Abstract
Among methicillin-resistant Staphylococcus aureus isolates, a staphylococcal chromosomal cassette containing the mecA gene (SCCmec) is integrated into the chromosome at a unique site. SCCmec also contains unique ccrAB recombinase genes mediating its integration and excision from the genome and is flanked by characteristic left and right direct- and inverted-repeat sequences. A few non-mecA-containing SCC elements that have the other molecular features described above have recently been described. The origin of these cassettes is not clear. We have identified two new members of the SCC family integrated within orfX in Staphylococcus epidermidis strain ATCC 12228, neither of which carries mecA. One is a 57-kb element flanked by a unique 28-bp SCC direct repeat. It was called the SCC composite island (SCC-CI) because it carries a 19-kb SCC element (SCCpbp4) nested within it. SCCpbp4 contains pbp4 and tagF genes, as well as one pair of ccrAB genes (allotype 2) flanked by classical SCC-specific terminal repeats. External to SCCpbp4, SCC-CI contains a second pair of ccrAB genes (allotype 4), three IS431 elements, and genes mediating resistance to heavy metals. Genes mediating restriction-modification that may facilitate horizontal transfer are also present within SCC-CI, both within and outside SCCpbp4. Several novel arrangements of the SCC direct and inverted repeats were identified. Several long stretches of homology with other SCCs were found within and outside SCCpbp4. In view of the fact that SCC-CI was found in a commensal species, it may represent a reservoir for sequences involved in genetic shuffling between staphylococci and may contribute to the diversity found in SCC elements.
Staphylococcus aureus is an important human pathogen that causes a variety of hospital- and community-acquired infectious syndromes. Comparative analysis of three S. aureus genomes (3, 14) has revealed that, within a relatively constant genetic background, plasticity in the species is conferred by horizontal transfer of large genetic elements of unknown origin that insert into the genome (11). Stabilization of such elements in the genome presumably occurs when the genes that encode the transfer or excision functions, or the cis-acting sites on which these recombinases act, are deleted or mutated.
Resistance to methicillin in S. aureus is an important clinical problem because few therapeutic options exist. The methicillin resistance phenotype results from mecA-directed production of the transpeptidase penicillin-binding protein 2′ or 2a (PBP 2′ or PBP 2a), which has decreased affinity for β-lactam antibiotics. A site-specific integrative genetic element called staphylococcal chromosomal cassette mec (SCCmec) carries the mecA gene complex, which consists of the mecA gene itself, its regulating genes, when present, and the insertion sequence IS431mec (10). It also contains unique cassette chromosome recombinase genes (ccr) that encode products responsible for the integration and excision of the SCCmec element (10).
In all methicillin-resistant S. aureus (MRSA) isolates, SCCmec elements are integrated into the S. aureus genome at a unique site (attBscc) located in frame at the 3′ end of an open reading frame (ORF) of unknown function called orfX (10, 16). When SCCmec integrates, it is flanked on either end with characteristic direct- and inverted-repeat sequences (DRSCC and IRSCC, respectively). It is not clear how these elements are horizontally transferred, since there are no known genes encoding bacteriophage structural proteins or proteins involved in the horizontal transfer of the elements identified to date (10, 16).
SCCmec elements have been classified into types I to IV according to the class of mecA gene complex and the type of ccr gene complex present (11). Three relatively large SCCmec elements, types I, II, and III, were initially described for MRSA isolates, which were mostly obtained from patients frequenting health care environments (10). A smaller SCCmec element, type IV, was first identified in community-acquired MRSA (CA-MRSA) isolates from Chicago (4, 16) and was subsequently found in MRSA isolates from other geographic locales (3, 6, 18) and in Staphylococcus epidermidis (22).
Two other SCC elements that contain the essential features of SCCmec but lack the mecA gene have been recognized recently. SCCcap1, which carries the gene cluster encoding the type 1 capsular polysaccharide, was identified in S. aureus strain M, a methicillin-susceptible strain (15). SCC12263 was recently identified in Staphylococcus hominis ATCC 27844. Because it carries a functional pair of ccr genes and other sequences with variable homology to those found in SCCmec elements (13), it has been proposed that SCC12263 may be ancestral to SCCmec elements.
The type IV SCCmec element has been of considerable interest because of its epidemiologic link to CA-MRSA and because its relatively small size makes it potentially amenable to interbacterial transfer, for example, on a bacteriophage. Molecular subdefinition of SCCmec type IV elements has been determined by DNA sequencing (4, 16). The DNA sequences of SCCmec type IV elements from two Chicago CA-MRSA isolates were highly similar to each other in all regions except the left extremity (L-C) region (16), whose sequence differences resulted in the designation of SCCmec IV subtypes IVa and IVb. Recently, a third subtype, IVc, also with L-C region polymorphism relative to other SCCmec type IV elements, was reported among MRSA isolates from France (9) and Japan (11).
It has been proposed that the L-C region contains unrelated genes and pseudogenes (9) and a number of unnecessary ORFs (16). Thus, this region has been termed a “junkyard” (9), a term suggesting functional insignificance.
We have hypothesized that the L-C region is useful for understanding the molecular epidemiology of SCC elements. While characterizing the L-C region in SCCmec elements in CA-MRSA isolates in our collection, we identified a ca. 9-kb sequence in one of these isolates that was found in its entirety in the sequenced genome of S. epidermidis ATCC 12228 and in the recently described SCCmec IVc element. Upon analysis of the S. epidermidis genome sequence flanking this ca. 9-kb homologous region, the sequence was found to be part of a unique 57-kb SCC composite island that lacked mecA but carried two pairs of complete ccr recombinase genes of different types, each contained within a region of DNA flanked by SCC-specific terminal repeat sequences. We have called this element the SCC composite island because we also found that it contained a second, smaller SCC element, SCCpbp4, that carried a homolog of the gene encoding PBP 4 (pbp4), a gene encoding a teichoic acid biosynthesis protein (tagF), and three stretches of DNA (>2 to 10 kb) that are highly homologous to those found in S. aureus SCCmec elements. Thus, these two new members of the SCC family found in S. epidermidis could be ancestral to SCCmec elements found in S. aureus by acting as a reservoir for donation of sequences to SCC elements in S. aureus by homologous recombination.
MATERIALS AND METHODS
Bacterial strains.
The characteristics of the strains used in this study are given in Table 1. CA-MRSA isolates CA05 and 8/6-3P, recovered from children in Chicago (16), served as the prototypes of SCCmec types IVa and IVb, respectively. CA-MRSA isolate 2314 was isolated in 1996 from a day care center attendee in Dallas (1).
TABLE 1.
