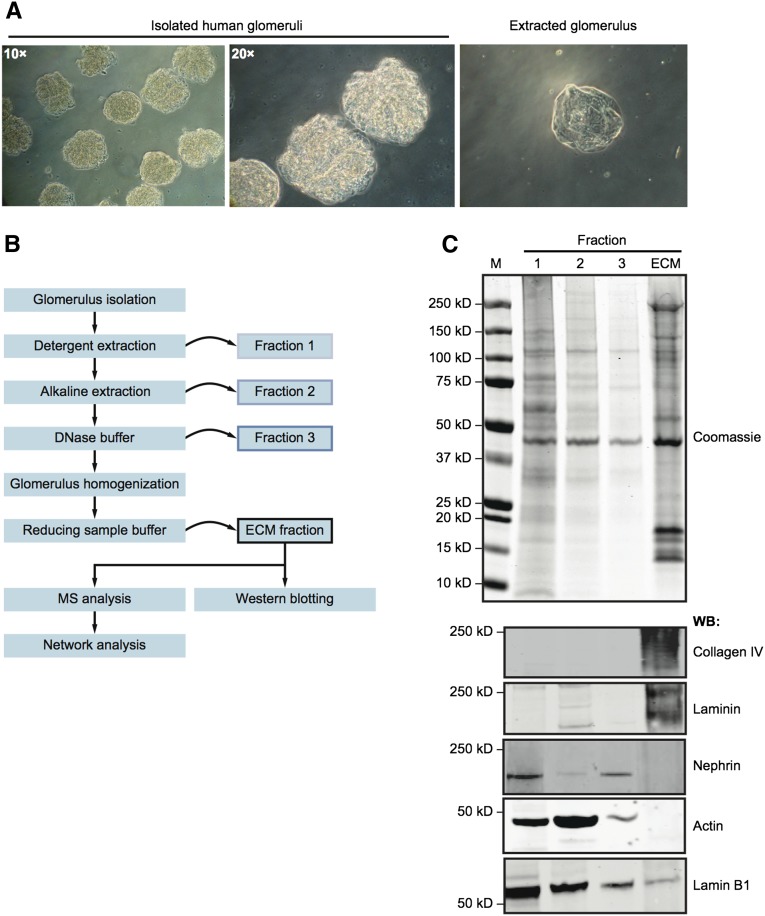
Using purified isolates of human glomeruli (Figure 1A), we developed a fractionation approach to collect glomerular ECM proteins (Figure 1B). Before homogenization and solubilization, extracted glomeruli appeared acellular, suggestive of successful ECM enrichment (Figure 1A). Western blotting confirmed enrichment of ECM proteins (collagen IV and laminin) and depletion of cytoplasmic and nuclear proteins (nephrin, actin, and lamin B1) in the ECM fraction (Figure 1C). Glomerular ECM fractions from three adult human kidneys were then extracted and analyzed by MS.
Figure 1.
Isolation of enriched glomerular ECM. (A) Human glomeruli were isolated by differential sieving, yielding >95% purity. Before homogenization, glomeruli appeared acellular (right). (B) A proteomic workflow for the isolation of enriched glomerular ECM by fractionation (details in Concise Methods). (C) Coomassie staining and Western blotting (WB) of fractions 1–3 and the ECM fraction probing for the extracellular proteins with pancollagen IV and panlaminin probes and the intracellular proteins nephrin, actin, and lamin B1. M, molecular mass marker.
Gene Ontology Enrichment Analysis of ECM Fractions
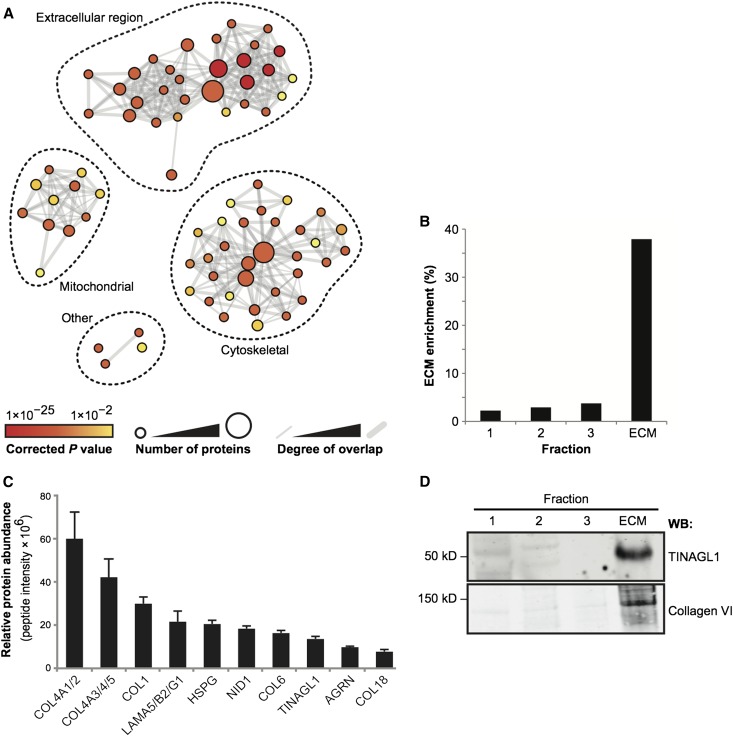
All proteins identified by MS were allocated to categories according to their gene ontology (GO) assignment, and the enrichment of GO terms in the dataset was assessed using GO enrichment analysis, which is described in Supplemental Methods. The network of enriched GO terms clustered into three subnetworks, and the largest, most confidently identified subnetwork was for ECM-related GO terms (Figure 2A). A large majority of other proteins were included in two additional subnetworks representing mitochondrial and cytoskeletal GO terms, and the presence of these proteins in the ECM fractions is likely to be caused by the strength of their intermolecular interactions with ECM proteins. Spectral counting was used to determine the enrichment of ECM in each of the four fractions collected from isolated human glomeruli. There was 38% enrichment of extracellular proteins in the glomerular ECM fractions (Figure 2B), and this result compares favorably with the 12%–30% enrichment of ECM proteins reported in comparable proteomic studies.15
Figure 2.
