Abstract
This study investigates the role of active efflux system MexXY in the emergence of aminoglycoside (AG) resistance among cystic fibrosis (CF) isolates of Pseudomonas aeruginosa. Three genotypically related susceptible and resistant (S/R) bacterial pairs and three other AG-resistant CF strains were compared to four non-CF strains moderately resistant to AGs. As demonstrated by immunoblot experiments, pump MexY was strongly overproduced in all of the resistant bacteria. This MexXY upregulation was associated with a 2- to 16-fold increase in the MICs of AGs in the S/R pairs and lower intracellular accumulation of dihydrostreptomycin. Alterations in mexZ, the repressor gene of operon mexXY, were found in all of the AG-resistant CF isolates and in one non-CF strain. Complementation of these bacteria with a plasmid-borne mexZ gene dramatically reduced the MICs of AGs, thus highlighting the role played by MexXY in the development of moderate resistance in CF patients. In contrast, complementation of the three non-CF strains showing wild-type mexZ genes left residual levels of resistance to AGs. These data indicate that a locus different from mexZ may be involved in overproduction of MexXY and that other nonenzymatic mechanisms contribute to AG resistance in P. aeruginosa.
Pseudomonas aeruginosa is a significant cause of morbidity and mortality in patients with cystic fibrosis (CF). Everyday clinical experience shows that, once established in the respiratory tree of CF patients, this opportunistic pathogen survives iterative cures of aggressive chemotherapy. Given intravenously or in aerosol, aminoglycosides (AGs) remain invaluable drugs in the management of chronic lung infection in CF (19). Their good in vitro bactericidal activity against P. aeruginosa is, however, notably altered in vivo, especially when bacteria grow as biofilms in the airways. The local anaerobiosis that resides inside biofilms tends to strongly reduce the driving force required by AGs to actively cross the cytoplasmic membrane of P. aeruginosa (21, 23). In sessile (planktonic) bacteria, intracellular accumulation of AGs is impaired by a multidrug active efflux system named MexXY, whose transporter protein (pump MexY) belongs to the RND family (for resistance-nodulation-cell division) (18, 22). Proteins MexXY that are coded by a single operon (mexXY) need to interact with a third component in order to form a functional tripartite efflux system. It has been suggested that the outer membrane proteins OprM (13, 18), OpmB (15), OmpG, and OmpI (9) may cooperate with MexXY in reference strain PAO1. Which one of these proteins is the preferential partner of MexXY in clinical strains is not yet known. As demonstrated previously (11), the natural resistance of P. aeruginosa to AGs in part relies on inducible expression of mexXY. The efflux system may also be involved in acquisition of higher resistance, in particular when bacteria are repeatedly exposed to subinhibitory doses of AGs (8). This reversible “adaptive” resistance described in vitro (6) and in CF patients (4) is concomitant to and dependent upon MexXY-induced production (8). Interestingly, Westbrock-Wadman et al. (22) showed recently that most AG-resistant CF isolates actually overproduce MexXY constitutively. The contribution of the efflux system to the resistance in these bacteria has, however, been questioned as mexXY overexpression in PAO1 mutants inactivated in repressor gene mexZ only results in a modest twofold increase in the MICs of AGs (22). In the present study, we demonstrate that, in the specific context of CF pulmonary infection, MexXY overproduction may provide P. aeruginosa with a significant resistance to these antibiotics.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions. P. aeruginosa PAO1 was used as a wild-type reference strain (B. W. Holloway). Mutant 11B is a mexX::Tn501 insertion derivative of strain PAO1 showing hypersusceptibility to aminoglycosides (AGs), erythromycin (ERY), and tetracycline (TET) (18). All of the clinical strains of P. aeruginosa described in the study were isolated from patients hospitalized either at the university-affiliated hospital of Besançon or at the Robert Debré Children's Hospital in Paris, France. The isolates were considered as clonally related when their RAPD [random(ly) amplified polymorphic DNA] profiles were strictly identical regarding the number and the intensity of DNA bands (10). Escherichia coli DH5α [F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ-thi-1 gyrA96 relA; Invitrogen] was the bacterial host in all DNA cloning experiments. Bacterial cultures were performed either in Luria-Bertani broth (2), in Mueller-Hinton broth with adjusted concentrations of Ca2+ and Mg2+ (MH broth; BBL), or on Mueller-Hinton agar plates (MHA plates; Bio-Rad). When necessary, growth media were made selective by the addition of the following antibiotics (with the final concentrations in parentheses): ampicillin (AMP; 100 μg/ml), kanamycin (KAN; 25 μg/ml), and TET (30 μg/ml) for E. coli and TET (100 μg/ml) and ticarcillin (TIC; 250 μg/ml) for P. aeruginosa. All of the bacterial cultures were incubated at 37°C. Bacterial matings were performed as described previously (18).