Molecular characteristics of CA-MRSA isolates
| Strain | Source | L-C region PCR product size (kb)
|
SCCmec subtypea | |
|---|---|---|---|---|
| cLs1-α5 | cL2′-α5 | |||
| CA05 | Chicago | 9.9 | No product | IVa |
| 8/6-3P | Chicago | No product | 9.1 | IVb |
| 2314 | Dallas | 3 | 9.6 | IV |
Classification by SCCmec IV subtype (a or b). The classification scheme used by Oliveira and de Lencastre, which uses the term SCCmec type IVA to refer to the presence of pUB110 in the I- R region of the SCCmec element (19), did not apply here.
Typing of the SCCmec element.
The mecA gene complex class, A or B, is assigned on the basis of the presence or absence of ψIS1272, mecI, and two functional regions of mecR1 (17). Sequence variability in the ccrA and ccrB genes determines the assignment to ccr gene complex types 1 to 4 (10, 20). Typing of mec and ccr complexes is performed by PCR amplification of the relevant regions with primers described previously (10). An SCCmec element was classified as type IV if the type 2 ccr gene complex and the class B mecA gene complex were both detected by PCR, as described previously (4, 16). The SCCmec type IVa and IVb elements contain four contiguous and overlapping structural regions (from left to right, L-C, C-M, M-I, and I-R) that were screened for by PCR with region-specific primers as reported previously.
Screening for subtypes IVa and IVb was conducted with primer pairs cLs1-α5 and cL2′-α5, which amplify the L-C regions from subtypes IVa and IVb, respectively (4, 16). The L-C region spans the left extremity of the SCCmec element from IRSCCmec-L to the ccr gene complex. We have further refined our SCCmec subtyping protocol by using restriction fragment length polymorphism (RFLP) to digest amplicons with the restriction endonuclease EcoRV as described elsewhere (16a). Digested fragments were separated in 0.8% agarose gels at 100 V, visualized by staining with ethidium bromide, and photographed under illumination from a UV light source. RFLP patterns were assigned by visual inspection of samples run on the same gel.
DNA sequencing.
Agarose gel-purified PCR products were sequenced by using a primer-walking strategy. (Primer sequences are available upon request.) Sequencing reactions were performed by using fluorescent dideoxy chain termination chemistry and an ABI Prism sequence detection system (Applied Biosystems, Foster City, Calif.) at the University of Chicago DNA Sequencing Facility. Homology searches were performed against all sequences in the GenBank database by using the BLAST search engine, available through the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov) (2). DNA sequence alignments were performed by using ClustalW alignment software (version 1.81; Silicon Graphics) (CMBI ClustalW server) (21). ORFs in the L-C region of isolate 2314 were identified by using ORF Finder, available on the NCBI website, or by comparison to annotated gene sequences in the S. epidermidis ATCC 12228 (GenBank accession no. AE016744) database.
DNA sequences of the other SCCmec types (I to III) were obtained from GenBank. NCTC 10442 (GenBank accession no. AB033763), N315 (D86934), and 85/2082 (AB037671) sequences were used as the prototypes of SCCmec types I, II, and III, respectively (10). SCCmec type IVa was from CA-MRSA isolates CA05 (GenBank accession no. AB063172) (16) and MW2 (3); SCCmec type IVc was from the health care-associated MRSA (HA-MRSA) isolate MR108 (GenBank accession no. AB096217). Sequences from coagulase-negative staphylococci were from S. epidermidis ATCC 12228 (GenBank accession no. AE016744) (23) and S. hominis ATCC 27844 (AB063171).
Nucleotide sequence accession numbers.
To annotate the SCC composite island and SCCpbp4 elements that we identified in the S. epidermidis ATCC 12228 sequence (GenBank accession no. AE016744), a third-party annotation (TPA BK001539) was added to GenBank. The sequence of the type IV SCCmec element of the Dallas CA-MRSA isolate 2314 was assigned GenBank accession no. AY271717.
RESULTS
SCCmec typing and subtyping of CA-MRSA isolate 2314 by PCR and EcoRV RFLP.
CA-MRSA isolate 2314, from a Dallas day care center attendee (1), contained an SCCmec type IV element with the characteristic combination of type 2 ccr and class B mecA gene complexes (ψIS1272-ΔmecR1-mecA-IS431mec).
PCR amplification of the L-C region of the SCCmec type IV element from isolate 2314 with the type IVa-specific primer pair cLs1-α5 produced a 3-kb product, which was not the expected size for type IVa (Table 1). By use of the primer pair cL2′-α5, a product similar in mobility to the expected 9.1-kb fragment obtained from the IVb prototype isolate 8/6-3P was obtained. However, the EcoRV restriction fragment pattern of the amplicon from isolate 2314 differed from that of type IVb (data not shown). Thus, this isolate could not be assigned to SCCmec type IVa or IVb.
We sequenced the ca. 9-kb PCR product (GenBank accession no. AY271717) to further characterize the L-C region of this SCCmec element. The amplified fragment had 9,612 bp and contained a 15-bp SCC-specific direct-repeat sequence (DRSCC-left [DRSCC-L]) (nucleotides [nt] 2691 through 2705; accession no. AY271717) juxtaposed to the left extremity, a feature that is characteristic of SCC elements. Adjacent to DRSCC-L was the 26-bp inverted-repeat sequence (IRSCC-L; nt 2706 through 2731), also characteristic of the left termini of SCCmec elements (Fig. 1).
FIG. 1.
Alignment of DNA sequences of DRSCC and IRSCC junctions from SCC and SCCmec elements. Shown are SCCpbp4 alignments with SCCmec types I, II, and IV and SCC12263 (A) and with SCCmec type III and SCCcap1 (B). Strains represented are isolate 2314 (SCCmec IV), ATCC 12228 (SCCpbp4), CA05 and MW2 (SCCmec IVa), 8/6-3P (SCCmec IVb), NCTC 10442 (SCCmec I), N315 (SCCmec II), 85/2082 (SCCmec III), ATCC 27844 (SCC12263), and S. aureus M (SCCcap1). DRSCC and IRSCC in the left junction were designated DRSCC-L and IRSCC-L, respectively; DRSCC and IRSCC in the right junction were designated DRSCC-R and IRSCC-R, respectively. Capital letters, nucleotides in SCC or SCCmec elements; lowercase letters, nucleotides in chromosomal regions. Black arrows, direct-repeat sequences; red arrows, inverted-repeat sequences. Asterisks indicate identical nucleotides in the comparison sequences; pairs of slashes indicate intervening omitted sequences.
Further analysis of the 9.6-kb L-C region sequence from strain 2314 revealed that it contained a ca. 6.9-kb sequence (from accession no. AY271717, nt 2678 through 9612) that was 99% identical to a ca. 6.9-kb stretch of DNA (GenBank accession no. AB096217, nt 6487 through 13421) in the left extremity region of the recently described SCCmec type IVc element of HA-MRSA strain MR108 (18). Also, the same region of DNA from strain 2314 (accession no. AY271717, nt 2689 through 9612) was 99% identical to a ca. 6.9-kb stretch of DNA in the recently reported genome sequence of methicillin-susceptible S. epidermidis isolate ATCC 12228 (complement of nt 51523 through 44600, GenBank accession no. AE016744) that was not annotated as lying within an SCC element.