MS analysis of enriched glomerular ECM fractions. (A) GO enrichment analysis of the full MS dataset. Nodes (circles) represent enriched GO terms, and edges (gray lines) represent overlap of proteins between GO terms. Node color indicates the significance of GO term enrichment; node diameter is proportional to the number of proteins assigned to each GO term. Edge weight is proportional to the number of proteins shared between connected GO terms. The full list of GO terms is detailed in Supplemental Figure 1. (B) All four protein fractions were analyzed by MS, and spectral counting was used to determine the enrichment of ECM proteins (identified by GO analysis). The mean ECM enrichment was 38% from three biologic replicates. (C) Relative quantification for the 10 most abundant ECM proteins detected by MS. Relative protein abundance was calculated using peptide intensity as described in Supplemental Methods. Gene names are shown for clarity, and collagen (COL) IV and laminin isoforms are combined as one value. (D) Western blotting (WB) confirmed enrichment of TINAGL1 and collagen VI in glomerular ECM.
Relative Abundance of Glomerular ECM Proteins
To determine the relative abundance of glomerular ECM proteins, we used MS to analyze additional glomerular ECM fractions from three human kidneys, and we quantified the proteins using a peptide intensity approach,20 which is described in Supplemental Methods. Using this analysis, the 10 most abundant proteins were a combination of collagens, laminin 521, and heparan sulfate proteoglycans (Figure 2C, Supplemental Table 1), which have all previously been reported in the glomerulus. In addition, we found collagen VI and the glycoprotein TINAGL1 among the 10 most abundant proteins, and the detection of these two components was confirmed by Western blotting (Figure 2D). Relative quantification was also performed using spectral counting, which was compared with the peptide intensity approach, and further showed that collagen VI and TINAGL1 were among the most abundant proteins detected in the glomerular ECM (Supplemental Figure 2).
Defining the Glomerular ECM Proteome
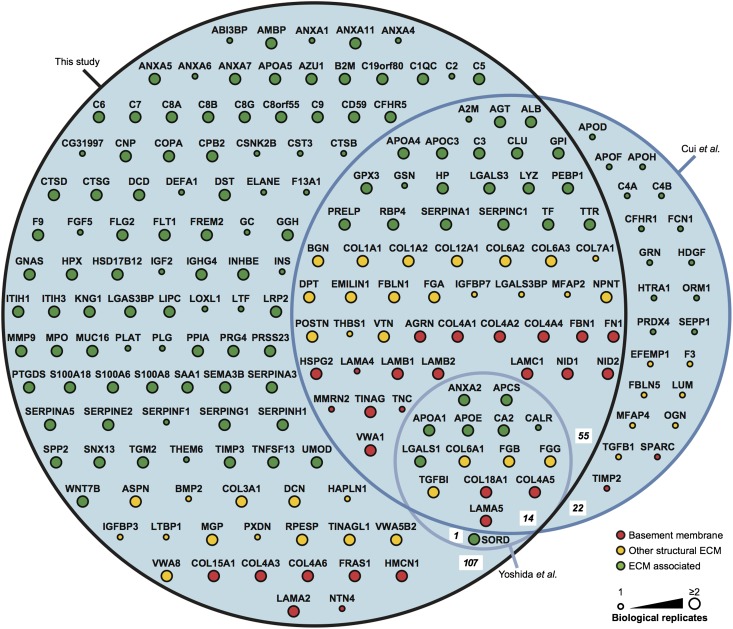
Using the GO classification of extracellular region proteins and crossreferencing with the human matrisome project3 (described in Supplemental Methods), we identified 144 extracellular proteins in the glomerular ECM, including all previously known glomerular ECM components. Only proteins identified in at least two of three biologic replicate analyses were included in the dataset, and identified ECM proteins were further categorized as basement membrane proteins (Table 1), other structural ECM proteins (Table 2), or ECM-associated proteins (Supplemental Table 2). There are a number of published glomerular proteomic studies,21–25 and a comparison was made with protein identifications in our dataset (Figure 3). Of two published studies for which complete MS datasets were available,22,25 neither study enriched for glomerular ECM or analyzed more than one biologic replicate. We detected all 15 ECM proteins reported by Yoshida et al.22 (which represents 8% of ECM proteins detected in this study) and 76% of 91 ECM proteins reported by Cui et al.25 (which represents 39% of ECM proteins detected in this study) based on identification in one biologic replicate, which was reported in these published studies. Furthermore, we identified 12 times as many ECM proteins as Yoshida et al.22 and 2 times as many ECM proteins as Cui et al.25 Therefore, our study yielded the greatest number of ECM proteins from glomeruli to date, likely owing to the use of effective ECM enrichment before state-of-the-art MS analysis.
Table 1.
Basement membrane proteins in the glomerular ECM proteome
| Basement Membrane Proteins | Gene Name | Molecular Mass (kDa) | Abundance (nSC) | Classification |
|---|---|---|---|---|
| Agrin | AGRN | 215 | 3.075 | Glycoprotein |
| Collagen α1(XV) chain | COL15A1 | 142 | 0.052 | Collagen |
| Collagen α1(XVIII) chain | COL18A1 | 154 | 5.230 | Collagen |
| Collagen α1(IV) chain | COL4A1 | 161 | 12.515 | Collagen |
| Collagen α2(IV) chain | COL4A2 | 168 | 17.709 | Collagen |
| Collagen α3(IV) chain | COL4A3 | 162 | 8.155 | Collagen |
| Collagen α4(IV) chain | COL4A4 | 164 | 8.961 | Collagen |
| Collagen α5(IV) chain | COL4A5 | 161 | 3.778 | Collagen |
| Collagen α6(IV) chain | COL4A6 | 164 | 0.589 | Collagen |
| Fibulin-1 | FBLN1 | 77 | 0.029 | Glycoprotein |
| Fibrillin-1 | FBN1 | 312 | 1.403 | Glycoprotein |
| Fibronectin | FN1 | 263 | 1.893 | Glycoprotein |
| ECM protein FRAS1 | FRAS1 | 443 | 0.039 | Glycoprotein |
| Hemicentin-1 | HMCN1 | 613 | 0.007 | Glycoprotein |
| Perlecan | HSPG2 | 467 | 7.012 | Proteoglycan |
| Laminin subunit α2 | LAMA2 | 343 | 0.043 | Glycoprotein |
| Laminin subunit α5 | LAMA5 | 400 | 9.482 | Glycoprotein |
| Laminin subunit β1 | LAMB1 | 198 | 0.977 | Glycoprotein |
| Laminin subunit β2 | LAMB2 | 196 | 14.745 | Glycoprotein |
| Laminin subunit γ1 | LAMC1 | 178 | 8.818 | Glycoprotein |
| Nidogen-1 | NID1 | 136 | 10.247 | Glycoprotein |
| Nidogen-2 | NID2 | 151 | 1.249 | Glycoprotein |
| Tubulointerstitial nephritis antigen | TINAG | 55 | 12.873 | Glycoprotein |
| von Willebrand factor A domain–containing protein 1 | VWA1 | 47 | 1.171 | Glycoprotein |
The glomerular ECM proteome was further categorized according to GO annotation. Twenty-four basement membrane proteins were identified by MS. Relative protein abundance is shown as normalized spectral counts (nSCs).
