Abstract
Topoisomerase IV and DNA gyrase are related bacterial type II topoisomerases that utilize the free energy from ATP hydrolysis to catalyze topological changes in the bacterial genome. The essential function of DNA gyrase is the introduction of negative DNA supercoils into the genome, whereas the essential function of topoisomerase IV is to decatenate daughter chromosomes following replication. Here, we report the crystal structures of a 43-kDa N-terminal fragment of Escherichia coli topoisomerase IV ParE subunit complexed with adenylyl-imidodiphosphate at 2.0-Å resolution and a 24-kDa N-terminal fragment of the ParE subunit complexed with novobiocin at 2.1-Å resolution. The solved ParE structures are strikingly similar to the known gyrase B (GyrB) subunit structures. We also identified single-position equivalent amino acid residues in ParE (M74) and in GyrB (I78) that, when exchanged, increased the potency of novobiocin against topoisomerase IV by nearly 20-fold (to 12 nM). The corresponding exchange in gyrase (I78 M) yielded a 20-fold decrease in the potency of novobiocin (to 1.0 μM). These data offer an explanation for the observation that novobiocin is significantly less potent against topoisomerase IV than against DNA gyrase. Additionally, the enzyme kinetic parameters were affected. In gyrase, the ATP Km increased ≈5-fold and the Vmax decreased ≈30%. In contrast, the topoisomerase IV ATP Km decreased by a factor of 6, and the Vmax increased ≈2-fold from the wild-type values. These data demonstrate that the ParE M74 and GyrB I78 side chains impart opposite effects on the enzyme's substrate affinity and catalytic efficiency.
Type II topoisomerases catalyze the interconversion of DNA topoisomers by transporting one DNA segment through another. Bacterial genomes encode two type II topoisomerases, DNA gyrase and topoisomerase IV (TopoIV), that function in DNA replication. DNA gyrase is unique in coupling the free energy of ATP hydrolysis to the introduction of negative supercoils into DNA. In the absence of the ATP substrate, DNA gyrase can relax negatively supercoiled plasmid DNA. These activities result from the enzyme's ability to wrap (≈150 bp) DNA (23, 31) around itself upon binding the DNA substrate. This DNA wrapping preferentially presents the T-segment (transported DNA segment) to the gyrase-DNA complex so that the introduction of negative supercoils is the primary outcome. In contrast, TopoIV and other eukaryotic type II topoisomerases only bind a ≈30-bp region of DNA (20, 35). TopoIV utilizes the energy of ATP hydrolysis to decatenate newly replicated chromosomal DNA but also has the ability to relax positive and negative DNA supercoils in an ATP-dependent manner (8, 43).
In prokaryotes, these type II topoisomerases are composed of two subunits. In Escherichia coli, the gyrase subunits are named A and B and the corresponding TopoIV subunits are named C and E. For each enzyme, these subunits combine into a heterotetrameric (gyrase, A2B2; and TopoIV, C2E2) complex to form the functional enzymes. In contrast, the eukaryotic type II topoisomerases are homodimers (6). In TopoIV, the E-subunit (ParE) and the corresponding gyrase B-subunit (GyrB) are functionally similar, as both contain the ATPase active site at the N-terminal domain (≈43 kDa), which is referred to as the ATP-operated clamp (27). This clamp closes (dimerizes) upon ATP binding, trapping the T-segment of the DNA. The GyrB (47-kDa) or ParE C-terminal domain is involved in the interaction with the other subunit (GyrA or ParC) and the DNA substrate.
The GyrA and ParC subunits are also organized in a similar fashion, with the N-terminal domain (64 kDa) providing the active-site tyrosine that covalently attaches to the cleaved DNA strands of the double-stranded DNA break (the DNA gate). It is through this DNA gate that the DNA T-segment must pass in order to change the superhelical density of the DNA substrate (27). The C-terminal domains (33 kDa) of GyrA and ParC differ both in sequence similarity and in the manner in which they bind to DNA (35, 37).
The crystal structure of the E. coli N-terminal 43-kDa fragment of GyrB bound with adenylyl-imidodiphosphate (ADPNP) suggests that a dimer is the functional unit (44) and is supported by biochemical evidence (3). Many of the contacts between these dimeric molecules occur at the N terminus, where residues 2 to 15 extend from the N terminus of each monomer to wrap around the other subunit in the dimer (44). The extending arms give rise to important contacts that stabilize the dimer (2, 3) and form a portion of the ATP binding site (44). Most of the remaining ATP binding residues arise from the first 220 amino acids, or domain 1, of the 43-kDa fragment. Domain 2 forms the sides of a large hole (≈20 Å) in the protein dimer that is believed to serve as the cavity in which the T-segment is captured for presentation to the DNA gate (44).
Domain 1 of E. coli (also referred to as the 24-kDa GyrB fragment) has been crystallized with coumarin antibiotics (22, 42) such as novobiocin. Two major structural differences occur when the inhibitor and ADPNP structures are compared: the inhibitor structures are all monomeric, and the flexible loop region (GyrB residues 97 to 120) forms a lid over the ATP binding site in the ADPNP structure (22, 42). (In the novobiocin structure, the loop is either disordered or adopts various conformations.) Recently, a crystal structure of novobiocin bound to the larger 43-kDa N-terminal fragment of GyrB of Thermus thermophilus was reported (18). In this structure, three major differences were apparent versus the 24-kDa GyrB novobiocin structure. First, the 43-kDa GyrB fragment is a dimer in the presence of novobiocin, and many of the dimerization contacts from the ADPNP structure are maintained. Second, domain 2 swivels 18 degrees from the twofold axis relative to domain 1 compared to the E. coli 43-kDa GyrB ADPNP structure. Finally, the flexible loop region adopts a more open conformation than is seen in the ADPNP structure.