We found that that the L-C region of the SCCmec element of isolate 2314 contained five tandem ORFs, each encoding >50 predicted amino acids, designated ORFs 1 through 5 in succession, beginning from the left extremity (Fig. 2) . The tandem architecture of the five ORFs was conserved in the L-C region of the type IVc SCCmec of HA-MRSA strain MR108 with 100% sequence identity. Downstream of ORF 5, there was one complete copy each of the ccrA and ccrB genes; these were determined by ClustalW sequence alignments (data not shown) to be of allotype 2. The chromosomal sequence flanking the left extremity of the SCCmec element of isolate 2314 (nt 1396 through 2690) was identical to the region in methicillin-susceptible S. aureus isolate 8325 immediately downstream of orfX (GenBank accession no. AB014440), data consistent with a perfect insertion into orfX. This was in contrast to the type IVc SCCmec element in HA-MRSA strain MR108, which was adjacent to a ca. 6.5-kb element termed IE25923, present to the left of the type IVc SCCmec (11).
FIG. 2.
Map depicting the regions of high nucleotide sequence similarity between the left portion of SCCpbp4 (L-C region), including both ccr genes, and the left extremities of SCCmec elements and non-mec-containing SCC elements, based on the nucleotide sequences deposited in the DDBJ/EMBL/GenBank database under accession no. AY271717 (Dallas CA-MRSA strain 2314, SCCmec type IV), AB096217 (strain MR108, SCCmec type IVc), AE016744 (S. epidermidis ATCC 12228, SCCpbp4), D86934 (strain N315, SCCmec type II), AB063172 (strain CA05, SCCmec type IVa), AP004822 (strain MW2, SCCmec type IVa), AB063173 (strain 8/6-3P, SCCmec type IVb), AB063171 (S. hominis ATCC 27844, SCC12263), AB033763 (NCTC 10442, SCCmec type I), and AB037671 (S. aureus strain 82/2082, SCCmec type III). Conserved ORFs of >99 bp are indicated by solid rectangles beneath the maps of the nucleotide sequences. Arrows above the rectangles for the map of SCCpbp4 indicate the direction of transcription. Identical colors for sequences from different elements indicate regions of >90% nucleotide sequence homology. Asterisk indicates a 28-bp sequence identity with SE0039 from SCCpbp4. Vertical arrowhead indicates the junction between the left SCC element and the chromosome.
Description of SCCpbp4, a novel element in S. epidermidis ATCC 12228.
The ca. 9-kb sequence from the SCCmec element of S. aureus isolate 2314, from the left extremity to the 3′ end of the ccrB gene (nt 2706 to 11771, GenBank accession no. AY271717) was compared with S. epidermidis ATCC 12228 sequences deposited in GenBank (nt 51506 to 42441, GenBank accession no. AE016744) (24), revealing 98 to 99% nucleotide sequence similarity. In S. epidermidis, this region contained five ORFs, designated SE0042, SE0041, SE0040, SE0039, and SE0038, from left to right, upstream of a pair of type 2 ccrA and ccrB genes (Fig. 2). ORFs SE0042 and SE0041 corresponded to ORFs 1 and 2 of strain 2314 (Fig. 2) and were annotated in accession no. AE016744 as encoding possible phage resistance proteins, similar to the abortive phage resistance protein of Lactococcus species and the Streptococcus thermophilus Abi-alpha protein. These likely represented type I restriction system enzymes. ORFs SE0040, SE0039, and SE0038 corresponded to ORFs 3, 4, and 5 of strain 2314 and to ORFs R002, R003, and R004 of strain MR108, and they encoded hypothetical proteins (Fig. 2).
Thus, the entire L-C region and the ccrA2 and ccrB2 genes of the type IV SCCmec of CA-MRSA strain 2314 were present in the genome of S. epidermidis ATCC 12228 as well as in the SCCmec type IVc element of the HA-MRSA strain MR108. We designated this ca. 9-kb homologous region in S. epidermidis “the SCCmec type IVc L-C homologous region” (SCCmec IVc-L-C HR) to underscore the shared homology between these two SCCmec type IV elements and the related region of S. epidermidis (Fig. 3).
FIG. 3.
Map of the 57-kb SCC composite island and the ca. 19-kb SCCpbp4 element (dark blue background) inserted into the 3′ end of orfX in S. epidermidis ATCC 12228. The map shows the DRSCC's and IRSCC's that mark the boundaries of the elements, as well as ORFs of interest (light blue arrows showing direction of transcription). Yellow rectangles, IS431 elements. Horizontal rectangles above the map, homologous regions (HR) of >2 kb that have 78% (lavender) to >90% (red) sequence identity to regions in the indicated SCCmec elements. The SCC composite island spans nt 32492 through 89965, and SCCpbp4 spans nt 32492 to 51506, of GenBank accession no. AE016744. In order to emphasize the SCC-specific repeat regions, the map is not drawn to scale. Nucleotide positions of all ORFs that correspond to those in GenBank accession no. AE016744 are given in supplemental Table 2 (http://www.ucch.org/daum-boyle_lab-composite_island_figures/table2.htm). Nucleotide positions of the SCC direct repeats and the newly annotated features (TPA BK001539) are detailed in Table 2. MerR is the ca. 10-kb region encoding genes for mercury resistance, flanked by two copies of IS431 (see Fig. 5). CadR is a region containing genes conferring resistance to cadmium (see Fig. 5). HDE288 HR is a 5.3-kb region with 92% nucleotide identity to a region containing type 4 ccrA and ccrB genes in the SCCmec element in strain HDE288 (see Fig. 6). SCCmec type IVc L-C HR is about 9 kb long, with 99 to 100% identity to the L-C region in type IVc SCCmec. The nucleotide sequence and structure of one of the three 28-bp DR-SCCcomposite island's are shown below the map, and the overlapping repeat sequences DR-SCCpbp4-R (right-pointing arrow) and IR-SCCpbp4-R (line with a diamond on the left), and IR-SCCpbp4-L (line with a Ball on the right) contained within it, are indicated. Also shown below the map are the sequence and structure of the novel SCC repeat sequence composed of overlapping DR/IR-R and IR/DR-L SCC sequences (SCC composite island-SCCpbp4 junction inverted repeat) that forms the 13-bp perfect inverted complementary repeat separated by 29 nt.
Our analysis revealed the presence of a pair of SCC-specific repeat sequences as summarized in Table 2 and annotated in our third-party annotation, accession no. BK001539. These were not annotated in the S. epidermidis ATCC 12228 genome sequence, but their presence suggested that an SCC-like element existed in the S. epidermidis isolate. No mecA or type 1 capsule gene sequences could be found within the flanking SCC-specific repeat sequences.
TABLE 2.