Table 2.
Other structural ECM proteins in the glomerular ECM proteome
| Other Structural ECM Proteins | Gene Name | Molecular Mass (kDa) | Abundance (nSC) | Classification |
|---|---|---|---|---|
| Asporin | ASPN | 44 | 1.199 | Proteoglycan |
| Biglycan | BGN | 42 | 2.268 | Proteoglycan |
| Collagen α1(XII) chain | COL12A1 | 333 | 0.073 | Collagen |
| Collagen α1(I) chain | COL1A1 | 139 | 0.253 | Collagen |
| Collagen α2(I) chain | COL1A2 | 129 | 0.594 | Collagen |
| Collagen α1(III) chain | COL3A1 | 139 | 0.143 | Collagen |
| Collagen α1(VI) chain | COL6A1 | 109 | 12.685 | Collagen |
| Collagen α2(VI) chain | COL6A2 | 109 | 5.889 | Collagen |
| Collagen α3(VI) chain | COL6A3 | 344 | 11.145 | Collagen |
| Decorin | DCN | 40 | 0.067 | Proteoglycan |
| Dermatopontin | DPT | 24 | 0.397 | Glycoprotein |
| EMILIN-1 | EMILIN1 | 107 | 0.699 | Glycoprotein |
| Fibrinogen α-chain | FGA | 95 | 0.954 | Glycoprotein |
| Fibrinogen β-chain | FGB | 56 | 1.965 | Glycoprotein |
| γ-A of fibrinogen γ-chain | FGG | 49 | 2.002 | Glycoprotein |
| Matrix Gla protein | MGP | 12 | 0.557 | Glycoprotein |
| Nephronectin | NPNT | 65 | 3.707 | Glycoprotein |
| Periostin, osteoblast-specific factor | POSTN | 90 | 0.191 | Glycoprotein |
| RPE-spondin | RPESP | 30 | 1.364 | Glycoprotein |
| TGF-β–induced protein ig-h3 | TGFBI | 75 | 0.369 | Glycoprotein |
| Tubulointerstitial nephritis antigen-like | TINAGL1 | 52 | 20.010 | Glycoprotein |
| Vitronectin | VTN | 54 | 9.015 | Glycoprotein |
| von Willebrand factor A domain–containing protein 5B2 | VWA5B2 | 133 | 0.024 | Glycoprotein |
| von Willebrand factor A domain–containing protein 8 | VWA8 | 211 | 0.076 | Glycoprotein |
The glomerular ECM proteome was categorized according to GO annotation. Twenty-four proteins were identified as having a structural role but without basement membrane association, and they were termed other structural ECM proteins. Relative protein abundance is shown as normalized spectral counts (nSCs).
Figure 3.
Comparison of the glomerular ECM proteome with published glomerular proteomic datasets. The glomerular ECM proteome identified in this study was compared with other glomerular proteomic studies for which full datasets were available (studies by Cui et al.25 and Yoshida et al.22). Numbers of proteins in each intersection set of the area proportional Euler diagram are in bold italics. ECM proteins were categorized as basement membrane, other structural ECM, or ECM-associated proteins, and they were colored and arranged accordingly. Nodes (circles) are labeled with gene names for clarity. ECM proteins detected in any of the three biologic replicates reported in this study were included in the comparison with other proteomic datasets; these published datasets each reported one biologic replicate. Large node size indicates proteins detected in at least two biologic replicates in this study.
Creation of a Protein Interaction Network
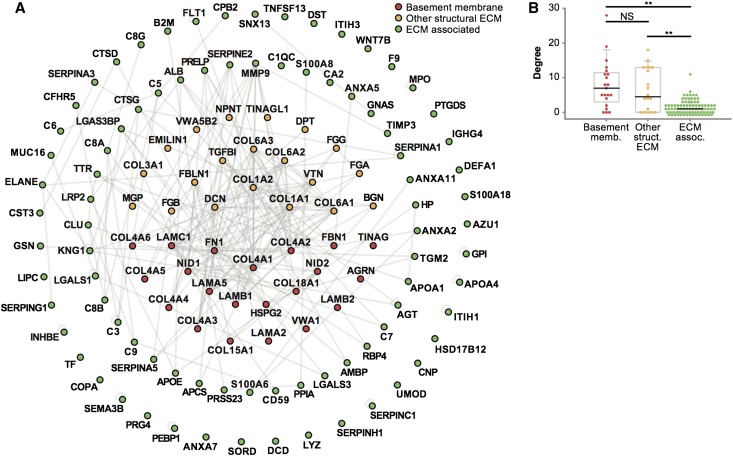
To visualize the components of glomerular ECM as a network of interacting proteins, the identified proteins were mapped onto a curated protein interaction database (Supplemental Methods) to generate an interaction network (Figure 4A). Statistical analysis showed that the network was more clustered than expected by chance, indicative of preferential interaction of proteins in a nonrandom network topology (Supplemental Table 3). Interestingly, basement membrane and structural ECM proteins were involved in more interactions with other proteins in the network than ECM-associated proteins (Figure 4B). Topological network analysis confirmed that basement membrane and other structural ECM proteins formed a highly connected core subnetwork, whereas ECM-associated proteins were less clustered in the network (Supplemental Figures 3 and 4). These data suggest that structural ECM proteins mediate multiple sets of protein–protein interactions and thus, have important roles in the assembly and organization of glomerular ECM.
Figure 4.
Interaction network analysis of human glomerular ECM. (A) Protein interaction network constructed from enriched glomerular ECM proteins identified by MS. Nodes (circles) represent proteins, and edges (gray lines) represent reported protein–protein interactions. ECM proteins were categorized as basement membrane, other structural ECM, or ECM-associated proteins, and they were colored and arranged accordingly. Nodes are labeled with gene names for clarity. (B) Distribution of degree (number of protein–protein interactions per protein) for basement membrane, other structural ECM, or ECM-associated proteins. Data points are shown as circles; outliers are shown as diamonds. **P<0.01. NS, P≥0.05.