Susceptibility tests.
Drug MICs were determined by the standard microdilution method in MH broth with inocula of ca. 2.5 × 105 CFU/ml (3). The antibiotics tested were obtained as titrated powders from Sigma-Aldrich or kindly provided by Bristol-Myers Squibb (amikacin [AMK] and cefepime [FEP]), GlaxoSmithKline (ceftazidime [CAZ]), Schering-Plough (gentamicin [GEN]), Lilly Laboratories (tobramycin [TOB]), Abbott Laboratories (ERY), or Bayer Pharma SA (ciprofloxacin [CIP]).
AG resistance phenotypes were determined by the Kirby-Bauer disk diffusion assay on MHA plates using the AG Resistance Test Kit provided by the Schering-Plough Institute. Mechanisms of resistance were deduced from the inhibition zone diameters around disks of 12 test antibiotics: AMK, apramycin, fortimicin, GEN, isepamicin, KAN, TOB, neomycin, netilmicin, 2′-N-ethylnetilmicin, 6′-N-ethylnetilmicin, and 5-episisomicin. According to Schering-Plough's guidelines, the so-called impermeability-type nonenzymatic resistance phenotype is characterized by a general decrease in susceptibility of bacteria to all of the 12 aforementioned compounds (12). No AG-modifying enzymes were detected phenotypically in the selected isolates by this method.
DNA methodology.
Chromosomal DNA used for PCR amplifications was extracted and purified following the procedure of Chen and Kuo (5). Plasmid DNA was prepared by the standard alkaline lysis method (2) or by using the Plasmid Midi Preps kit from Qiagen S.A. Selected restriction fragments were purified from agarose gels with a GenElute gel extraction kit (Sigma-Aldrich). Other reagents for molecular biology were from Roche, Stratagene, or Sigma-Aldrich. Electrotransformation of strains of E. coli (7) and P. aeruginosa (20) with plasmid DNA has been described in detail elsewhere.
Sequencing of gene mexZ.
A 1,252 nucleotide fragment carrying the repressor gene mexZ and the intergenic region between mexZ and mexX was amplified by using Taq DNA polymerase (Perkin-Elmer) with the primers p819 (5′-GCA CCT GAT GGC GGA CGA-3′), which anneals at the 3′ end of mexX, and p2071 (5′-GCA GCC CAG CAG GAA TAG-3′), which anneals at the 5′ end of mexZ. The PCR mixture was heated for 5 min at 94°C, followed by 30 cycles of 30 s at 94°C, 40 s at 50°C, and 50 s at 72°C, before a final elongation step of 10 min at 72°C. The PCR products were purified with the GenElute PCR cleanup kit (Sigma-Aldrich) and sequenced twice on both strands by Qiagen SA (Hilden, Germany). Nucleotide and deduced amino acid sequences were analyzed with the BLAST Align algorithm available at the web site of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) (1).
Complementation with the gene mexZ.