Gyrase and TopoIV have similarities in their conserved active sites, subunit organization, and antibiotic sensitivity to coumarins and quinolones (17, 34, 41). Both enzymes are also independently essential for bacterial growth. These similarities suggest that small-molecule inhibitors of the active sites might simultaneously target both type II prokaryotic topoisomerases. For some quinolones, dual targeting of type II topoisomerases (GyrA and ParC) has been achieved (33, 40). The prokaryotic type II topoisomerases have been successful antibacterial targets because they differ sufficiently from eukaryotic type II topoisomerases and are highly conserved enzymes within many clinically relevant bacteria. These properties permit the design of specific bacterial inhibitors with the possibility for a broad spectrum of action. A prospectively designed dual-target inhibitor with equal potency has the potential to reduce the development of target-based resistance because two independent resistant mutations from each target would be required to confer resistance (9).
To date, no structures of TopoIV have been reported. To aid us in our structure-based drug design approach for novel antibacterials, we sought to determine the structure of TopoIV bound with ADPNP and novobiocin. Novobiocin is an ATP-competitive inhibitor of both DNA gyrase and TopoIV and occupies a portion of the ATP binding site (13, 22; this study). In all literature reports and under our own assay conditions, the TopoIV novobiocin 50% inhibitory concentration (IC50) is always greater than the DNA gyrase IC50 (Table 1) (12, 14, 34, 40), suggesting that a structural difference exists between the TopoIV and gyrase ATPase active sites. Here we report the first crystal structures of an E. coli TopoIV 43-kDa ParE N-terminal fragment complexed with ADPNP and a 24-kDa N-terminal fragment crystal structure complexed with novobiocin. We provide evidence that in DNA gyrase and TopoIV, a single residue (GyrB I78 and ParE M74) is responsible for the differences in novobiocin potency between these type II topoisomerases.
TABLE 1.
Kinetic parameters and novobiocin IC50s for the ATPase activity of wild-type and mutant proteins
| E. coli enzyme | Mutation | Novobiocin IC50 (nM)a | IC50 ratiob | V/K (% of wild-type value)c | ATP Km (μM)d |
|---|---|---|---|---|---|
| GyrA2B2 | None | 46 | 100 | 830 | |
| GyrA2B2 | I78Ae | 1,400 | 30 | 3 | >5,000 |
| GyrA2B2 | I78Le | 640 | 14 | 17 | >5,000 |
| GyrA2B2 | I78M | 1,000 | 22 | 14 | 4,200 |
| GyrA2B2 | I78Ve | 72 | 2 | 56 | 1,600 |
| GyrA2B2 | G81A | 95 | 990 | ||
| GyrA2B2 | G81D | 75 | 2 | 60 | 1,100 |
| GyrA2B2 | EcTopoIV loopf | 70 | 2 | 170 | 480 |
| GyrA2B2 | SaTopoIV loopf | 53 | 1 | 180 | 830 |
| TopoIV C2E2 | None | 210 | 100 | 700 | |
| TopoIV C2E2 | M74I | 12 | 0.06 | 934 | 120 |
IC50s were determined as decribed in Materials and Methods. The IC50 standard error was typically 15% of the IC50.
IC50 for mutant/IC50 for wild type.
The V/K ratio is expressed as the percentage of that for the wild-type enzyme, which was 0.5 mM−1 s−1 for gyrase and 1.0 mM−1 s−1 for TopoIV.
Km values are apparent.
Values are from Gross et al. (13).
Loop substitutions are described in Materials and Methods.
MATERIALS AND METHODS
Cloning of E. coli ParE and ParC subunits.
The parE and parC genes from E. coli (ATCC 10798D) were amplified by PCR from purified chromosomal DNA with Taq polymerase (Qiagen). Oligonucleotide primers complementary to the 5′ and 3′ ends of the open reading frames were designed to introduce NdeI and EcoRI restriction sites for parE and BfaI and EcoRI for parC. The PCR products were digested with appropriate restriction endonucleases and ligated to an E. coli-baculovirus dual expression vector, pBEV (5). The resulting plasmids, pBEV10-parE and pBEV10-parC, produced Par proteins with amino-terminal hexahistidine (His6) tags (MGSSHHHHHHSSGLVPRGSH for ParE and MGSSHHHHHHSGLVPRGSHS for ParC). For the 43-kDa construct, the ParE coding sequence contained residues 1 through 390 plus the His6 tag. For the 24-kDa construct, the ParE coding sequence contained residues 1 to 217 and the N-terminal His6 tag but with deletion of residues 93 to 116 (the flexible loop region). The parE and parC genes were sequenced to ensure their integrity and were identical to the GenBank sequences, accession numbers D65090 and P20082, respectively.
Mutagenesis of E. coli GyrB and ParE subunits.
The DNA gyrase constructs have been described (13). Site-directed mutations and flexible loop mutations were introduced into gyrB and parE genes by PCR. For the gyrase flexible loop mutations, E. coli gyrase B residues D105 to V111 (DDNSYKV) were replaced with sequences from E. coli ParE (SNKNYQF) and from Staphylococcus aureus GrlB (ParE homologue) (GQGGYKT). All fragments were sequenced to confirm the presence of the mutations and the absence of unwanted mutations.