Localization of Glomerular ECM Proteins
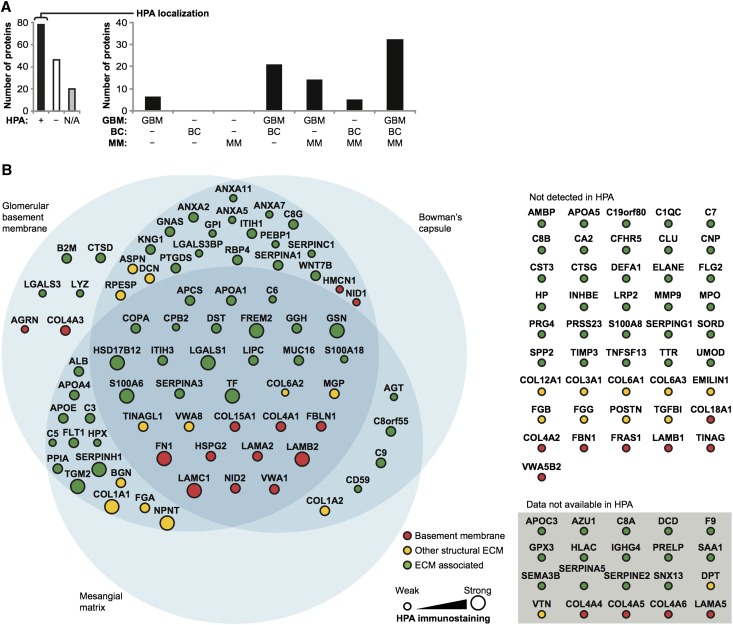
Validation of protein expression was performed by searching the Human Protein Atlas (HPA) database26 for 144 glomerular ECM proteins identified in this study (Figure 5). Glomerular immunostaining was not available for 20 proteins, including the known glomerular ECM proteins collagen IV α3, α4, and α5 and laminin α5. Protein expression was confirmed for 78 ECM components (63% of proteins with available HPA data) and reported as negative for 46 ECM components, although there were notable false negatives, including collagen IV α2, collagen XVIII, and laminin β1. Immunohistochemistry relies on antibody specificity, and therefore, combining expression data from antibody-based investigations with MS data has the potential to significantly increase the number of protein identifications. Indeed, our proteomic dataset increased the number of ECM proteins detected in the glomerulus by 59% compared with HPA data alone. We also evaluated the HPA database to determine the pattern of immunostaining as GBM, mesangial matrix, Bowman’s capsule, or a combination of compartments (Supplemental Methods). Most components were present in more than one ECM compartment (Figure 5B), suggesting a common core of protein components between the glomerular ECM compartments.
Figure 5.
Localization of glomerular ECM proteins in the HPA database. (A) The HPA was searched for glomerular ECM proteins identified in at least two biologic replicates in this study. (Left) Glomerular immunostaining was reviewed (+, detected; −, not detected; N/A, data not available in the HPA), and (right) localization was determined as GBM, mesangial matrix (MM), Bowman’s capsule (BC), or a combination of these ECM compartments. (B) ECM proteins were categorized as basement membrane, other structural ECM, or ECM-associated proteins, and (right) they were colored and arranged accordingly. Proteins not detected or without data in glomeruli in the HPA are shown separately. Nodes (circles) are labeled with gene names for clarity. (Left) Node diameter (proteins localized in glomeruli in the HPA only) is proportional to the intensity of glomerular immunostaining in the HPA.
Colocalization of Novel Glomerular ECM Proteins
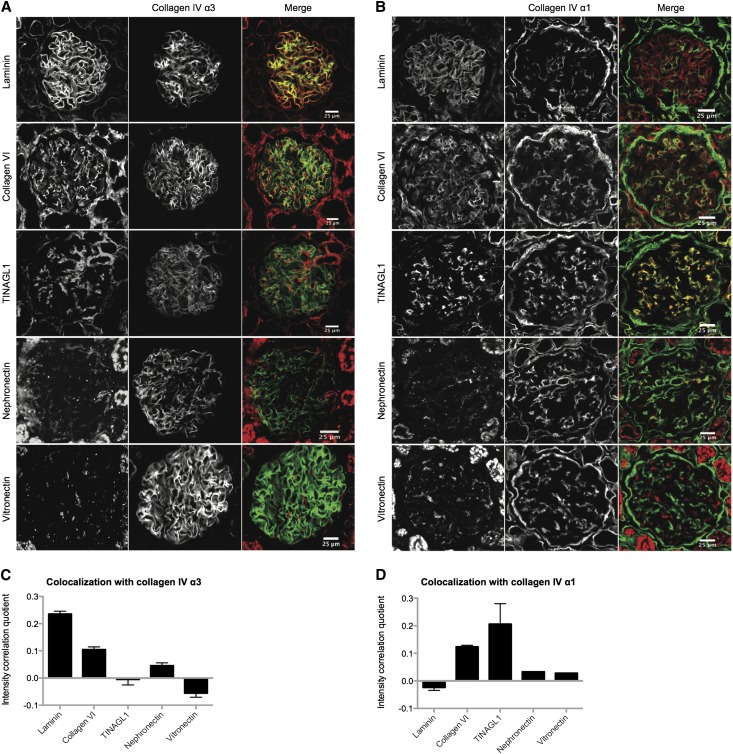
To confirm the expression of new glomerular ECM proteins, which were either abundant in our MS analysis or not detected in the HPA database, we conducted colocalization studies. Collagen VI and TINAGL1 were among the most abundant proteins detected in this study (Supplemental Figure 2), and they were validated by Western blotting (Figure 2D). Collagen VI was also present in the highly connected subnetwork of ECM proteins (Supplemental Figures 3 and 4). Immunohistochemical and correlation intensity analysis of human renal cortex revealed colocalization of TINAGL1 with collagen IV α1, which has a mesangial pattern of immunostaining, whereas collagen VI overlapped with both collagen IV α1 and collagen IV α3/laminin, which both have a GBM pattern of immunostaining (Figure 6, A and C). MS analysis also detected nephronectin and vitronectin (Table 2). Correlation intensity analysis showed that nephronectin localized in the GBM and mesangial matrix, whereas vitronectin localized in the mesangial matrix alone (Figure 6, B and D). This objective and quantitative method of colocalization allows protein expression to be correlated with reliable markers predominating in distinct ECM compartments. With antibodies of suitable specificity, this approach could be extended to map the full proteome into glomerular ECM compartments.
Figure 6.