The genes mexZ and mexX from reference strain PAO1 were previously cloned on a 4.1-kbp BamHI-NotI fragment in phagemid pBluescript II KS(+) to yield plasmid pJR41 (18). In the present study, a 1.7-kbp SalI-SalI DNA fragment from pJR41 containing the entire sequence of mexZ and the first 801 nucleotides of mexX was subcloned in plasmid vector pUC18 (AMP resistant [Ampr]) (24). The new construct named pUS17 was transferred into E. coli DH5α by electroporation. The insert of pUS17 was subsequently exised by digestion with enzymes BamHI and HindIII which cleaved at sites in the polylinker of pUC18, and recloned into the broad-host-range plasmid pAK1900 (Ampr, TIC resistant [Ticr]) (17). The recombinant plasmid (pAZ17) was introduced and propagated in E. coli DH5α. The strains of P. aeruginosa were finally electrotransformed with pAZ17 or pAK1900 purified from strain DH5α, and the transformants were selected on MHA plates containing TIC at concentrations equal to four MICs.
Obtention of a MexXY-overproducing mutant.
Mutant MutGr1 was obtained by cultivating strain PAO1 for 2 h in MH broth with one MIC of GEN (2 μg/ml), and plating the bacteria on MHA plates containing one MIC of FEP (2 μg/ml). One resistant clone (MutGr1) exhibiting a stable phenotype of resistance through serial subcultures in antibiotic-free medium was analyzed further by Western blotting and DNA sequencing. This mutant was found to constitutively hyperproduce protein MexY as a result of a nucleotide substitution at position 307 of repressor gene mexZ (C→T) (see Table 2).
TABLE 2.
Mutational events in the clinical isolates of P. aeruginosa
| Straina | Mutation in mexZb | Protein MexZ |
|---|---|---|
| PAO1 | Wild type | Wild type |
| MutGr1 | 307c (CAG→TAG) | Q103c → AMBd |
| 615S* | Wild type | Wild type |
| 615R* | Δ409 nt | Aberrant |
| 2112S* | 400b (CAA→TAA) | Q134c → OCHd |
| 2112R* | 400b (CAA→TAA) | Q134c → OCH |
| 2114S* | Wild type | Wild type |
| 2114R* | Δ1 nt (386b) | Aberrant |
| 2116R* | Δ14 nt (328-341b) | Aberrant |
| 2117R* | 614b (CAG→CCG) | L305→P |
| 2119R* | Δ22 nt (595-616b) | Aberrant |
| 1724R | 400b (CAA→TAA) | Q134c→OCH |
| 1764R | Wild type | Wild type |
| 1800R | Wild type | Wild type |
| 1803R | Wild type | Wild type |
CF isolates are indicated by an asterisk.
Nucleotide (nt) position compared to mexZ sequence in the wild-type susceptible strain PAO1.
Stop codon within mexZ.
AMB and OCH are nonsense codons (UAG and UAA, respectively).
Mutation rates to RIF.
The CF strains were cultured overnight in MH broth, centrifuged, and resuspended in 1:20 volume of fresh medium. Fractions (50 μl) of serial twofold dilutions in saline buffer of this suspension were plated onto MHA plates supplemented or not with rifampin (RIF; 300 μg/ml) by using a Spiral Plater inoculator (AEL Laboratoire, Combourg, France). Colony counts were determined after 36 h of incubation. All experiments were performed in duplicate. According to Oliver et al. (16), a strain was considered as hypermutable when giving rise to RIF-resistant mutants at rates at least 20-fold higher than those observed for the PAO1 strain.
RESULTS AND DISCUSSION
Selection of CF isolates.