Locations of features in SCC composite island documented in TPA BK001539
| Feature | nt location in:
|
Characteristic or DNA sequence (5′-3′) complement | |
|---|---|---|---|
| BK 001539 | AE 016744 | ||
| SCC elements | |||
| SCC composite island | 1-57474 | 32492-89965 | |
| SCCpbp4 element | 1-19015 | 32492-51506 | |
| L-C homologous region | 9950-19015 | 42441-51506 | Similar to L-C regions of SCCmec type IV |
| ccrAB4 homologous region | 24565-29904 | 57056-62395 | Similar to nt 6986-12345 of strain HDE288 (accession no. AF411935) |
| CadR homologous region | 38639-41107 | 73585-76053 | 78% similarity to CadR region in SCCmec type III |
| MerR homologous region | 57475-66754 | 80621-89900 | 92% similarity to MerR region in SCCmec type III |
| SCC repeat sequences | |||
| DRSCC composite island-R (III) | 1-29 | 32492-32520 | AAAAACCGCATCATTTATGATATGCTTCG |
| DRSCCpbp4-R | 1-15 | 32492-32506 | TTATGATATGCTTCG |
| DRSCC composite island (II) | 19016-19044 | 51507-51535 | AAAAACCGCATCACTTATGATATGCTTCT |
| IRSCCpbp4-L | 18990-19015 | 51481-51506 | GCTTATCAGTTGATGATGCGGTTTTT |
| DRSCCpbp4-L | 19016-19030 | 51507-51521 | TTATGATATGCTTCT |
| SCC composite island-SCCpbp4 junction inverted repeat | 18990-19044 | 51481-51535 | AAAAACCGCATCACTTATGATATGCTTCTGCTTATCAGTTGATGATGCGGTTTTT |
| DRSCC-L (1 bp mm) | 57448-57460 | 89939-89951 | TTATGATATGCTTCA |
| IRSCC-L | 57450-57474 | 89941-89965 | AAAAACCGCATCATTTATGATATGC |
| DRSCC composite island-L (I) | 57446-57474 | 89937-89965 | AAAAACCGCATCATTTATGATATGCTTCA |
| IS431 elements | |||
| IS431-3 | 37850-38638 | 70341-71129 | |
| IS431-2 | 48131-48919 | 80622-81410 | |
| IS431-1 | 56620-57409 | 89111-89900 | |
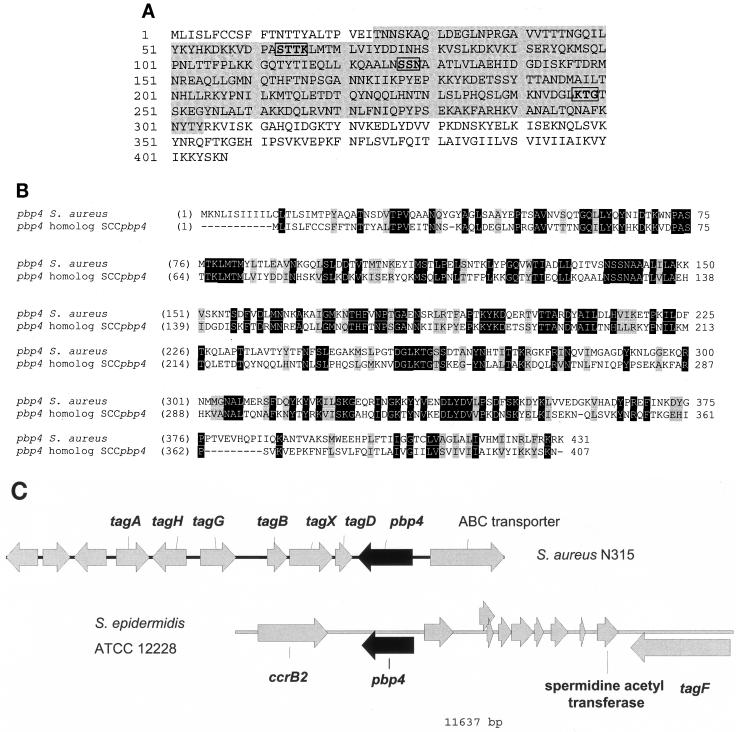
However, an ORF that was annotated as pbp4 (SE0035; GenBank accession no. AE016744) was found immediately to the right of the type 2 ccrB gene (Fig. 3). The translated protein had conserved domains from β-lactamase class A enzymes and d-Ala-d-Ala carboxypeptidases that belonged to pfam groups 00144 and 00768 (Fig. 4A), respectively, from the conserved database at the NCBI. The predicted amino acid sequence of this ORF was 34 to 37% homologous to that of PBP 4 from S. aureus (Fig. 4B). This pbp4 homolog, located between the characteristic SCCmec left and right terminal DRSCC and IRSCC sequences, led to our designation of this novel element in the S. epidermidis ATCC 12228 sequence as SCCpbp4. No other pbp4 homolog could be found in the S. epidermidis ATCC 12228 sequence. Interestingly, the architecture of the ORFs surrounding pbp4 from S. epidermidis was unlike that surrounding pbp4 in S. aureus (Fig. 4C). Downstream of pbp4 in S. epidermidis, there was one gene encoding a teichoic acid synthesis function (tagF), whereas downstream of pbp4 in S. aureus, there were other genes for teichoic acid synthesis (tagA, tagH, tagG, tagB, tagX, and tagD) (Fig. 4C). Also, the ATP binding cassette protein upstream of, and in reverse orientation to, the pbp4 gene in S. aureus (5, 8) was not present upstream of pbp4 in S. epidermidis.
FIG. 4.
Analysis of the pbp4 homolog of SCCpbp4. (A) Theoretical translation of the pbp4 homolog of S. epidermidis ATCC 12228, highlighting functional domain architecture. Shading indicates the peptidase S_1 domain, starting at aa 24 and ending at aa 304. The penicillin-binding domains, SXXK, SSN, and KTG, are boxed. (B) ClustalW alignment of the pbp4 genes of S. aureus (GenBank accession no. SAPBP4GEN) and SCCpbp4 of S. epidermidis ATCC 12228. White letters on solid background, identical residues; letters on shaded background, conservative replacements. Overall there were 46.2% similarity and 32.7% identity between the two sequences. Pairwise alignments were performed with the Vector NTI suite, version 8 (Informax, Inc.), by using the blosum62mt2 matrix. (C) Architecture of ORFs in the DNA sequences surrounding and including the pbp4 genes (solid arrows) in S. aureus strain N315 (nt 91873 to 93737; accession no. AP003131) and the SCCpbp4 element of S. epidermidis ATCC 12228. Arrows indicate ORFs and their direction of transcription.