Colocalization of novel and known glomerular ECM proteins. (A and B) Immunohistochemistry of human renal cortex was used to examine the colocalization of collagen VI, TINAGL1, nephronectin, and vitronectin with laminin, (A) collagen IV α3, and (B) collagen IV α1. (C and D) Bar charts show intensity correlation quotients calculated from immunohistochemistry images (n=6–10 images for each analysis), showing (C) colocalization of laminin, collagen VI, and nephronectin with collagen IV α3 and (D) colocalization of collagen VI, TINAGL1, nephronectin, and vitronectin with collagen IV α1.
Discussion
We used an ECM enrichment strategy coupled with an unbiased proteomics approach to confirm the presence of all known glomerular ECM proteins in addition to many potentially novel components. Moreover, using immunohistochemistry, we have confirmed the MS identification of TINAGL1, collagen VI, nephronectin, and vitronectin within specific glomerular compartments. These findings show that the composition of glomerular ECM is far more complex than previously appreciated and imply that many more ECM components may contribute to glomerular development and disease processes.
This unbiased, global approach to define the glomerular ECM proteome shows that this specialized ECM has a core of highly connected extracellular components. In adult human glomeruli, 144 ECM proteins, including all previously described components, were identified. In addition, we found many more structural and regulatory ECM proteins, revealing the complexity of the glomerular ECM. This proteome is comparable in size with ECM profiles recently reported for vasculature-, lung-, and bowel-derived ECMs.3,27,28 In our analysis, several novel glomerular proteins were highly abundant, including collagen VI and TINAGL1. Collagen VI is a structural component that forms microfibrils, and it is important for muscle function29,30; however, its role in the glomerulus has not been investigated. Our study shows that collagen VI is present in a highly connected core network of proteins in the glomerular ECM, and it is localized within both the GBM and mesangial matrix. TINAGL1 was also abundantly expressed in glomerular ECM, where it predominantly localized to the mesangial matrix. TINAGL1 (also known as TIN-Ag_RP, lipocalin-7, oxidised LDL-responsive gene 2 and androgen regulated gene 1) is a glycoprotein, and it is structurally related to TINAG, a tubular basement membrane component, which is the antigenic target in autoimmune anti–tubular basement membrane disease.31 TINAGL1 has a proteolytically inactive cathepsin domain,32 and it has been shown to have a role in angiogenesis.33 However, its function within the glomerulus is undefined. Both of these highly abundant proteins may have roles in barrier function or glomerular disease.
To assess the relative abundance of ECM proteins, we used two distinct methods of quantification (peptide intensity analysis20 and spectral counting), and both approaches gave very similar results. Peptide intensity analysis revealed collagen IV and laminin isoforms to be the most abundant proteins in the glomerular ECM, and both analyses revealed abundant collagen VI and TINAGL1. There were some differences between the methods for the quantification of collagen isoforms, but the stoichiometry of different proteins is difficult to determine by any global proteomic methodology, including antibody-based techniques, which rely on probe immunoaffinity. To perform absolute protein quantification by MS, labeled peptide standards for each protein could be used; however, it would be a significant undertaking for a large protein dataset and would not permit the discovery of unknown ECM proteins. Therefore, the findings presented in this study provide relative patterns of glomerular ECM protein enrichment and pave the way for additional in-depth studies of the ECM components identified.
Compared with other large-scale glomerular proteomic studies, which have identified up to a total of 1800 proteins,22,25 this investigation had the greatest yield of ECM proteins, thus highlighting the importance of sample fractionation for improving protein identification. However, the large-scale requirement for the analysis prohibited the separation of glomerular ECM components, and therefore, protein localization was determined by immunohistochemistry. Although laser microdissection studies have the ability to precisely define the anatomic structure for analysis, the total number of proteins identified in the most recent of these studies, using a variety of tissue samples, ranged from 114 to 340, which is fewer proteins than the large-scale glomerular studies and significantly fewer ECM proteins compared with this study.13,34,35 Future developments in technologies, which may couple tissue imaging with improved sensitivity of molecular analysis, may allow direct compositional analysis of ECM within distinct tissue compartments.36,37
The profile of glomerular ECM that we have described is likely to represent a snapshot of a highly dynamic extracellular environment that changes under the influence of cellular, physical, and chemical environmental cues. Nonetheless, the datasets provide a valuable resource for additional investigation of the composition and complexity of glomerular ECM. Our methodology can now be applied to the comparison of glomerular ECMs in vivo in the context of development or disease. Systems-level analysis will be integral to the downstream interrogation of data to identify informative, predictive networks of ECM composition and function. Combined with the methodologies and datasets described herein, such analyses will enable the construction of a dynamic glomerular ECM interactome and help to build understanding of how this network may alter during glomerular development and disease.
Concise Methods
Antibodies
Monoclonal antibodies used were against actin (clone AC-40; Sigma-Aldrich, Poole, UK), nephronectin (ab64419; Abcam, Cambridge, UK), TINAGL1 (ab69036; Abcam), vitronectin (ab11591; Abcam), and collagen IV chain–specific antibodies (provided by B. Hudson, Vanderbilt University Medical Center, Nashville, TN). Polyclonal antibodies used were against pancollagen IV (ab6586; Abcam), panlaminin (ab11575; Abcam), pancollagen VI (ab6588; Abcam), lamin B1 (ab16048; Abcam), and nephrin (ab58968; Abcam). Secondary antibodies against rabbit IgG conjugated to tetramethylrhodamineisothiocyanate and mouse or rat IgG conjugated to FITC (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) were used for immunofluorescence; secondary antibodies conjugated to Alexa Fluor 680 (Life Technologies, Paisley, UK) or IRDye 800 (Rockland Immunochemicals, Glibertsville, PA) were used for Western blotting.
Isolation of Human Glomeruli
Normal renal cortex from human donor kidneys technically unsuitable for transplantation was used with full ethical approval (reference 06/Q1406/38). Three adult men donors between 37 and 63 years of age were chosen to reduce the influence of age and sex. Normal renal function was determined by prenephrectomy serum creatinine values. At 4°C, renal cortex (2.5 g) was finely diced and pressed onto a 250-μm sieve (Endecotts, London, UK) using a 5-ml syringe plunger. Glomeruli were rinsed through sieves (250 and 150 μm) with cold PBS and separated from tubular fragments by collection on both 150- and 100-μm sieves. Retained glomeruli were retrieved into 10 ml PBS and washed another three times with PBS and interval centrifugation. Glomerular purity was consistently >95% as determined by counting whole glomeruli and nonglomerular fragments using phase-contrast light microscopy.