Expression and purification of wild-type and recombinant proteins.
DNA gyrase expression and purification have been described (13). Full-length parE and parC constructs were expressed in baculovirus-infected Sf9 cells by standard methods. Full-length ParE and ParC proteins were purified with Talon resin (Clonetech). Fractions containing the His6-tagged TopoIV subunits were further purified over a Sephacryl S-200 column (16 by 600 mm; Pharmacia). Protein fractions were concentrated in Ultrafree-15 Biomax-100 centrifugal filter units (Millipore). Protein concentrations were determined by absorbance measurements at 280 nm with the calculated extinction coefficients (http://us.expasy.org [ProtParam]). The protein samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a known standard, followed by Coomassie blue staining to confirm that the protein concentration was accurate (not shown).
To form the functional gyrase A2B2 or TopoIV C2E2 heterotetrameric enzymes, equal molar amounts of the subunits were mixed together at a high protein concentration (>15 μM) 15 min prior to the start of the assay. The DNA gyrase enzyme used for this study has been characterized (13). For the TopoIV enzymes (C2E2), ATPase active site titration were performed with subnanomolar compounds (e.g., coumermycin A1 [Sigma]) (not shown) and agreed with the determined protein concentration, suggesting that all of the TopoIV enzyme molecules were active.
Truncated 43-kDa and 24-kDa parE expression plasmids were transformed into the recombination-deficient E. coli (recA) expression strain BLR(DE3) (Novagen) to avoid gene rearrangements. Recombinant protein production and purification were carried out as described before (13). Protein-containing imidazole fractions were further purified over a Sephacryl S-100 column (16 by 600 mm; Pharmacia). Protein fractions were concentrated in Ultrafree-15 Biomax-10 centrifugal filter units (Millipore) to 10 mg/ml.
ATPase assay.
ATP hydrolysis rates were monitored spectrophotometrically on a Molecular Devices plate reader by monitoring the decrease in NADH absorbance during the coupled enzyme reaction as described (13). The standard coupled enzyme reactions were carried out at 30°C (reaction volume, 100 μl) with the following components for TopoIV: 100 mM Tris-HCl (pH 7.6), 6 mM MgCl2, 20 mM KCl, 2.5 mM phosphoenolpyruvate, 0.2 mM NADH, 10 mM dithiothreitol, 0.03 mg of pyruvate kinase per ml, 0.01 mg of lactate dehydrogenase per ml, 4% dimethyl sulfoxide, 0.05 mg of bovine serum albumin per ml, 5.25 μg of HindIII-linearized pBR322 plasmid DNA per ml, 700 μM ATP, and 56.4 nM TopoIV enzyme. The specific activity of the wild-type Topo IV enzyme was 1.3 μmol of ATP/min/μg of enzyme. DNA gyrase conditions were identical to those reported before (13).
Determination of kinetic parameters and data analysis.
For enzymes, the V and K values were derived from the linear portion of an ATPase rate versus ATP substrate concentration plot (i.e., the slope of the line). This value is a good approximation of each enzyme's overall catalytic efficiency (i.e., Vmax/Km) (11). The apparent Km for ATP was determined when apparent saturation had been achieved, by fitting the rectangular hyperbolae plot of linear rates of ATP hydrolysis versus increasing ATP concentration to the Michaelis-Menten equation. IC50 values for the ATP-competitive inhibitor novobiocin (Sigma) were obtained by measuring the decrease in linear rates of ATP hydrolysis at increasing concentrations of inhibitor and fitting the data with a two-parameter hyperbolic fit: y = IC50/(B * [X + IC50]), where B is proportional to 1/Vmax, y is the rate of reaction, and X is the concentration of the inhibitor. For all enzymes, the ATP concentration did not exceed the apparent Km. For all IC50 determinations, the enzyme concentration was less than or equal to two times the reported IC50.
ParE crystallization and X-ray data collection.
For the 43-kDa ParE fragment in complex with ADPNP, a concentration of 10 mg/ml was used for crystallization trials. The protein solution (with 20 mM HEPES [pH 7.0], 5% glycerol, and 20 mM NaCl) was added in equal ratio, 1.0 μl of each, to the reservoir liquid of 20% (wt/vol) polyethylene glycol 3350, 0.2 M KCl, and sodium acetate, pH 6.2. At room temperature, single crystals were obtained in hanging droplets after 2 weeks of equilibration. A single crystal with dimensions of 0.15 by 0.10 by 0.35 mm was transferred to a cryoprotectant solution of mother liquor containing 20% glycerol shortly prior to being flash-cooled to 100 K in a nitrogen gas stream. X-ray data were collected to 2.1-Å resolution at the PX14.2 station in Daresbury, United Kingdom. The data images were indexed and integrated with MOSFLM (21) and Scalepack software (32). The crystals belonged to space group P212121 with unit cell dimensions of a = 94.9 Å, b = 120.8 Å, c = 136.8 Å, α = 90.0o, β = 90.0o, and γ = 90.0o. Five percent of the data was assigned for testing free R-factor in the later refinements. The Matthew's specific-volume calculation (26) suggested that there were two monomers in the asymmetric unit, giving a solvent content of 54%. Table 2 presents the crystallographic parameters.
TABLE 2.