The SCC-specific DR-L (nt 51507 to 51521 in accession no. AE016744) and IR-L (nt 51506 through 51481 in accession no. AE016744) sequences adjacent to the left of the gene encoding the abortive phage resistance protein defined the left extremity of SCCpbp4 and thus were designated DRSCCpbp4-L and IRSCCpbp4-L, respectively (Fig. 1 and 3; Table 2). The right extremity of SCCpbp4 was defined by a pair of IRSCC and DRSCC sequences, positioned ca. 19 kb to the right of DRSCCpbp4-L and IRSCCpbp4-L, that were similar to the SCCmec IRSCC-R and DRSCC-R (labeled IRSCCpbp4-R and DRSCCpbp4-R, respectively) (Fig. 1 and 3). In SCCpbp4, the IRSCCpbp4-R and DRSCCpbp4-R sequences were in opposite orientations to each other, like the corresponding sequences at the right extremities of most S. aureus SCCmec elements, and overlapped by 11 bp (Fig. 1 and 3). Thus, SCC-specific repeat sequences defined the boundaries of SCCpbp4; we have annotated these in our third-party accession no. TPA BK001539 (Table 2).
To the right of pbp4 were ORFs encoding eight hypothetical proteins, spermidine acetyltransferase, TagF, and an additional hypothetical protein abutting the IRSCCpbp4-R sequence. Thus, SCCpbp4 carries ORFs that encode cell wall biosynthesis proteins in addition to PBP 4.
SCCpbp4 was found to be inserted into the same chromosomal location as SCCcap1 in S. aureus and SCC12263 in S. hominis. The DRSCCpbp4-R of SCCpbp4 was located in an orfX homolog that had 91% amino acid similarity to orfX of S. aureus (Fig. 3).
Description of the SCC composite island, a second, novel element in S. epidermidis ATCC 12228 that encompasses SCCpbp4.
SCCpbp4 was found to be contained within a larger element that contained identical copies of the 28-bp sequence (5′-AAAAACCGCATCATTTATGATATGCTTC-3′) referred to as DRSCC composite island at both its left and right extremities (Table 2; Fig. 3). Interestingly, this 28-bp sequence was composed of the overlapping IRSCCpbp4-R and DRSCCpbp4-R sequences found at the right extremities of SCCpbp4 and other SCC elements (see sequences in Fig. 1 and 3). Thus, the right extremity of the SCC composite island was also the right extremity of SCCpbp4. Interestingly, this 28-bp sequence has not previously been observed at the left extremity of an SCC element.
To the left of the DRSCCpbp4-L repeat, we found an IR/DRSCC-R sequence that overlapped the DRSCCpbp4-L sequence (see Fig. 3 and Table 2). Previously, the right and left DR/IRSCC regions have been found only at the right and left extremities of SCC elements, separated by at least 20 to ca. 60 kb. In this novel arrangement, the overlapping right and left SCC terminal-repeat sequences formed a 55-bp sequence (AAAAACCGCATCACTTATGATATGCTTCTGCTTATCAGTTGATGATGCGGTTTTT) that contained a terminal 13-bp perfect complementary inverted repeat separated by 29 nt (AAAAACCGCATCA-N29-TGATGATGCGGTTTTT). Since this repeat sequence was found at the left junction of the SCCpbp4 element within the SCC composite island, it has been designated the SCC composite island-SCCpbp4 junction inverted repeat. This inverted repeat was also present in the noncoding region downstream of a gene encoding abortive phage resistance protein and upstream of the hsdSR operon, encoding type I restriction proteins (Fig. 3), suggesting that it could contain a transcriptional terminator for the former.
Further defining the SCC composite island was a second pair of ccrA and ccrB recombinase genes to the left of SCCpbp4 (Fig. 3) that were closely related to the newly identified type 4 ccrA and ccrB allotypes (see Fig. 6) found in the SCCmec element of MRSA strain HDE288 (20), the so-called pediatric clone. A truncated copy of another ccrA gene, which was too short to assign to an allotype, was also present. In addition, there were 3 copies of a transposase for an insertion sequence-like element that we have annotated as IS431 (TPA BK001539) and ORFs (see supplemental Table 2 at http://www.ucch.org/daum-boyle_lab-composite_island_figures/table2.htm) that encode cadmium resistance, mercury and heavy-metal transport proteins, the type I restriction system proteins HsdR and HsdS, truncated transposases, another copy of spermidine acetyltransferase, and 20 hypothetical proteins. As shown in Fig. 3, many of these features in the SCC composite island are present in other members of the SCC family.
FIG. 6.
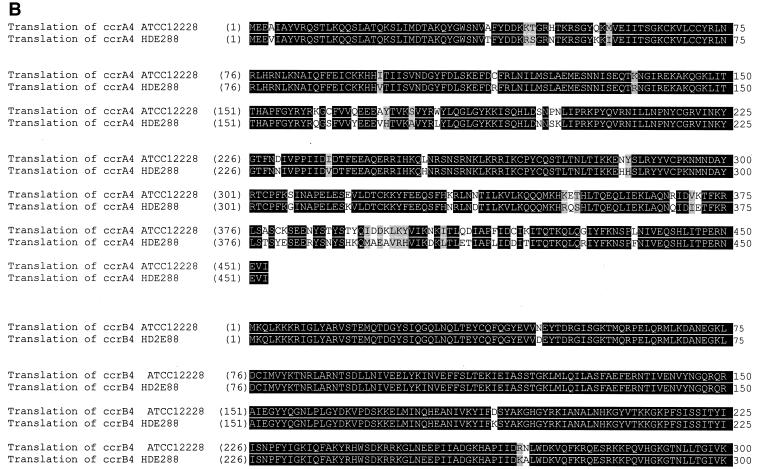
(A) ClustalW alignment of the ccrB4 ORF of the SCC composite island of S. epidermidis ATCC 12228 (nt 58592 to 60,220 complement) with that of SCCmec of strain HDE288 (the pediatric clone) (nt 9184 to 10812, GenBank accession no. AF411935). The missing adenine at position 1326 (shaded dash) of the ccrB4 ORF of strain HDE288 creates a putative premature stop codon (shaded box). Open box, putative stop codon of the ccrB4 ORF of strain ATCC 12228. Asterisks indicate identical residues. SCC-CI, SCC composite island. (B) Pairwise comparison of the translated amino acid sequences from the ccrA4 and ccrB4 genes of the SCC composite island of S. epidermidis ATCC 12228 (see supplemental Table 2 [http://www.ucch.org/daum-boyle_lab-composite_island_figures/table2.htm]) and of strain HDE288 SCCmec (accession no. AF411935). These strains showed 91.2% similarity and 87.2% identity for CcrA4 and 80.4% similarity and 80.1% identity for CcrB4. The pairwise alignments were performed by using the Vector NTI suite, version 8 (Informax, Inc.), with the blosum62mt2 matrix. A pairwise ClustalW alignment of the DNA sequences from the entire 5.3-kb region of homology surrounding the ccrAB4 genes of the SCC composite island of S. epidermidis ATCC 12228 (nt 57056 to 62395 complement, accession no. AE016744) and SCCmec of strain HDE288 (the pediatric clone) (nt 6986 to 12345, GenBank accession no. AF411935) is shown in supplemental Fig. 6D (available at http://www.ucch.org/daum-boyle_lab-composite_island_figures/fig_6D.htm). (C) Map of the region of nucleotide conservation between the SCC composite island and strain HDE288 that encompasses the ccrA4 and ccrB4 genes, highlighting the larger size of the ccrB4 ORF in the SCC composite island (1,629 versus 1,329 bp, encoding 542 versus 453 aa, respectively; vertical line indicates premature stop codon in strain HDE288). Solid arrows, unique ORFs in the SCC composite island.