Isolation of Enriched Glomerular ECM
This method was adapted from previously published methods38 and used to reduce the complexity of protein samples for MS analysis by removing cellular components and enriching for ECM proteins. All steps were carried out at 4°C to minimize proteolysis. Pure glomerular isolates from three human kidneys were incubated for 30 minutes in extraction buffer (10 mM Tris, 150 mM NaCl, 1% [vol/vol] Triton X-100, 25 mM EDTA, 25 μg/ml leupeptin, 25 μg/ml aprotinin, and 0.5 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride) to solubilize cellular proteins, and samples were then centrifuged at 14,000×g for 10 minutes to yield fraction 1. The remaining pellet was incubated for 30 minutes in alkaline detergent buffer (20 mM NH4OH and 0.5% [vol/vol] Triton X-100 in PBS) to further solubilize cellular proteins and disrupt cell–ECM interactions. Samples were then centrifuged at 14,000×g for 10 minutes to yield fraction 2. The remaining pellet was incubated for 30 minutes in a deoxyribonuclease buffer (10 μg/ml deoxyribonuclease I [Roche, Burgess Hill, UK] in PBS) to degrade DNA. The sample was centrifuged at 14,000×g for 10 minutes to yield fraction 3, and the final pellet was resuspended in reducing sample buffer (50 mM Tris⋅HCl, pH 6.8, 10% [wt/vol] glycerol, 4% [wt/vol] SDS, 0.004% [wt/vol] bromophenol blue, and 8% [vol/vol] β-mercaptoethanol) to yield the ECM fraction. Samples were heat denatured at 70°C for 20 minutes.
Western Blotting
After SDS-PAGE, resolved proteins were transferred to nitrocellulose membrane (Whatman, Maidstone, UK). Membranes were blocked with casein blocking buffer (Sigma-Aldrich) and probed with primary antibodies diluted in blocking buffer containing 0.05% (vol/vol) Tween 20. Membranes were washed with Tris-buffered saline (10 mM Tris⋅HCl, pH 7.4, and 150 mM NaCl) containing 0.05% (vol/vol) Tween 20 and incubated with species-specific fluorescent dye–conjugated secondary antibodies diluted in blocking buffer containing 0.05% (vol/vol) Tween 20. Membranes were washed in the dark and then scanned using the Odyssey infrared imaging system (LI-COR Biosciences, Cambridge, UK) to visualize bound antibodies.
MS Data Acquisition
Protein samples were resolved by SDS-PAGE and visualized by Coomassie staining. Gel lanes were sliced and subjected to in-gel trypsin digestion as described previously39 Liquid chromatography–tandem MS analysis was performed using a nanoACQUITY UltraPerformance liquid chromatography system (Waters, Elstree, UK) coupled online to an LTQ Velos mass spectrometer (Thermo Fisher Scientific, Waltham, MA) or offline to an Orbitrap Elite analyzer (Thermo Fisher Scientific) for experiments incorporating analysis with Progenesis (Supplemental Methods). Peptides were concentrated and desalted on a Symmetry C18 preparative column (20-mm length, 180-μm inner diameter, 5-μm particle size, 100-Å pore size; Waters). Peptides were separated on a bridged ethyl hybrid C18 analytical column (250-mm length, 75-μm inner diameter, 1.7-μm particle size, 130-Å pore size; Waters) using a 45-minute linear gradient from 1% to 25% (vol/vol) acetonitrile in 0.1% (vol/vol) formic acid at a flow rate of 200 nl/min. Peptides were selected for fragmentation automatically by data-dependent analysis.
MS Data Analyses
Tandem mass spectra were extracted using extract_msn (Thermo Fisher Scientific) executed in Mascot Daemon (version 2.2.2; Matrix Science, London, UK). Peak list files were searched against a modified version of the International Protein Index Human Database (version 3.70; release date, March 4, 2010) containing 10 additional contaminant and reagent sequences of nonhuman origin using Mascot (version 2.2.03; Matrix Science).40 Carbamidomethylation of cysteine was set as a fixed modification; oxidation of methionine and hydroxylation of proline and lysine were allowed as variable modifications. Only tryptic peptides were considered, with up to one missed cleavage permitted. Monoisotopic precursor mass values were used, and only doubly and triply charged precursor ions were considered. Mass tolerances for precursor and fragment ions were 0.4 and 0.5 Da, respectively. MS datasets were validated using rigorous statistical algorithms at both the peptide and protein levels41,42 implemented in Scaffold (version 3.00.06; Proteome Software, Portland, OR). Protein identifications were accepted on assignment of at least two unique validated peptides with ≥90% probability, resulting in ≥99% probability at the protein level. These acceptance criteria resulted in an estimated protein false discovery rate of 0.1% for all datasets.
MS Quantification and Proteomic Analyses
MS quantification and proteomic data analyses were performed as previously described39,43 with the modifications described in Supplemental Methods. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD000456 (http://www.ebi.ac.uk/pride).
Immunohistochemistry and Image Analysis
Formalin-fixed, paraffin-embedded tissue blocks were sectioned at 5 μm. Sections were dewaxed and treated with recombinant proteinase K (Roche Diagnostics, Indianapolis, IN) for 15 minutes. Sections were blocked with 5% (vol/vol) donkey serum (Sigma-Aldrich) and 1.5% (vol/vol) BSA (Sigma-Aldrich) for 30 minutes and primary antibodies overnight at 4°C. Sections were washed three times with PBS, incubated with secondary antibodies, mounted with polyvinyl alcohol mounting medium (Fluka 10981; Sigma-Aldrich), and imaged using a BX51 upright microscope (Olympus, Southend on Sea, UK) equipped with a 20× UPlan Fln 0.50 objective and controlled through MetaVue software (Molecular Devices, Wokingham, UK). Images were collected using a CoolSnap HQ camera (Photometrics, Tucson, AZ) and separate 4′,6-diamidino-2-phenylindole, FITC, and Cy3 filters (U-MWU2, 41001, and 41007a, respectively; Chroma, Olching, Germany) to minimize bleed through between the different channels. Images were processed and analyzed using Fiji/ImageJ software (version 1.46r; National Institutes of Health, Bethesda, MD). Raw images were subjected to signal rescaling using linear transformation for display in the figures. For calculation of the Pearson’s correlation coefficient, a region of interest was drawn, and a threshold was set to restrict analysis to a single glomerulus. The coefficient was measured using the Intensity Correlation Analysis plugin for Fiji/ImageJ,44 and the subsequent plotting steps were performed using MATLAB (version R2012a; MathWorks, Natick, MA).