To assess the relevance of MexXY as a resistance mechanism in CF, we selected three pairs of isolates susceptible and resistant (S/R) to AGs from three different CF patients (615S/R, 2112S/R, and 2114S/R). The clonal identity of the S/R paired bacteria was established by RAPD analysis (data not shown). The resistant bacteria proved to be less susceptible than PAO1 and their susceptible counterparts to a panel of 12 AGs (Schering Plough kit; data not shown), thus strongly suggesting the acquisition of nonenzymatic resistance to these antibiotics in vivo. Three other CF strains (2116R, 2117R, and 2119R) and four non-CF isolates (1724R, 1764R, 1800R, and 1803R) showing similar “nonenzymatic” profiles of resistance to AGs were also selected for further analysis. The bacteria of this second group displayed very different RAPD profiles and were considered as genotypically distinct. As indicated in Table 1, the emergence of resistance in the CF strains (615R, 2112R, and 2114R) was characterized by a significant increase in the MICs of AGs (up to sixteenfold) associated with a lower (in most cases from two- to fourfold) concomitant increase in the MICs of other known MexXY substrates, including ERY, TET, CIP, and FEP (11, 13, 18). Clinical strains 2116R, 2117R, 2119R, 1724R, 1764R, 1800R, and 1803R all exhibited moderate resistance levels to AGs (Table 1). In agreement with previous results (22), an in vitro-selected mutant (MutGr1) overproducing MexXY constitutively was found to be only twofold more resistant to AGs than its wild-type parent PAO1.
TABLE 1.
Antibiotic susceptibilities of P. aeruginosa strainsa
| Strain(plasmid) | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| AMK | GEN | TOB | ERY | TET | CIP | FEP | CAZ | |
| PAO1 | 4 | 2 | 0.5 | 256 | 32 | 0.12 | 2 | 1 |
| PAO1(pAZ17) | 0.5 | 0.25 | 0.12 | 64 | 16 | 0.12 | 1 | 0.5 |
| 11B mexX::Tn501 | 1 | 0.25 | 0.12 | 64 | 8 | 0.06 | 1 | 1 |
| MutGr1 | 8 | 4 | 1 | 512 | 32 | 0.5 | 4 | 1 |
| 615S* | 2 | 1 | 0.25 | 256 | 32 | 1 | 2 | 0.5 |
| 615R* | 16 | 8 | 1 | 256 | 32 | 2 | 4 | 1 |
| 615R(pAZ17)* | 0.5 | 0.25 | 0.12 | 32 | 8 | ≤0.03 | 2 | 0.5 |
| 2112S* | 16 | 4 | 2 | 512 | 32 | 2 | 64 | >128 |
| 2112R* | 64 | 32 | 4 | >1,024 | 64 | 4 | >64 | >128 |
| 2114S* | 2 | 2 | 0.5 | 256 | 16 | 0.06 | 1 | 4 |
| 2114R* | 32 | 16 | 8 | 256 | 32 | 1 | 16 | 4 |
| 2114R(pAZ17)* | 0.5 | 0.25 | 0.12 | 16 | 8 | 0.06 | 4 | 4 |
| 2116R* | 16 | 8 | 4 | >1,024 | 32 | 0.25 | 8 | 16 |
| 2116R(pAZ17)* | 0.5 | 0.25 | 0.25 | 512 | 16 | 0.12 | 4 | 16 |
| 2117R* | 32 | 8 | 4 | 1,024 | 32 | 1 | 4 | 8 |
| 2119R* | 8 | 4 | 0.5 | 512 | 32 | 1 | 8 | 1 |
| 2119R(pAZ17)* | 0.5 | 0.25 | 0.12 | 256 | 8 | 0.5 | 8 | 1 |
| 1724R | 16 | 8 | 2 | 256 | 16 | 1 | 4 | 2 |
| 1724R(pAZ17) | 0.5 | 0.25 | 0.12 | 128 | 8 | 0.5 | 2 | 2 |
| 1764R | 8 | 4 | 1 | 512 | 64 | 0.5 | 4 | 2 |
| 1764R(pAZ17) | 2 | 1 | 0.5 | 256 | 32 | 0.25 | 2 | 2 |
| 1800R | 16 | 4 | 2 | 512 | 32 | 0.25 | 16 | 4 |
| 1800R(pAZ17) | 4 | 2 | 0.5 | 256 | 16 | 0.12 | 8 | 2 |
| 1803R | 8 | 8 | 2 | 512 | 32 | 0.25 | 16 | 2 |
| 1803R(pAZ17) | 2 | 1 | 0.5 | 256 | 16 | 0.12 | 8 | 2 |
Drug susceptibilities of isolates transformed with pAZ17 (cloned mexZ, Ticr) were determined in the presence of 250 μg of TIC/ml for plasmid maintenance. Preliminary experiments with strains PAO1, 615S, 615R, 1724R, 1764R, 1800R, and 1803R transformed with parental vector pAK1900 (Ticr) showed no influence of TIC on the MIC values compared to nontransformed strains. Similar MIC values were obtained for strain 615S harboring pAZ17. Values in boldface or in italics indicate that the bacteria were at least fourfold more or less resistant than their susceptible or resistant counterparts, respectively. CF isolates are indicated by an asterisk.