Shown in supplemental Table 2 (at the URL given above) are the locations of all ORFs present in the SCC composite island and the degrees of similarity of their translated proteins to the products of similar ORFs found in other SCC elements. Of particular interest was a ca. 9.3-kb region at the left extremity of the SCC composite island (nt 89900 to 80621, GenBank accession no. AE016744) encoding mercury resistance proteins. This region contained six ORFs encoding hypothetical proteins and was flanked by the first and second copies of IS431. The region was 91.6% identical to the mer operon region in SCCmec type III of strain 85/2082 (nt 36066 to 44580, GenBank accession no. AB037671) (Fig. 5). The main difference found between the Mer regions of SCCmec type III and the SCC composite island was the presence of two additional ORFs (SE0089 and SE0088), located to the right of the first IS431 element in the SCC composite island, encoding hypothetical proteins that were not present in type III SCCmec of strain 85/2082 (Fig. 5). Furthermore, cadmium resistance proteins (CadR region) were encoded between the second and third IS431 elements (Fig. 3 and 5). The products of ORFs SE0075 and SE0073 (Fig. 5), encoding a putative cadmium resistance transporter and a cadmium efflux protein, respectively, were 90% similar to that encoded by ORF Z020 and 80% similar to that encoded by a portion of ORF Z019 from SCCmec type III (Fig. 3 and 5). Also, the CadR region contains a 2,469-bp stretch (nt 73585 through 76053, GenBank accession no. AE016744) with 75% nucleotide identity to the CadR region encompassing ORFs Z020 and Z019 in the SCCmec III element of strain 85/2082 (Fig. 3 and 5).
FIG. 5.
Map showing the region of the SCC composite island containing three IS431 elements (hatched arrows) and the intervening genes encoding proteins conferring heavy metal resistance (also shown, in less detail, in Fig. 3). Directions of arrows indicate the orientations of the ORFs. Shaded arrows, ORFs within a ca. 9-kb region with 99% similarity to the Mer resistance region (mer R) in SCCmec type III of strain 85/2082 (nt 36002 to 44579, GenBank accession no. AB037671); the region is indicated below the map. Open arrows, ORFs encoding cadmium resistance proteins. The ca. 2.3-kb region with 78% nucleotide identity with the cadmium resistance region (cad R) in SCCmec type III is indicated below the map. The two unique ORFs (SE0089 and SE0088, encoding hypothetical proteins with no homolog in the mer R region of SCCmec type III) are indicated by solid black arrows.
In a pairwise alignment, we also identified a 5.3-kb stretch of 92% sequence identity (Fig. 6A) between nt 57056 to 62395 (complement, GenBank accession no. AE016744) of the SCC composite island and nt 6986 to 12345 (GenBank accession no. AF411935) of SCCmec of strain HDE288 (the so-called pediatric MRSA clone). This region contained the type 4 ccrA and ccrB genes and flanking nucleotide sequences mentioned above (Fig. 6).
A third important stretch of similarity between other SCC elements and the SCC composite island was the ca. 9-kb region at the left extremity of SCCpbp4 referred to as the L-C HR, mentioned above (Fig. 3).
In summary, a large, novel composite island was found in the genome sequence of S. epidermidis ATCC 12228. This composite island lacks mecA but contains two pairs of ccr recombinases of different allotypes (ccrA2/ccrB2 and ccrA4/ccrB4) described previously in SCCmec elements in S. aureus. Nested within the composite island was a smaller (19-kb) element, SCCpbp4, that had a ca. 9-kb stretch of sequence homology 98 to 99% similar to a region in the SCCmec element of a CA-MRSA strain that harbors an SCCmec type IV element and of an HA-MRSA isolate that harbors type IVc SCCmec (11). Furthermore, outside the SCCpbp4 region, at the left extremity, genes that encoded resistance determinants against toxic heavy metals, identical to those found in the large S. aureus type III SCCmec, were identified.
Conservation of repeat sequences and L-C region ORFs in the SCC composite island and other members of the SCC family among staphylococci. (i) Repeat elements.
The IRSCC-R sequences of SCCpbp4 of S. epidermidis were more closely related to those in SCCmec type I, II, and IV (Fig. 1A) than those in SCCmec type III and SCCcap1 (Fig. 1B). In contrast, the IRSCC-L of SCCpbp4 was closely related to that of SCCmec type III as well as types I, II, and IV but was unrelated to the IR-L of SCCcap1.
The 15-bp DRSCC-L in CA-MRSA strain 2314 contained two mismatches with the 15-bp DRSCCpbp4-L contained in SCCpbp4 in S. epidermidis ATCC 12228. The 26-bp IRSCCpbp4-Lof S. epidermidis ATCC 12228 was 100% identical to the IRSCC-L's in the SCCmec elements of strain 2314 and similar to other MRSA isolates (N315, CA05, MW2, 8/6-3P) (3, 10, 16).
(ii) Comparison of ORFs with those in other SCC elements.
Inspection of the aligned regions from the left extremities of many SCC elements depicted in Fig. 2 and the data in supplemental Table 2 (available at the URL given above) suggests a model by which SCCmec elements may have evolved from the SCCpbp4 element in the 57-kb SCC composite island of S. epidermidis. As noted, all five ORFs (shown as solid rectangles in Fig. 2) present in the L-C HR of the SCC composite island were conserved in the L-C region of two SCCmec type IV elements in strain 2314 and SCCmec IVc in strain MR108. ORFs SE0042 and SE0041, encoding abortive phage resistance protein and Abi-alpha protein, annotated in the sequence of S. epidermidis ATCC 12228, were not found in any other SCC element except SCCpbp4, SCCmec IV in strain 2314, and SCCmec IVc in strain MR108. It is possible that these two ORFs were either lost by deletion from a primordial element such as SCCpbp4 or gained by homologous recombination. An ORF with a high degree of similarity to SE0040 was found in five SCC elements: the type II SCCmec elements of N315 and Mu50, the type IV SCCmec elements of strains 2314 and MR108, and the SCC element found in S. hominis. ORFs homologous to ORF SE0039 were found in nine SCC elements: SCCmec type II of strains Mu50 and N315, SCCmec type IV of strains 2314 and MR108, SCCmec IVa of strains CA05 and MW2, SCCmec type I of strain NCTC 10442, SCCmec type III of strain 85/2082, and SCC12263 of S. hominis. ORFs similar to SE0040, SE0039, and SE0038 were present together in only four of the SCC elements identified to date.