Statistical Analyses
All measurements are shown as mean±SEM. Box plots indicate 25th and 75th percentiles (lower and upper bounds, respectively), 1.5× interquartile range (whiskers), and median (black line). Numbers of protein–protein interactions were compared using Kruskal–Wallis one-way ANOVA tests with post hoc Bonferroni correction. GO enrichment analyses were compared using modified Fisher’s exact tests with Benjamini–Hochberg correction. P values <0.05 were deemed significant.
Disclosures
None.
Acknowledgments
Mass spectrometry was performed in the Biological Mass Spectrometry Core Facility, Faculty of Life Sciences, University of Manchester, and we thank Stacey Warwood for advice and technical support. We thank Julian Selley for bioinformatic support. Microscopy was performed in the Bioimaging Core Facility, Faculty of Life Sciences, University of Manchester. The collagen IV chain-specific antibodies were kindly provided by the Billy Hudson research group, Department of Medicine, Vanderbilt Medical Center.
This work was supported by Wellcome Trust Intermediate Fellowship Award Reference 090006 (to R.L.) and Wellcome Trust Grant 092015 (to M.J.H.). R.Z. is supported by Veterans Affairs Merit Awards 1I01BX002196-01, DK075594, DK069221, and DK083187, and an American Heart Association Established Investigator Award. The mass spectrometer and microscopes used in this study were purchased with grants from the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, and the University of Manchester Strategic Fund.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013030233/-/DCSupplemental.
References
- 1.Pozzi A, Jarad G, Moeckel GW, Coffa S, Zhang X, Gewin L, Eremina V, Hudson BG, Borza DB, Harris RC, Holzman LB, Phillips CL, Fassler R, Quaggin SE, Miner JH, Zent R: Beta1 integrin expression by podocytes is required to maintain glomerular structural integrity. Dev Biol 316: 288–301, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Has C, Spartà G, Kiritsi D, Weibel L, Moeller A, Vega-Warner V, Waters A, He Y, Anikster Y, Esser P, Straub BK, Hausser I, Bockenhauer D, Dekel B, Hildebrandt F, Bruckner-Tuderman L, Laube GF: Integrin α3 mutations with kidney, lung, and skin disease. N Engl J Med 366: 1508–1514, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO: The matrisome: In silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics 11: M111.014647, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yurchenco PD: Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb Perspect Biol 3: pii: a004911, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miner JH: The glomerular basement membrane. Exp Cell Res 318: 973–978, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miner JH: Renal basement membrane components. Kidney Int 56: 2016–2024, 1999 [DOI] [PubMed] [Google Scholar]
- 7.Courtoy PJ, Kanwar YS, Hynes RO, Farquhar MG: Fibronectin localization in the rat glomerulus. J Cell Biol 87: 691–696, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker DF, Hostikka SL, Zhou J, Chow LT, Oliphant AR, Gerken SC, Gregory MC, Skolnick MH, Atkin CL, Tryggvason K: Identification of mutations in the COL4A5 collagen gene in Alport syndrome. Science 248: 1224–1227, 1990 [DOI] [PubMed] [Google Scholar]
- 9.Zenker M, Aigner T, Wendler O, Tralau T, Müntefering H, Fenski R, Pitz S, Schumacher V, Royer-Pokora B, Wühl E, Cochat P, Bouvier R, Kraus C, Mark K, Madlon H, Dötsch J, Rascher W, Maruniak-Chudek I, Lennert T, Neumann LM, Reis A: Human laminin beta2 deficiency causes congenital nephrosis with mesangial sclerosis and distinct eye abnormalities. Hum Mol Genet 13: 2625–2632, 2004 [DOI] [PubMed] [Google Scholar]
- 10.Fogo AB: Mesangial matrix modulation and glomerulosclerosis. Exp Nephrol 7: 147–159, 1999 [DOI] [PubMed] [Google Scholar]
- 11.Spiro RG: Studies on the renal glomerular basement membrane. Preparation and chemical composition. J Biol Chem 242: 1915–1922, 1967 [PubMed] [Google Scholar]
- 12.van den Heuvel LP, van den Born J, van de Velden TJ, Veerkamp JH, Monnens LA, Schroder CH, Berden JH: Isolation and partial characterization of heparan sulphate proteoglycan from the human glomerular basement membrane. Biochem J 264: 457–465, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shapiro JP, Biswas S, Merchant AS, Satoskar A, Taslim C, Lin S, Rovin BH, Sen CK, Roy S, Freitas MA: A quantitative proteomic workflow for characterization of frozen clinical biopsies: Laser capture microdissection coupled with label-free mass spectrometry. J Proteomics 77: 433–440, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakatani S, Wei M, Ishimura E, Kakehashi A, Mori K, Nishizawa Y, Inaba M, Wanibuchi H: Proteome analysis of laser microdissected glomeruli from formalin-fixed paraffin-embedded kidneys of autopsies of diabetic patients: Nephronectin is associated with the development of diabetic glomerulosclerosis. Nephrol Dial Transplant 27: 1889–1897, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Byron A, Humphries JD, Humphries MJ: Defining the extracellular matrix using proteomics [published online ahead of print February 19, 2013]. Int J Exp Pathol 10.1111/iep.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eremina V, Sood M, Haigh J, Nagy A, Lajoie G, Ferrara N, Gerber HP, Kikkawa Y, Miner JH, Quaggin SE: Glomerular-specific alterations of VEGF-A expression lead to distinct congenital and acquired renal diseases. J Clin Invest 111: 707–716, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welsh GI, Hale LJ, Eremina V, Jeansson M, Maezawa Y, Lennon R, Pons DA, Owen RJ, Satchell SC, Miles MJ, Caunt CJ, McArdle CA, Pavenstädt H, Tavaré JM, Herzenberg AM, Kahn CR, Mathieson PW, Quaggin SE, Saleem MA, Coward RJ: Insulin signaling to the glomerular podocyte is critical for normal kidney function. Cell Metab 12: 329–340, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokoi H, Mukoyama M, Mori K, Kasahara M, Suganami T, Sawai K, Yoshioka T, Saito Y, Ogawa Y, Kuwabara T, Sugawara A, Nakao K: Overexpression of connective tissue growth factor in podocytes worsens diabetic nephropathy in mice. Kidney Int 73: 446–455, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Kahsai TZ, Enders GC, Gunwar S, Brunmark C, Wieslander J, Kalluri R, Zhou J, Noelken ME, Hudson BG: Seminiferous tubule basement membrane. Composition and organization of type IV collagen chains, and the linkage of alpha3(IV) and alpha5(IV) chains. J Biol Chem 272: 17023–17032, 1997 [DOI] [PubMed] [Google Scholar]