X-ray experimental data and refinement summary
| Parameter | 43-kDa fragment | 24-kDa fragment |
|---|---|---|
| General | ||
| Temp (K) | 100 | 100 |
| Radiation source | PX14.2 | 5.02 ALS |
| Wavelength (Å) | 1.00 | 1.00 |
| Space group | P212121 | P3121 |
| a (Å) | 94.9 | 74.9 |
| b (Å) | 120.8 | 74.9 |
| c (Å) | 136.8 | 138.2 |
| α (°) | 90.0 | 90.0 |
| β (°) | 90.0 | 90.0 |
| γ (°) | 90.0 | 120.0 |
| No. of molecules in asymmetric unit | 2 | 2 |
| Recording device | ADSC QR4 | ADSC Q210 |
| Resolution (Å) | 2.10 | 2.00 |
| Total observation | 287,282 | 176,669 |
| Unique reflection | 88,735 | 30,716 |
| Completeness (%) | 96.8 | 99.4 |
| Rsyma (%) | 7.7 | 5.2 |
| Structure solution | ||
| Method | M.R.e | M.R. |
| Search modelb | 43-kDa GyrB (1EI1) | 24-kDa GyrB (1AJ6) |
| Resolution range (Å) | 10.0-4.0 | 10.0-4.0 |
| Software | AMoRe | AMoRe |
| R-factor (%) | 54.1 | 41.4 |
| Correlation coefficient factor | 21.8 | 54.9 |
| Refinement | ||
| Resolution range (Å) | 20.0-2.1 | 20.0-2.0 |
| Sigma cut off (σ) | 1.0 | 3.0 |
| Program | CNX | CNX |
| No of reflections | 88,698 | 29,580 |
| R-factorc (%) | 22.4 | 21.9 |
| Free R-factorc (%) | 24.1 | 27.8 |
| Stereochemical parameters | ||
| R.m.s.d bond distance (Å) | 0.010 | 0.009 |
| R.m.s. bond angle (°) | 1.31 | 1.29 |
| R.m.s. dihedral angle (°) | 22.5 | 21.4 |
| No. of nonglycine residues | ||
| Most favored (%) | 92.0 | 92.6 |
| Additional allowed (%) | 7.8 | 7.3 |
| Generously allowed (%) | 0.2 | 0.1 |
| Disallowed (%) | 0.0 | 0.0 |
Rsym = Σ|Ii − <I>|/ΣIi, Where Ii and I are the intensities for the ith observation and mean of the reflection, respectively.
The RCSB Protein Data Bank identifier for each search model is given in parentheses.
R-factor = Σ|Fobs − Fcal|/Σ|Fobs, where Fobs and Fcal are the observed and calculated model structure factors, respectively.
R.m.s., root mean square.
M.R., molecular replacement.
For the 24-kDa ParE fragment complexed with novobiocin, a protein sample of 10 mg/ml in 20 mM HEPES (pH 7.0)-10 mM NaCl-5% (vol/vol) glycerol was prepared for the slow vapor diffusion experiment. For each hanging droplet, 1.0 μl of protein solution was added to an equal volume of reservoir solution (20% polyethylene glycol 3350, 0.2 M potassium sodium tartrate [pH 7.2], and 0.1 M NaCl). At 298 K, crystals were harvested after approximately 2 weeks, with typical dimensions of 0.07 by 0.04 by 0.25 mm. Diffraction extended beyond 2.0-Å resolution with an ADSC Q210 detector on the 5.02 beam line at the Advanced Light Source synchrotron station in Berkeley, Calif. The diffraction images were recorded at 100 K by cryostreaming the crystal in mother liquid plus 20% (vol/vol) glycerol. The data reduction was done with Denzo and Scalepack software (32). Five percent of the data was set aside for free R-factor calculations. The crystals were determined to have unit cell dimensions of a = 74.9 Å, b = 74.9 Å, c = 138.2 Å, α = 90.0o, β = 90.0o, and γ = 120.0o, with a space group P3121. Assuming that there are two independent protein molecules in the asymmetric unit, the calculated Matthew's specific volume is 2.57 Å3/Da.
Structure determination and refinement.
For the 43-kDa ParE fragment with ADPNP, the correct orientation and translation solution were calculated by molecular replacement methods with AMoRe and a closely related homolog molecule, 43-kDa GyrB, as the search model (PDB accession code 1EI1). The correlation coefficient and crystallographic R-factor for the MR solution in the resolution range of 10 to 4 Å were 21.8 and 54.1%, respectively. Rigid-body and positional refinement of the polyalanine model was initially carried out with CNX (Accelrys Inc., 2000). The side chains, insertions, and deletions of the protein molecule were modeled into the electron density map by several runs of manual rebuilding with Quanta programs (Accelrys Inc., 2000). A calculated Fo-Fc map was used to locate the ADPNP molecule at the active site. Further refinements at a resolution of 2.1 Å were performed, and the ADPNP molecules were included in the calculations. Several cycles of refinements and subsequent manual fitting improved the quality of the model. Further inclusion of solvent molecules in the refinement and individual B-factor refinement reduced the R-factor and free R-factor to 22.4 and 24.1%, respectively. Residuals included in the refined model ranged from 4 to 383 for both crystallographically independent molecules. The final model also contains two ADPNP molecules, 2 Mg2+, 2 SO4−2 anions, and 247 water molecules. Table 2 shows the refinement indicators and the model quality.
