Abstract
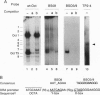
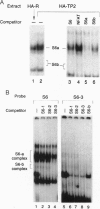
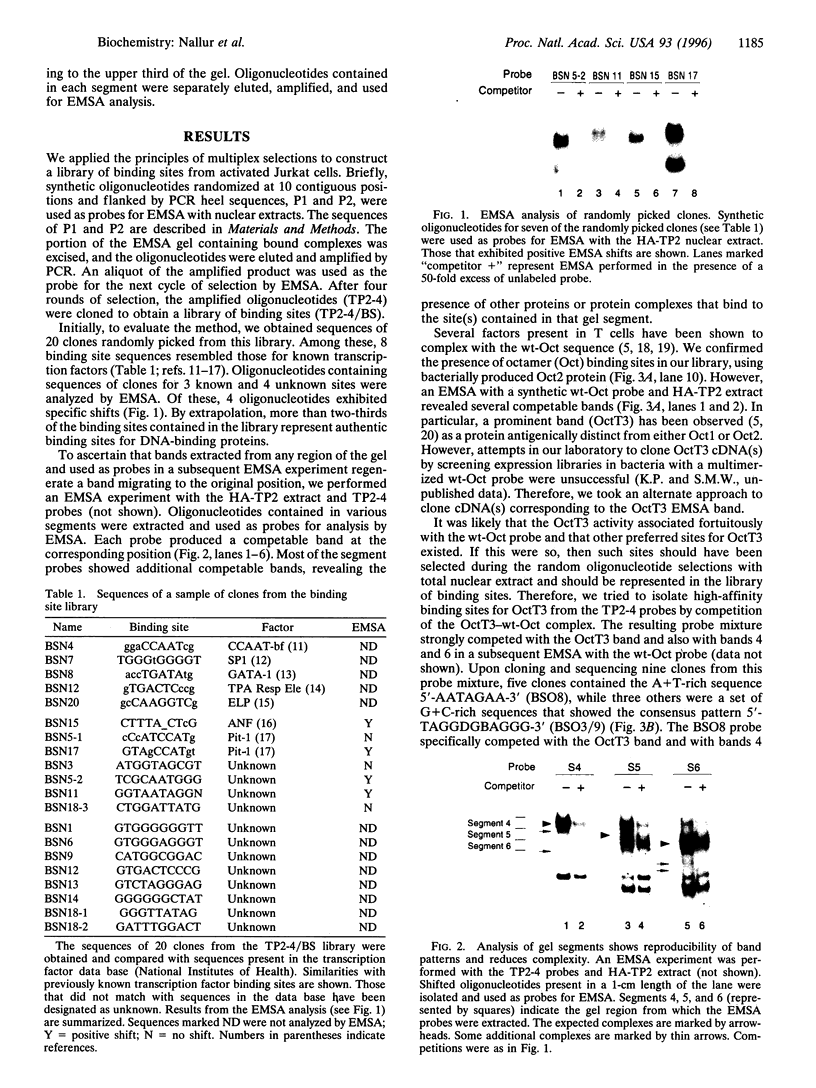
We have used a multiplex selection approach to construct a library of DNA-protein interaction sites recognized by many of the DNA-binding proteins present in a cell type. An estimated minimum of two-thirds of the binding sites present in a library prepared from activated Jurkat T cells represent authentic transcription factor binding sites. We used the library for isolation of "optimal" binding site probes that facilitated cloning of a factor and to identify binding activities induced within 2 hr of activation of Jurkat cells. Since a large fraction of the oligonucleotides obtained appear to represent "optimal" binding sites for sequence-specific DNA-binding proteins, it is feasible to construct a catalog of consensus binding sites for DNA-binding proteins in a given cell type. Qualitative and quantitative comparisons of the catalogs of binding site sequences from various cell types could provide valuable insights into the process of differentiation acting at the level of transcriptional control.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Bhargava A. K., Li Z., Weissman S. M. Differential expression of four members of the POU family of proteins in activated and phorbol 12-myristate 13-acetate-treated Jurkat T cells. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10260–10264. doi: 10.1073/pnas.90.21.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Buckingham M. E. Muscle: the regulation of myogenesis. Curr Opin Genet Dev. 1994 Oct;4(5):745–751. doi: 10.1016/0959-437x(94)90142-p. [DOI] [PubMed] [Google Scholar]
- Chang C. P., Shen W. F., Rozenfeld S., Lawrence H. J., Largman C., Cleary M. L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995 Mar 15;9(6):663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- Delhase M., Vergani P., Malur A., Hooghe-Peters E. L., Hooghe R. J. The transcription factor Pit-1/GHF-1 is expressed in hemopoietic and lymphoid tissues. Eur J Immunol. 1993 Apr;23(4):951–955. doi: 10.1002/eji.1830230428. [DOI] [PubMed] [Google Scholar]
- Desprez P. Y., Hara E., Bissell M. J., Campisi J. Suppression of mammary epithelial cell differentiation by the helix-loop-helix protein Id-1. Mol Cell Biol. 1995 Jun;15(6):3398–3404. doi: 10.1128/mcb.15.6.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier D. K., Schiffenbauer J., Woulfe S. L., Zacheis M., Schwartz B. D. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Elsholtz H. P., Albert V. R., Treacy M. N., Rosenfeld M. G. A two-base change in a POU factor-binding site switches pituitary-specific to lymphoid-specific gene expression. Genes Dev. 1990 Jan;4(1):43–51. doi: 10.1101/gad.4.1.43. [DOI] [PubMed] [Google Scholar]
- Evans T., Reitman M., Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisst S., Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992 Jan 11;20(1):3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan W. M., Corthésy B., Bram R. J., Crabtree G. R. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991 Aug 29;352(6338):803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- Funk W. D., Wright W. E. Cyclic amplification and selection of targets for multicomponent complexes: myogenin interacts with factors recognizing binding sites for basic helix-loop-helix, nuclear factor 1, myocyte-specific enhancer-binding factor 2, and COMP1 factor. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9484–9488. doi: 10.1073/pnas.89.20.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruffat H., Sergeant A. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 1994 Apr 11;22(7):1172–1178. doi: 10.1093/nar/22.7.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C. V., Graves B. J. Identification of ETS domain proteins in murine T lymphocytes that interact with the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1994 Nov;14(11):7569–7580. doi: 10.1128/mcb.14.11.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R. S., Boczko E. M., Darnell J. E., Jr, Babiss L. E. The mouse albumin enhancer contains a negative regulatory element that interacts with a novel DNA-binding protein. Mol Cell Biol. 1990 Aug;10(8):3896–3905. doi: 10.1128/mcb.10.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989 Dec 14;342(6251):749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Irvine D., Tuerk C., Gold L. SELEXION. Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J Mol Biol. 1991 Dec 5;222(3):739–761. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- John S., Reeves R. B., Lin J. X., Child R., Leiden J. M., Thompson C. B., Leonard W. J. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-kappa B family proteins. Mol Cell Biol. 1995 Mar;15(3):1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston R. E., Schuetz T. J., Larin Z. Heat-inducible human factor that binds to a human hsp70 promoter. Mol Cell Biol. 1987 Apr;7(4):1530–1534. doi: 10.1128/mcb.7.4.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Hollenberg S. M., Snider L., Turner D. L., Lipnick N., Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995 May 12;268(5212):836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987 Jun 19;49(6):741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- Lew A. M., Elsholtz H. P. Cloning of the human cDNA for transcription factor Pit-1. Nucleic Acids Res. 1991 Nov 25;19(22):6329–6329. doi: 10.1093/nar/19.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald G. H., Itoh-Lindstrom Y., Ting J. P. The transcriptional regulatory protein, YB-1, promotes single-stranded regions in the DRA promoter. J Biol Chem. 1995 Feb 24;270(8):3527–3533. doi: 10.1074/jbc.270.8.3527. [DOI] [PubMed] [Google Scholar]
- Messier H., Brickner H., Gaikwad J., Fotedar A. A novel POU domain protein which binds to the T-cell receptor beta enhancer. Mol Cell Biol. 1993 Sep;13(9):5450–5460. doi: 10.1128/mcb.13.9.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parslow T. G., Blair D. L., Murphy W. J., Granner D. K. Structure of the 5' ends of immunoglobulin genes: a novel conserved sequence. Proc Natl Acad Sci U S A. 1984 May;81(9):2650–2654. doi: 10.1073/pnas.81.9.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. A., Rogers A. E., Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET). EMBO J. 1995 Jun 1;14(11):2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E., Matthias P., Müller M. M., Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989 Aug 11;17(15):6419–6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen H. J., Bach C. Target Detection Assay (TDA): a versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res. 1990 Jun 11;18(11):3203–3209. doi: 10.1093/nar/18.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T., Niwa O., Yokoro K. Mechanism of suppression of the long terminal repeat of Moloney leukemia virus in mouse embryonal carcinoma cells. Mol Cell Biol. 1989 Nov;9(11):4670–4676. doi: 10.1128/mcb.9.11.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymms M. J., Kola I. Regulation of gene expression by transcription factors Ets-1 and Ets-2. Mol Reprod Dev. 1994 Oct;39(2):208–214. doi: 10.1002/mrd.1080390214. [DOI] [PubMed] [Google Scholar]
- Ullman K. S., Flanagan W. M., Edwards C. A., Crabtree G. R. Activation of early gene expression in T lymphocytes by Oct-1 and an inducible protein, OAP40. Science. 1991 Oct 25;254(5031):558–562. doi: 10.1126/science.1683003. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., LaMarco K. L., Johnson P. F., Landschulz W. H., McKnight S. L. In situ detection of sequence-specific DNA binding activity specified by a recombinant bacteriophage. Genes Dev. 1988 Jul;2(7):801–806. doi: 10.1101/gad.2.7.801. [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Petryniak B., Ho I. C., Thompson C. B., Leiden J. M. Evolutionarily conserved Ets family members display distinct DNA binding specificities. J Exp Med. 1992 May 1;175(5):1391–1399. doi: 10.1084/jem.175.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederrecht G., Shuey D. J., Kibbe W. A., Parker C. S. The Saccharomyces and Drosophila heat shock transcription factors are identical in size and DNA binding properties. Cell. 1987 Feb 13;48(3):507–515. doi: 10.1016/0092-8674(87)90201-7. [DOI] [PubMed] [Google Scholar]
- Wright W. E., Funk W. D. CASTing for multicomponent DNA-binding complexes. Trends Biochem Sci. 1993 Mar;18(3):77–80. doi: 10.1016/0968-0004(93)90156-h. [DOI] [PubMed] [Google Scholar]
- Wurster A. L., Siu G., Leiden J. M., Hedrick S. M. Elf-1 binds to a critical element in a second CD4 enhancer. Mol Cell Biol. 1994 Oct;14(10):6452–6463. doi: 10.1128/mcb.14.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. S., Hanke J. H., Carayannopoulos L., Craft C. M., Capra J. D., Tucker P. W. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol Cell Biol. 1993 Sep;13(9):5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]