Overproduction of efflux system MexXY.
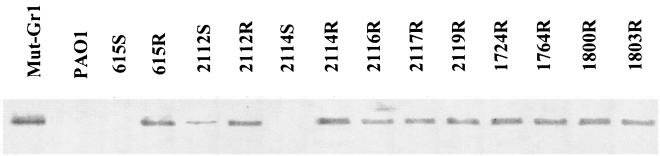
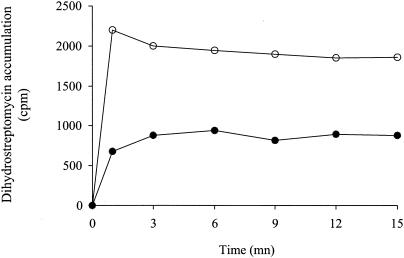
The whole (outer and inner) membranes of the selected strains were isolated and analyzed by Western blotting with a MexY-specific antiserum (8). As shown in Fig. 1, the emergence of resistance to AGs in the three S/R CF pairs was associated with a dramatic increase in the production of pump MexY. However, in contrast to 615S and 2114S showing no detectable MexY bands on immunoblots, strain 2112S appeared to produce substantial amounts of the protein. Interestingly, this partially derepressed strain was comparatively more resistant to AGs than 615S and 2114S but more susceptible than 615R and 2114R (Table 1). All of the other strains selected for their panaminoglycoside resistance also turned out to be strong MexY overproducers. Uptake assays with [3H]dihydrostreptomycin were carried out as reported previously (18) to determine whether this MexXY upregulation was correlated with a reduced accumulation of AGs in the resistant bacteria. Two bacterial pairs (615S/R and 2114S/R) were investigated and gave similar results. As shown in Fig. 2, resistant isolate 615R accumulated significantly less AG (2.1- to 2.5-fold at the steady state) than its susceptible counterpart 615S, thus supporting the implication of an efflux-mediated mechanism in the emergence of AG resistance in vivo. Accumulation experiments in the presence of proton conductor CCCP were not attempted since disruption of the cytoplasmic membrane potential by this product inhibits both efflux and active transport of AGs and gives uninterpretable results (14, 18).
FIG. 1.
Immunodetection of MexY in bacterial membranes. Total membrane proteins were isolated, subjected to electrophoresis (20 μg per lane) in a sodium dodecyl sulfate-12% polyacrylamide gel, electrotransferred to nylon membrane, and labeled with a MexY-specific antiserum as described previously (8).
FIG. 2.
Uptake of [3H]dihydrostreptomycin by intact cells of P. aeruginosa. Susceptible strain 615S (○) and its resistant counterpart 615R (•) were evaluated. As described previously (18), the cells were grown in MH broth, harvested, washed (50 mM phosphate buffer, 100 mM LiCl), and resuspended in 50 mM phosphate buffer (pH 7) containing 1 mM MgSO4 and 0.2% (wt/vol) glucose as a source of carbon. After 5 min at 37°C, a mixture of dihydrostreptomycin and [3H]dihydrostreptomycin (20 Ci/mmol; American Radiolabeled Chemicals, Inc., St. Louis, Mo.) was added to a final concentration of 8 μg/ml. Samples were removed and filtered, and the radioactivity was counted. The data are the means of three independent experiments.
Alterations in mexZ.