Homologs of the ORF (SE0038) closest to the ccrA2 genes were present in all SCC elements, including SCCmec types I, II, III, and IV, SCCpbp4, and SCC12263, with various degrees of conservation. ORF SE0038 of S. epidermidis ATCC 12228 was highly conserved (97% identity) in strains 2314 (ORF 5), MR108 (ORF R004), and N315 (ORF N031). In contrast, the nucleotide and predicted amino acid sequences of these ORFs in strain 2314 and strain MR108 differed considerably at the 5′ end of the gene (from the start codon to bp 450 to 550) and the amino terminus of the translated protein from the corresponding ORFs in the type IVa SCC elements of isolates MW2 (ORF MW0040) and CA05 (ORF Q003), and the type IVb SCC elements of isolate 8/6-3P (ORF M002). Nevertheless, the 3′ terminus of this ORF in CA-MRSA strain 2314 was 95 to 96% identical to those of CA-MRSA isolates MW2, CA05, and 8/6-3P. Thus, surprisingly, ORF 5 of CA-MRSA strain 2314 was more closely related to the corresponding ORFs of S. epidermidis ATCC 12228 and HA-MRSA strains with type II SCCmec (N315 and Mu50) than to those in CA-MRSA strains with either SCCmec subtype IVa (MW2 and CA05) or IVb (strain 8/6-3P) or SCCmec type I or III.
Interestingly, the 3′ terminus of ORF SE0038 in S. epidermidis ATCC 12228 is conserved among SCC elements that have the type 2 ccr gene complex but is less similar to the corresponding ORFs contained in SCC elements carrying the type 1 or type 3 ccr gene complex. The 3′ terminus of ORF 5 of CA-MRSA strain 2314 was 89% similar to the corresponding ORF 13 in S. hominis ATCC 27844 and SCCmec type I in the MRSA strain NCTC 10442. In summary, there was a tendency for an ORF from the L-C HR to be conserved in a greater number of SCC elements when the position of the ORF in S. epidermidis was closer to the ccr genes.
(iii) ccr gene comparisons.
An alignment was performed between the two sets of ccrAB genes in CA-MRSA strain 2314 and SCCpbp4 and genes of ccrAB allotypes 1, 2, 3, and 4. The ccrA gene identified in isolate 2314 was 98% identical to those of SCCpbp4 and CA-MRSA strain CA05 and was 96 to 97% identical to the ccrA2 alleles of isolates MW2, N315, and 8/6-3P. The ccrB gene was also 96 to 97% identical to the ccrB2 alleles of isolates 8/6-3P, N315, MW2, and CA05, in contrast to 92% homology to that of ATCC 12228. Thus, the ccrA and ccrB genes in CA-MRSA strain 2314 and SCCpbp4 are related to the type 2 ccr gene allotype, a feature of type IV and type II SCCmec elements.
Pairwise comparison of the translated amino acid sequences from the ccrA4 and ccrB4 genes of the SCC composite island and SCCmec of strain HDE288 (accession no. AF411935) revealed that ccrA4 had 91.2% similarity and 87.2% identity, and ccrB4 had 80.4% similarity and 80.1% identity, between these strains (Fig. 6B). In the SCC composite island, the ccrB4 ORF was larger than that of strain HDE288 (1,629 versus 1,329 bp, encoding 542 versus 453 amino acid [aa] residues, respectively) due to the deletion of a single nucleotide (adenine) at position 1326 of the latter ORF (Fig. 6A).
DISCUSSION
We found a novel 57-kb composite island in S. epidermidis strain ATCC 12228 (the SCC composite island) that may represent a primordial genetic element contributing DNA sequences to the SCCmec elements found in S. aureus.The island has two sets of ccr recombinases and a DRSCC composite island sequence at either end closely resembling the IRSCC-R and DRSCC-R sequences found in SCCmec elements in S. aureus. Moreover, a smaller (19-kb) SCC element called SCCpbp4 is nested within the right side of the SCC composite island. The mecA gene itself and the genes responsible for its regulation were notably absent.
The SCC composite island had several large regions of substantial homology with SCCmec elements (summarized in Fig. 3). A 6.9-kb area in the L-C region of SCCpbp4 was nearly identical to the L-C region of SCCmec type IV elements of CA-MRSA strain 2314 and HA-MRSA strain MR108. This region also shared homology with the SCC element of S. hominis. Also, the left side of the island contained a ca. 10-kb stretch of DNA with 92% similarity to a region within SCCmec type III (10) that has mercury resistance genes flanked on either end by IS431. The CadR region of the SCC composite island contained a 2.5-kb stretch of similarity to the type III element. Also, a highly homologous region of ca 5.3 kb that included the ccrAB4 genes was present in both the SCC composite island and the SCCmec element of MRSA isolate HDE288. Thus, >25 kb of the 57-kb DNA sequence of the SCC composite island was conserved in other SCCmec elements. The presence of these regions of virtual identity to two distinct SCCmec element types suggests that there has been extensive horizontal genetic exchange between this SCC composite island in S. epidermidis and the SCCmec elements found in MRSA isolates.
The SCC composite island is unique within this family in several regards. First, within the SCC composite island are two pairs of ccrA and ccrB genes of different allotypes (types 2 and 4). One pair was within the nested element SCCpbp4. Second, several novel arrangements of the DRSCC and IRSCC sequences were present. The 28-bp direct repeat sequence (DRSCC composite island) that flanks the SCC is composed of exact copies of the overlapping composite island DRSCCpbp4-R and IRSCCpbp4-R sequences, whereas in SCCmec elements, the structures of the DRSCC and IRSCC sequence junctions differ slightly at the left and right extremities. Moreover, a third copy of this DRSCC composite island was present overlapping with the SCCpbp4 left terminal repeat sequences. This presence of a direct repeat at the margins of the SCC composite island is similar to the architecture of the ends of pathogenicity islands in bacteria (7). The terminal-repeat sequences of the nested SCCpbp4 element were more typical of SCCmec elements. However, the DRSCC composite island sequence (i.e., an IR/DRSCC-R terminal-repeat sequence) overlapping the DR/IRSCCpbp4-L sequence was atypical (Fig. 3). Previously, the left and right terminal repeats had been detected at least 20 kb apart at the left and right termini of SCC elements, respectively. The overlap created a novel 55-bp sequence flanked by a B-bp complementary inverted repeat in the intergenic region between SCCpbp4 and the left portion of the SCC composite island, downstream of an ORF encoding a phage resistance protein (a probable restriction protein). The significance of this novel juxtaposition of these repeats at the junction of SCCpbp4 remains to be determined. It is possible that this sequence contains a transcriptional terminator for the phage resistance protein.