The structure of 24-kDa ParE in complex with novobiocin was solved by molecular replacement methods with AMoRe (29) with the 24-kDa GyrB molecule as the search model (PDB accession code 1AJ6). Table 2 lists the solution parameters and the refinement statistics. Initial rigid-body and minimization refinements were performed to improve the model quality. The difference in protein sequences was manually built into the electron density map in several cycles of fitting and refinement. The novobiocin molecules were introduced into the model based on interpretation of the Fo-Fc difference density map. After several cycles of positional and B-factor refinements, 531 solvent molecules were added to the model. The improved model had an R-factor and free R-factor of 21.9 and 27.8%, respectively. The model quality was assessed with Procheck (19); 92.6 and 7.3% of nonglycine amino acids were mapped in the most favored and additional allowed areas of the Ramachandran plot. The accession codes with the Protein Data Bank for the atomic coordinates of 43-kDa ParE with ADPNP and that of 24-kDa ParE in complex with novobiocin have been assigned as 1S16 and 1S14, respectively.
RESULTS AND DISCUSSION
E. coli N-terminal 43-kDa ParE domain is a dimer in the presence of ADPNP.
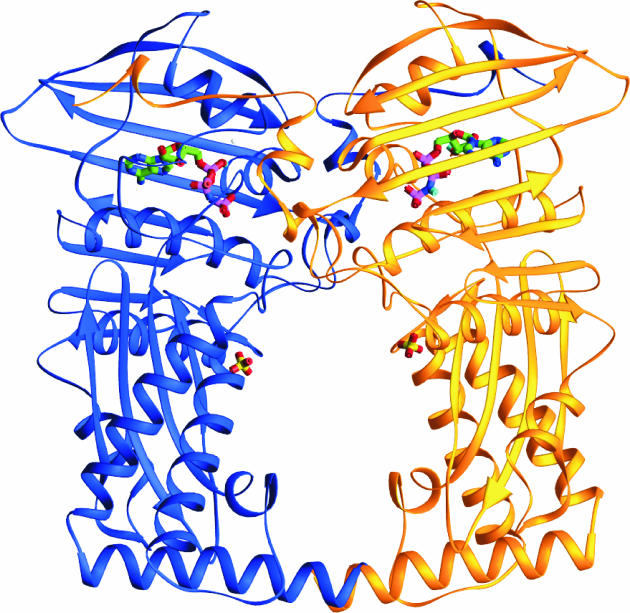
The crystal structure of the E. coli ParE complexed with ADPNP was solved to 2.1-Å resolution. The N-terminal 43-kDa domain of E. coli ParE (residues 1 to 390) contained the ATP binding site. Amino acid residues 4 to 383 were ordered in the 43-kDa ParE crystal structure. This structure, shown in Fig. 1, consisted of a dimer of the 43-kDa fragment, with one ADPNP molecule bound per monomer. As with E. coli GyrB, each ParE monomer has two distinct subdomains: an N-terminal subdomain (residues 1 to 217) comprising an eight-stranded β-sheet and five α-helices, and a C-terminal subdomain (residues 218 to 390) comprising a four-stranded β-sheet and four α-helices. The dimer is stabilized by an N-terminal arm (residues 1 to 15) that wraps around the other monomer of the dimer (Fig. 1), and, as in gyrase, the conserved Tyr5 residue interacts with ATP (Fig. 3). At the C-terminal subdomain, the contacts are less extensive; however, this protein has been truncated, and as result, the contacts may differ in the full-length protein. The two C-terminal subdomains of the dimer form a 20-Å-diameter cavity that is surrounded by long α-helices that are present at the C terminus of each monomer (Fig. 1). As in gyrase, the cavity is lined with arginine residues and is thought to be involved in the transport of the T-segment DNA to the DNA gate that is located at the heterotetrameric interface.
FIG. 1.
E. coli N-terminal 43-kDa ParE dimer cocomplexed with ADPNP. One monomer is shown in blue, and the other is in yellow. The two bound ADPNP molecules are depicted in stick representation, as are the two crystallographic sulfate anions, where green is carbon, blue is nitrogen, red is oxygen, pink is phosphate, and yellow is sulfur. This figure was produced with the Ribbons program (4).
FIG. 3.
Stereo view of selected amino acid side chains of the E. coli ParE ATP binding site. The ParE side chains shown and labeled and the corresponding E. coli GyrB residues (in parentheses) are Y5 (Y5), E38 (E42), N42 (N46), E46 (E50), D69 (D73), M74 (I78), K99 (K103), Y105 (Y109), T163 (T165), Q332 (Q335), and K334 (K337). The E. coli GyrB ADPNP-bound ligand is colored gray.