The involvement of repressor gene mexZ in the development of AG resistance among the CF isolates was first investigated by nucleotide sequencing analysis. All of the resistant strains demonstrated strong alterations in mexZ, the product of which downregulates mexXY expression (C. Vogne, D. Hocquet, J. Ramos Aires, F. El Garch, and P. Plésiat, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-434, 2002) (Table 2). Gene mexZ was inactivated in strains 615R, 2114R, 2116R, and 2119R as a result of frameshift mutations caused by deletions of 1 to 409 nucleotides. Of note, the 409-nucleotide deletion seen in 615R appeared to be generated by a homologous recombination between two distant, identical 14-bp sequences (GGTGCCGGCGCTGG): one located within mexZ (PA2020) and the other one located at the 3′ end of adjacent putative gene PA2021. A nucleotide substitution in isolates 2112S (moderately resistant to AGs) and 2112R (highly resistant) introduced a stop codon at position 400 (C→T) of gene mexZ leading to a MexZ truncated peptide lacking 77 amino acid residues at the C-terminal end. No additional mutations in mexZ or in the mexZ-mexX intergenic region were detected in 2112R, which could account for its increased resistance to AGs compared to 2112S (Table 1). In CF strain 2117R, a single mutation, A614→C, created a Leu205→Pro substitution of unknown significance at the C terminus of MexZ. Of the four non-CF strains, only one (1724R) appeared to be defective in MexZ because of a stop codon at position 134, as in the isolates 2112S and 2112R. Interestingly, a similar genetic event resulting in a truncated MexZ product was also identified in in vitro mutant MutGr1 (C307→T substitution in gene mexZ). With the exception of strain 2119R, AG resistance in mutant MutGr1 was constantly lower (most often twofold) than in clinical strains producing truncated (2112S and 1724R) or aberrant (615R, 2114R, and 2116R) MexZ peptides (Table 1). Overall, these data suggested that gene mexZ is involved in the development of stable AG resistance among CF strains of P. aeruginosa.
Recently, Oliver et al. (16) reported that resistant hypermutable P. aeruginosa strains are frequent in the CF lung. Alterations of genes mutS and mutY result in an increased rate of polymerase errors and/or defect in the proofreading and mismatch repair mechanisms. Surprisingly, only one of our CF strains (isolates 2112S and 2112R) exhibited a hypermutator phenotype with RIF-resistant mutants occurring at frequencies (2.4 × 10−6 and 1 × 10−6, respectively) at least 20-fold higher than in wild-type control strain PAO1 (3.9 × 10−9) (data not shown). So we found no evidence that CF P. aeruginosa strains with deletions in mexZ are hypermutable.
Complementation of the clinical strains.
To better evaluate the contribution of MexXY-mediated efflux in the emergence of AG resistance, we complemented the selected strains with the intact mexZ gene from PAO1. The gene was cloned into broad-host-range plasmid pAK1900 (Ticr). Despite multiple attempts, Ticr strains 2112R and 2117R remain refractory to transformation with the new construct named pAZ17. In all of the strains showing alterations in mexZ (615R, 2114R, 2116R, 2119R, and 1724R), operon mexXY was very responsive to repression by MexZ since introduction of pAZ17 dramatically reduced the MICs of AGs, to levels similar to that for the MexXY-null mutant 11B (18) or PAO1(pAZ17) (Table 1). Overproduction of MexZ from pAZ17 also lowered the resistance to other MexXY substrates such as ERY, TET, and CIP. The lack of residual resistance to AGs in the pAZ17-complemented CF isolates indicated unambiguously that the efflux system MexXY plays a major role in the emergence of moderate AG resistance in vivo. In contrast, in the three non-CF isolates harboring intact mexZ genes (1764R, 1800R, and 1803R), complementation with pAZ17 reduced the MICs of AGs only two- to eightfold. The resistance levels of these complemented strains remained well above that of the MexXY-null mutant 11B or PAO1(pAZ17) (Table 1). A first explanation for these results would be that the plasmid-borne mexZ gene partially turned down mexXY expression because of unknown mutations in the bacteria. This was ruled out by Western blot experiments that showed the complete disappearance of the MexY band in all of the strains harboring pAZ17 (data not shown). A more likely hypothesis is that one or several additional nonenzymatic mechanisms of resistance coexist with derepressed MexXY in 1764R, 1800R, and 1803R. Since these non-CF strains have intact mexZ genes, it is clear that genetic events independent of mexZ may also lead to MexXY overproduction in clinical strains. Identification of the mutated loci would provide useful information on the regulation of the mexXY operon and on how the emergence of resistance to AGs occurs in vivo.