The SCC composite island and the nested SCCpbp4 element are the third and fourth members of the family of SCC elements identified to date that have intact ccrA and ccrB genes and the characteristic direct- and inverted-repeat sequences. These are features previously shown to allow excision and integration of the element (12). SCCcap1 (15) is another member of this family, but it lacks a ccrA gene homolog, limiting its potential for excision and spread. In this regard, the ccrAB2 allotype is the only allotype to date shown to mediate excision activity. The functionality of the ccrAB4 allotype has not yet been investigated, but the ccrB4 gene in the SCC composite island of ATCC 12228 appears to be more intact than that of HDE288, which has a truncated ccrB gene (see Fig. 6).
The presence of cadmium and mercury resistance genes, also present in other SCCmec elements, suggests that SCC elements are also used by staphylococci for horizontal transfer of genes encoding resistance to heavy metals. The presence of restriction system genes in the SCC composite island both within and outside SCCpbp4 suggests that at some time, both portions of the element were transferable between bacterial species.
It remains to be determined whether the SCC composite island is a feature only of S. epidermidis ATCC 12228 or whether it can be found among other S. epidermidis isolates. If it is common, it is possible that S. epidermidis, a skin commensal, acts as a reservoir of DNA sequences that have been horizontally transferred and recombined within SCCmec elements by homologous recombination. It was recently proposed that an SCC element found in S. hominis was an ancestral element providing sequences for SCCmec elements (13). Our data describe two additional possible ancestral SCC elements present in a commensal staphylococcal species that could also act as a source of DNA for SCCmec elements. In this regard, the SCC12263 element in S. hominis carries type 1 ccr genes, whereas the SCC composite island in S. epidermidis carries type 2 and 4 ccr genes. Thus, S. epidermidis and S. hominis would be able to provide diversity to SCCmec elements by contributing to their different types of ccr genes. In this regard, the GC content of SCC elements is very close to that of staphylococcal genomes, supporting the notion that the elements have indeed likely originated from staphylococcal species.
It is not known how SCC elements are horizontally transferred between S. aureus strains and other Staphylococcus species. The SCCpbp4 and SCCmec elements both carry important cell wall synthesis genes, and SCCcap1 carries genes responsible for a capsular polysaccharide. These data suggest that SCC elements are specialized elements in staphylococci that carry genes encoding cell wall synthesis enzymes. In this scenario, SCCmec elements have been exploited for the transfer of antibiotic resistance by importation of a PBP gene with low affinity for β-lactams. The biological importance of carrying PBP genes such as these on presumably mobile genetic elements remains to be determined but could account for the functional redundancy of PBPs in staphylococci.
Inspection of the aligned maps of tandem ORFs located adjacent to the ccr genes in SCCmec elements suggests a model in which SCCmec elements evolved from the SCCpbp4 element in S. epidermidis (Fig. 2). Isolated in 1996, Dallas CA-MRSA isolate 2314 was one of the first CA-MRSA isolates to be recognized. All five ORFs positioned between the ccr genes and the DRSCCpbp4-L sequence were present in the L-C region of this isolate and the HA-MRSA strain MR108. The type IV SCCmec elements from more-recent CA-MRSA isolates such as MW2, CA05, and 8/6-3P have remnants of ORF 5 but do not contain ORFs 1, 2, 3, and 4. Furthermore, ORFs 3, 4, and 5, upstream of ccrA in CA-MRSA isolate 2314, were more closely related to the corresponding ORFs found in HA-MRSA strains bearing SCCmec type II. Thus, the ORFs in this region that were closer to the ccr genes were more likely to be conserved among a greater range of SCC element types than those more distal to the ccr genes. These data also suggest that recombination events have resulted in gradual elimination of ORFs 1, 2, 3, and 4 from SCCmec as SCCmec type II evolved into SCCmec type IVc and ultimately types IVa and IVb.
The left portion of the novel SCCpbp4 element in SCCmec type IVc provides evidence that, at some point, there was horizontal genetic exchange between the progenitors of CA-MRSA strain 2314 and S. epidermidis ATCC 12228. It is not clear whether isolate 2314 obtained the L-C portion of SCCmec IVc from the SCCpbp4 element of ATCC 12228 or vice versa. A model whereby homologous pairing occurred between the DRSCC and ccr gene sequences of the two SCC elements, resulting in gene replacement in the L-C region of a previously extant SCCmec cassette, is one plausible explanation.
Acknowledgments
S.B. and R.S.D. are the recipients of R01 AI40481-01A1 from NIAID, RO1 CCR523379 from the CDC, and support from the Grant Health Care Foundation. S.B. was also supported by R03 AI44999-01 from NIAID. K.M. was supported by a grant from the Children's Research Foundation, Northbrook, Ill.
REFERENCES
- 1.Adcock, P. M., P. Pastor, F. Medley, J. E. Patterson, and T. V. Murphy. 1998. Methicillin-resistant Staphylococcus aureus in two child care centers. J. Infect. Dis. 178:577-580. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 5.Domanski, T. L., and K. W. Bayles. 1995. Analysis of Staphylococcus aureus genes encoding penicillin-binding protein 4 and an ABC-type transporter. Gene 167:111-113. [DOI] [PubMed] [Google Scholar]
- 6.Fey, P. D., B. Said-Salim, M. E. Rupp, S. H. Hinrichs, D. J. Boxrud, C. C. Davis, B. N. Kreiswirth, and P. M. Schlievert. 2003. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 8.Henze, U. U., and B. Berger-Bachi. 1995. Staphylococcus aureus penicillin-binding protein 4 and intrinsic β-lactam resistance. Antimicrob. Agents Chemother. 39:2415-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiramatsu, K., K. Okuma, X. X. Ma, K. Yamamoto, S. Hori, and M. Kapi. 2002. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr. Opin. Infect. Dis. 15:407-413. [DOI] [PubMed] [Google Scholar]
- 10.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec in the chromosome of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updat. 6:41-52. [DOI] [PubMed] [Google Scholar]
- 12.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus casette chromosome mec, encodes methicillin resistance. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama, Y., F. Takeuchi, T. Ito, X. X. Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 185:2711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K.-I. Aoki, Y. Nagai, J.-Q. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R.-I. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genomic sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 15.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184:3623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. A novel type of staphylococcal cassette chromosome mec (SCCmec) identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Mongkolrattanothai, K., S. Boyle, M. D. Kahana, and R. S. Daum. 2003. Severe Staphylococcus aureus infections caused by clonally related community-acquired methicillin-susceptible and methicillin-resistant isolates. Clin. Infect. Dis. 37:1050-1058. [DOI] [PubMed] [Google Scholar]
- 17.Niemeyer, D. M., M. J. Pucci, J. A. Thanassi, V. K. Sharma, and G. L. Archer. 1996. Role of mecA transcriptional regulation in the phenotypic expression of methicillin resistance in Staphylococcus aureus. J. Bacteriol. 178:5464-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant S. aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira, D. C., and H. de Lencastre. 2002. Multiple PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 21.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wisplinghoff, H., A. E. Rosato, M. C. Enright, M. Noto, W. Craig, and G. L. Archer. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science 291:1962-1965. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]