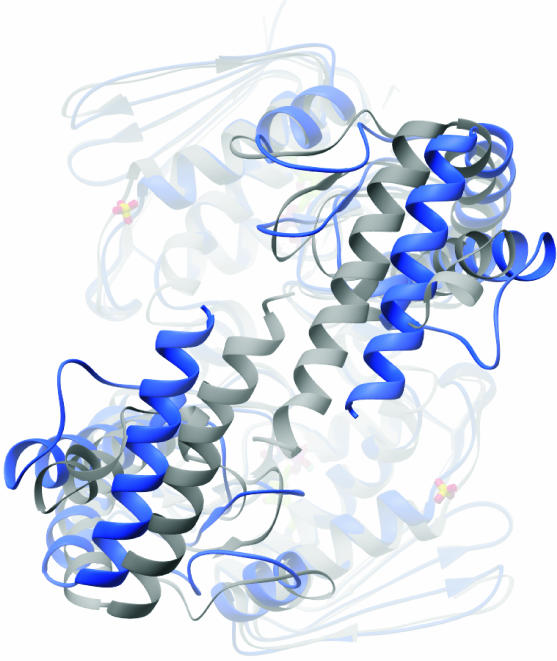
The 43-kDa E. coli ParE structure overall is remarkably similar to the 43-kDa E. coli GyrB structure. The largest difference between the two structures occurs in the long α-helices that are present at the C terminus of each monomer. In gyrase, the long α-helices from each monomer stack themselves in close proximity (shown in gray in Fig. 2), whereas in the ParE dimer these long α-helices display greater openness than in the GyrB structure (colored blue in Fig. 2). The distance between the Cα carbons of the last ordered residue A383 and its equivalent (A383′) in the ParE dimer is 22.8 Å, whereas for the two equivalent structural residues (R386-R386′) in GyrB the distance between the Cα carbons is 9.8 Å. In ParE these structural differences begin at the D217 residue and proceed to the end of the protein. In gyrase, the equivalent residue (G220) is a glycine, which could explain the shift in the α-helices. Whether α-helical openness has relevance for TopoIV function is not known; it could be an artifact of crystal formation, or the extent of openness may be different in full-length ParE.
FIG. 2.
Greater openness of ParE C-terminal dimer interface in TopoIV. The superimposition of the GyrB (grey) and ParE (blue) structures is shown.
The structural resemblance of ParE to GyrB places both proteins into the GHKL phosphotransferase superfamily (10). Members of the GHKL superfamily have a similar three-dimensional arrangement of secondary structural elements that form the ATP binding site (15). Family members include GyrB, bacterial MutL and mammalian MLH and PMS (mismatch repair proteins), human Hsp90, and bacterial (CheA) and mitochondrial (BCK) protein kinases. While the proteins are similar at the ATP binding site, they also have significant structural and activity differences that allow a diverse set of biological functions to be performed. The Hsp90 and MutL proteins have very poor rates of ATP hydrolysis (kcat ≈ 0.1 to 0.5 min−1) (1, 28) compared to GyrB and ParE (kcat = 72 to 240 min−1) (13), suggesting that structural similarities between these proteins are not sufficient to retain similar ATP utilization rates. Hsp90 and MutL may utilize ATP better in higher-order protein complexes; these individual proteins have diverged from the ability to hydrolyze ATP as efficiently as the bacterial type II topoisomerases. Additionally, Hsp90 has been reported to bind to novobiocin (24, 25, 39); however, the novobiocin binding site appears to be present in the C-terminal ATP binding domain of Hsp90 and not in the N-terminal ATP binding domain, whose solved structure (36) resembles the members of the superfamily. Furthermore, the binding affinity of novobiocin for Hsp90 and MutL is significantly weaker (i.e., in the millimolar range) (1, 25) than it is for TopoIV and DNA gyrase. Despite the ATP binding site similarities, the differences in inhibitor potencies and rates of ATPase activity argue that very significant structural differences remain among the GHKL protein members.
ATP active-site comparison of the E. coli ParE and GryB 43-kDa structures.
The ParE and GyrB ATP binding sites are strikingly similar in that all essential amino acid side chains (13) overlap each other when the two ADPNP structures are superimposed. Selected ParE ATP active-site residues Y5, E38, N42, E46, D69, M74, K99, Y105, T163, Q332, and K334 are shown in Fig. 3. The superimposition of the ADPNP ligands from the 43-kDa structures of E. coli GyrB (colored grey, Fig. 3) and E. coli ParE (colored, Fig. 3) illustrates that the ligands bind in a very similar conformation. The striking similarity of the structures was not obvious from the protein sequence alignments of the E. coli 43-kDa domains, as they have only 35% identity and 55% similarity to each other.
Recently, we completed an extensive structure-function relationship study of the residues in the E. coli GyrB ATP active site (13). This study, together with those of others (2, 3, 7, 16, 30, 38), provides an understanding of the functional role and the essentiality of each side chain in the GyrB active site. The remarkable similarity in the active site suggests that the specific roles of the residues identified in gyrase are transferable to the TopoIV enzyme. Our GyrB data (13) suggest that two ParE residues, P75 (Fig. 4A) and K99 (Fig. 3), may be required in coupling ATP hydrolysis to DNA relaxation in TopoIV (an ATP-dependent activity). The P75 side chain is ≈4.7 Å from the N-3 adenine ring, whereas the K99 side chain forms a salt bridge with the β-phosphate of ADPNP. As suggested for gyrase, the ParE K99 side chain may act as a sensor for the detection of when ATP hydrolysis has occurred. However, for some residues, the relative contribution to ATP binding and/or hydrolysis activity may be altered by subtle differences in ATP binding or in overall ParE structure.
FIG. 4.
Novobiocin binding site in the 24-kDa E. coli ParE structure. (A) Stereo view of selected amino acid side chains of the E. coli ParE novobiocin binding site. The ParE side chains shown and labeled and the corresponding E. coli GyrB residues (in parentheses) are E46 (E50), D69 (D73), R72 (R76), M74 (I78), D77 (G81), I90 (I94), R132 (R136), and T163 (T165). The E. coli GyrB novobiocin-bound ligand is colored gray. (B) Fo-Fc omit map of novobiocin contoured at the three-sigma level. Apparent in panel B are the interactions that novobiocin has with side chains E46 (3.7 Å), D77 (3.1 and 2.7 Å), M74 (3.5 Å), R72 (3.3 Å), I90 (3.8 Å), and the backbone carbonyl N42 (2.7 Å).