Although the present study shows that MexXY constitutive overproduction may increase the MIC of AGs up to 16-fold, one may wonder whether such a moderate resistance is clinically significant in the context of CF. The observation that a high proportion of AG-resistant CF isolates are actually mexXY-overexpressing mutants (22) suggests that this efflux pump provides a selective advantage, allowing P. aeruginosa to better adapt to the hostile environment of the CF lung. In support of this, follow-up analysis of sputum samples from the patient colonized with 615R demonstrated persistence of the isolate with unchanged susceptibility profile for more than 7 years, despite iterative aerosol and intravenous treatments with AGs.
Acknowledgments
We thank Gérard Couetdic, Florence Giachetti, Colette Godard, and Angelin Fontaine for excellent technical assistance. We are also grateful to Patricia Mariani-Kurkdjian and Edouard Bingen for providing strains 2112S/R, 2114S/R, 2116R, 2117R, and 2119R.
C.V. was sponsored by the French association against cystic fibrosis Vaincre la Mucoviscidose.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Balows, A., W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy. 1991. Manual of clinical microbiology, 5th ed. ASM Press, Washington, D.C.
- 4.Barclay, M. L., E. J. Begg, S. T. Chambers, P. E. Thornley, P. K. Pattemore, and K. Grimwood. 1996. Adaptive resistance to tobramycin in Pseudomonas aeruginosa lung infection in cystic fibrosis. J. Antimicrob. Chemother. 37:1155-1164. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W. P., and T. T. Kuo. 1993. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 21:2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daikos, G. L., G. G. Jackson, V. T. Lolans, and D. M. Livermore. 1990. Adaptive resistance to aminoglycoside antibiotics from first-exposure down-regulation. J. Infect. Dis. 162:414-420. [DOI] [PubMed] [Google Scholar]
- 7.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hocquet, D., C. Vogne, F. El Garch, A. Vejux, A. Lee, O. Lomovskaya, N. Gotoh, and P. Plésiat. 2003. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47:1371-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jo, J. T., F. S. Brinkman, R. E. Hancock, M. L. Chiu, M. Folcher, P. Griffin, T. Holt, T. Klatt, and C. J. Thompson. 2003. Aminoglycoside efflux in Pseudomonas aeruginosa: involvement of novel outer membrane proteins. Antimicrob. Agents Chemother. 47:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller, G. H., F. J. Sabatelli, R. S. Hare, Y. Glupczynski, P. Mackey, D. Shlaes, K. Shimizu, and K. J. Shaw. 1997. The most frequent aminoglycoside resistance mechanisms change with time and geographic area: a reflection of aminoglycoside usage patterns? Clin. Infect. Dis. 24(Suppl. 1):S46-S62. [DOI] [PubMed] [Google Scholar]
- 13.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murata, T., N. Gotoh, and T. Nishino. 2002. Characterization of outer membrane efflux proteins OpmE, OpmD and OpmB of Pseudomonas aeruginosa: molecular cloning and development of specific antisera. FEMS Microbiol. Lett. 217:57-63. [DOI] [PubMed] [Google Scholar]
- 16.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blasquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 289:391-392. [DOI] [PubMed] [Google Scholar]
- 17.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 18.Ramos Aires, J., T. Köhler, H. Nikaido, and P. Plésiat. 1999. Involvement of an efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramsey, B. W. 1996. Management of pulmonary disease in patients with cystic fibrosis. N. Engl. J. Med. 335:179-188. [DOI] [PubMed] [Google Scholar]
- 20.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters, M. C., III, F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Döring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13 mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]