- 20.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ: Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol Cell Proteomics 5: 144–156, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Xu BJ, Shyr Y, Liang X, Ma LJ, Donnert EM, Roberts JD, Zhang X, Kon V, Brown NJ, Caprioli RM, Fogo AB: Proteomic patterns and prediction of glomerulosclerosis and its mechanisms. J Am Soc Nephrol 16: 2967–2975, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Yoshida Y, Miyazaki K, Kamiie J, Sato M, Okuizumi S, Kenmochi A, Kamijo K, Nabetani T, Tsugita A, Xu B, Zhang Y, Yaoita E, Osawa T, Yamamoto T: Two-dimensional electrophoretic profiling of normal human kidney glomerulus proteome and construction of an extensible markup language (XML)-based database. Proteomics 5: 1083–1096, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto M, Yoshida Y, Taguchi I, Nagasaka Y, Tasaki M, Zhang Y, Xu B, Nameta M, Sezaki H, Cuellar LM, Osawa T, Morishita H, Sekiyama S, Yaoita E, Kimura K, Yamamoto T: In-depth proteomic profiling of the normal human kidney glomerulus using two-dimensional protein prefractionation in combination with liquid chromatography-tandem mass spectrometry. J Proteome Res 6: 3680–3690, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Yoshida Y, Xu B, Magdeldin S, Fujinaka H, Liu Z, Miyamoto M, Yaoita E, Yamamoto T: Comparison of human glomerulus proteomic profiles obtained from low quantities of samples by different mass spectrometry with the comprehensive database. Proteome Sci 9: 47, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Z, Yoshida Y, Xu B, Zhang Y, Nameta M, Magdeldin S, Makiguchi T, Ikoma T, Fujinaka H, Yaoita E, Yamamoto T: Profiling and annotation of human kidney glomerulus proteome. Proteome Sci 11: 13, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F: Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28: 1248–1250, 2010 [DOI] [PubMed] [Google Scholar]
- 27.Didangelos A, Yin X, Mandal K, Baumert M, Jahangiri M, Mayr M: Proteomics characterization of extracellular space components in the human aorta. Mol Cell Proteomics 9: 2048–2062, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angel PM, Nusinow D, Brown CB, Violette K, Barnett JV, Zhang B, Baldwin HS, Caprioli RM: Networked-based characterization of extracellular matrix proteins from adult mouse pulmonary and aortic valves. J Proteome Res 10: 812–823, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bönnemann CG: The collagen VI-related myopathies Ullrich congenital muscular dystrophy and Bethlem myopathy. Handb Clin Neurol 101: 81–96, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonaldo P, Braghetta P, Zanetti M, Piccolo S, Volpin D, Bressan GM: Collagen VI deficiency induces early onset myopathy in the mouse: An animal model for Bethlem myopathy. Hum Mol Genet 7: 2135–2140, 1998 [DOI] [PubMed] [Google Scholar]
- 31.Miyazato H, Yoshioka K, Hino S, Aya N, Matsuo S, Suzuki N, Suzuki Y, Sinohara H, Maki S: The target antigen of anti-tubular basement membrane antibody-mediated interstitial nephritis. Autoimmunity 18: 259–265, 1994 [DOI] [PubMed] [Google Scholar]
- 32.Wex T, Lipyansky A, Brömme NC, Wex H, Guan XQ, Brömme D: TIN-ag-RP, a novel catalytically inactive cathepsin B-related protein with EGF domains, is predominantly expressed in vascular smooth muscle cells. Biochemistry 40: 1350–1357, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Brown LJ, Alawoki M, Crawford ME, Reida T, Sears A, Torma T, Albig AR: Lipocalin-7 is a matricellular regulator of angiogenesis. PLoS One 5: e13905, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kley RA, Maerkens A, Leber Y, Theis V, Schreiner A, van der Ven PF, Uszkoreit J, Stephan C, Eulitz S, Euler N, Kirschner J, Müller K, Meyer HE, Tegenthoff M, Fürst DO, Vorgerd M, Müller T, Marcus K: A combined laser microdissection and mass spectrometry approach reveals new disease relevant proteins accumulating in aggregates of filaminopathy patients. Mol Cell Proteomics 12: 215–227, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mu Y, Chen Y, Zhang G, Zhan X, Li Y, Liu T, Li G, Li M, Xiao Z, Gong X, Chen Z: Identification of stromal differentially expressed proteins in the colon carcinoma by quantitative proteomics. Electrophoresis 34: 1679–1692, 2013 [DOI] [PubMed] [Google Scholar]
- 36.Seeley EH, Caprioli RM: 3D imaging by mass spectrometry: A new frontier. Anal Chem 84: 2105–2110, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angel PM, Caprioli RM: Matrix-assisted laser desorption ionization imaging mass spectrometry: In situ molecular mapping. Biochemistry 52: 3818–3828, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todorović V, Desai BV, Eigenheer RA, Yin T, Amargo EV, Mrksich M, Green KJ, Patterson MJ: Detection of differentially expressed basal cell proteins by mass spectrometry. Mol Cell Proteomics 9: 351–361, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphries JD, Byron A, Bass MD, Craig SE, Pinney JW, Knight D, Humphries MJ: Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci Signal 2: ra51, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS: Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567, 1999 [DOI] [PubMed] [Google Scholar]
- 41.Keller A, Nesvizhskii AI, Kolker E, Aebersold R: Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392, 2002 [DOI] [PubMed] [Google Scholar]
- 42.Nesvizhskii AI, Keller A, Kolker E, Aebersold R: A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658, 2003 [DOI] [PubMed] [Google Scholar]
- 43.Rashid ST, Humphries JD, Byron A, Dhar A, Askari JA, Selley JN, Knight D, Goldin RD, Thursz M, Humphries MJ: Proteomic analysis of extracellular matrix from the hepatic stellate cell line LX-2 identifies CYR61 and Wnt-5a as novel constituents of fibrotic liver. J Proteome Res 11: 4052–4064, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF: A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: Analysis by quantitative immunocolocalization. J Neurosci 24: 4070–4081, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]