Novobiocin binding site comparison of the E. coli ParE and GyrB 24-kDa structures.
In gyrase, novobiocin is an ATP-competitive inhibitor but only partially overlaps the ATP ligand binding site (13, 22). Novobiocin is also an inhibitor of E. coli TopoIV activity (33). We solved the structure of the N-terminal 24-kDa domain of E. coli ParE (residues 1 to 217) that comprises most of the ATP/novobiocin binding site to beyond 2.0-Å resolution. Amino acid residues 27 to 217 and 17 to 217 are ordered in the first and second molecules of the asymmetric unit of the 24-kDa ParE structure. Nearly all amino acid side chains (not shown) overlap each other when the two E. coli novobiocin structures are superimposed. Selected ParE novobiocin binding site residues (E46, D69, R72, M74, D77, I90, R132, and T163) are shown in Fig. 4A. The superimposition of the novobiocin ligand from the 24-kDa structures of E. coli GyrB (colored grey, Fig. 4A) and E. coli ParE (colored, Fig. 4A) illustrate that the novobiocin binds in a very similar conformation in the two proteins. The electron density of the novobiocin ligand is shown in Fig. 4B. Apparent in Fig. 4B are the interactions that novobiocin has with the following side chains: E46 (3.7 Å), D77 (3.1 and 2.7 Å), M74 (3.5 Å), R72 (3.3 Å), I90 (3.8 Å), and the backbone carbonyl N42 (2.7 Å). Examination of all the residues surrounding novobiocin in the ParE and GyrB structures reveals two ParE residues (M74 and D77) (Fig. 4) that are different in the GyrB structure (I78 and G81, respectively). The significance of these differences was examined in both the TopoIV and the DNA gyrase enzymes with a mutational approach.
Single residue dictates differences in novobiocin potency between DNA gyrase and topoisomerase IV.
In the literature (12, 14, 34, 40) and under our own assay conditions, the TopoIV novobiocin IC50 is always greater than the DNA gyrase novobiocin IC50 (Table 1). The ≈5-fold difference suggested that a structural difference exists between the TopoIV and the gyrase ATPase active sites. Comparison of the ParE and GyrB structures and sequences revealed three possible differences that may affect the IC50. The first difference is that ParE has a methionine residue at position 74. In GyrB, the equivalent residue is isoleucine (I78). The second distinction in the ParE structure is the presence of an aspartic acid at position 77 that forms a solvent-exposed hydrogen bond (i.e., weak interaction) with the hydroxyl benzoate group (2.7 and 3.1 Å) of novobiocin (Fig. 4B). In contrast, GyrB equivalent residue G81 cannot form a hydrogen bond with novobiocin. Intuitively, the hydrogen bond interaction in the ParE-novobiocin structure might work against a greater IC50 for TopoIV. However, the opposite effect cannot be discounted, and therefore the residue was examined. The third distinction not seen in the E. coli X-ray structures but evident from sequence alignments of the flexible-loop region suggested that other amino acid differences between GyrB and ParE proteins may account for the IC50 discrepancy. We examined each difference experimentally via mutagenesis and assayed the purified full-length heterotetrameric TopoIV and DNA gyrase to determine their effects on the novobiocin IC50.
The novobiocin potency difference is most easily explained by the change from isoleucine to methionine in GyrB (I78) versus ParE (M74). X-ray analysis of the gyrase-novobiocin complex places the I78 side chain in proximity to the oxygen of the methoxy group on the novobiose sugar (the I78 γ1-carbon is ≈3.5 Å from the oxygen) and the isopentenyl group (the I78 γ2-carbon is ≈4.4 Å from the distal carbon of the isopentenyl) as it wraps back in the direction of the sugar (22). In the ParE-novobiocin complex, the M74 side chain is located in a nearly identical position (the γ-carbon on the M74 side chain is ≈3.7 Å from the oxygen of the methoxy group) (Fig. 4). However, in ParE the M74 side chain lacked the γ2-carbon interaction that is seen with the GyrB I78 side chain, suggesting a reason for the potency difference.
In our previous GyrB mutagenesis work, when the GyrB I78 residue was changed to alanine or leucine, the result was an increase in the novobiocin IC50 by factors of 30 and 14, respectively (Table 1) (13). In contrast, when valine, a more conserved substitution, is introduced, only a twofold increase in IC50 is found (Table 1), suggesting that a branching of the hydrophobic side chain at the β-carbon is important for novobiocin affinity. From these results, an I78M mutation in GyrB would also be expected to increase the novobiocin IC50, and indeed it did increase by 22-fold (Table 1). The I78M mutation also resulted in a fivefold increase in the ATP Km for GyrB enzyme (Table 1) and was primarily responsible for the lower V/K value (Table 1), although a ≈30% diminution in the ATPase rate was determined. Previously, we have shown that the E. coli GyrB I78 side chain is not essential for bacterial growth (13), and therefore, changes in this side chain may lead to target-based resistance in bacteria that are novobiocin susceptible (e.g., S. aureus).
We then performed the converse experiment by changing the M74 residue of the ParE protein to isoleucine. We found a corresponding decrease in the novobiocin IC50 from 210 nM for the wild type to 12 nM for the M74I enzyme (Table 1). The IC50 ratio (mutant IC50/wild-type IC50) was 0.06, reflecting that the M74I enzyme is ≈18 times more sensitive to novobiocin than the wild-type enzyme (Table 1). The magnitude of the decrease in IC50 for the ParE M74I mutation is comparable to the increase in IC50 for the GyrB I78M mutation and suggests that the side chains have similar effects on the ability of both proteins to bind novobiocin. In contrast to the GyrB I78M mutation, the ParE M74I mutation resulted in a lower ATP Km than a methionine side chain (Table 1). The ParE M74I mutation had a V/K value that was an order of magnitude greater than the wild-type value (Table 1), primarily the result of a lower ATP Km, although a nearly twofold increase in the rate of ATP hydrolysis (Vmax) was also evident in the enzyme.
Interestingly, amino acid alignments of more than 30 ParE and GyrB proteins showed that isoleucine and methionine are the only two amino acids represented at this position (not shown). The alignments also showed a trend for GyrB proteins to have an isoleucine whereas most ParE proteins frequently have a methionine; the significance is not clear. Perhaps the nearly order of magnitude increase in TopoIV ATPase catalytic efficiency as result of a methionine-to-isoleucine change is not necessarily desired in vivo. Here we have provided evidence that in TopoIV and DNA gyrase, a single amino acid residue (GyrB I78 and TopoIV M74) is responsible for novobiocin potency differences.
Although the difference in novobiocin affinity between gyrase and TopoIV is explained by the isoleucine-to-methionine change, we were still interested in determining whether the glycine-to-aspartic acid change or flexible-loop sequence differences affected the IC50. For the GyrB G81D substitution, only a twofold increase in the IC50 was found (Table 1); from this we infer that the D77 residue in ParE has a minimal effect on the IC50.
The gyrase flexible-loop region spans amino acid residues 97 to 120 and is strongly conserved at the N-terminal and C-terminal ends; however, a seven-amino-acid stretch (D105 to V111) in the center lacks sequence conservation. For the gyrase flexible-loop mutations, E. coli gyrase B residues D105 to V111 (DDNSYKV) were replaced with the E. coli ParE (SNKNYQF) or S. aureus GrlB (ParE homologue) (GQGGYKT) sequence. Neither S. aureus nor E. coli ParE loop mutations in E. coli GyrB had any dramatic effect on enzyme activity or IC50 (Table 1), suggesting that these nonconserved side chains lack a significant role in novobiocin affinity. The T. thermophilus 43-kDa GyrB-novobiocin structure, having an observable flexible loop, positions only four side chains within ≈5 Å of the bound novobiocin (18). These residues in the E. coli GyrB numbering are K103, F104, K110, and V118. Only E. coli GyrB K110 and E. coli ParE Q106 (position-equivalent residues) are not completely conserved in the prokaryotic type II topoisomerases. In the T. thermophilus structure (18), the GyrB K109 amine is ≈3.8 Å from the ring oxygen and the number one carbon in novobiose sugar and is also ≈4.2 Å from the hydroxyl on the novobiose sugar. We assayed the E. coli GyrB K110A enzyme (13) and found that the IC50 did not change (not shown). These mutagenesis data demonstrate that beyond the I78 and M74 residues, no other nonconserved active-site side chain has a significant effect on novobiocin affinity. The remaining fourfold IC50 disparity between the wild-type GyrB protein (46 nM) and TopoIV M74I (12 nM) protein (Table 1) may be from residue differences beyond the active site that impart subtle changes to the novobiocin binding residues.
In summary, our data demonstrate that a methionine in E. coli ParE at position 74 or in E. coli GyrB at position 78 is detrimental to novobiocin affinity in bacterial type II topoisomerases. We have shown that isoleucine in place of methionine in either of the two E. coli type II enzymes results in a ≈18- to 22-fold decrease in novobiocin IC50. The sensitivity of novobiocin potency to single-amino-acid substitutions in both targets suggests that they are independently vulnerable to resistance mutations. Therefore, targeting both type II topoisomerases will yield more durable therapies. Finally, we believe that the ParE structures and the mutagenesis results presented provide a better understanding of ligand binding in the prokaryotic type II topoisomerases, and the addition of these structures to the gyrase structures will assist in the prospective design of novel inhibitors of gyrase and TopoIV as potent antibacterial agents with the potential for low rates of resistance.
Acknowledgments
We thank Scott A. Raybuck, Trudy H. Grossman, Nagraj Mani, Dean Stamos, Subramania Pazhanisamy, Eric Olson, Jonathan Moore, Michael Partridge, and John Thomson for discussions and comments on the manuscript. A special thank you to Matthew Fitzgibbon for technical assistance.
REFERENCES
- 1.Ban, C., and W. Yang. 1998. Crystal structure and ATPase activity of MutL: Implications for DNA repair and mutagenesis. Cell 95:541-552. [DOI] [PubMed] [Google Scholar]
- 2.Brino, L., C. Bronner, P. Oudet, and M. Mousli. 1999. Isoleucine 10 is essential for DNA gyrase B function in Escherichia coli. Biochimie 81:973-980. [DOI] [PubMed] [Google Scholar]
- 3.Brino, L., A. Urzhumtsev, M. Mousli, C. Bronner, A. Mitschler, P. Oudet, and D. Moras. 2000. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 275:9468-9475. [DOI] [PubMed] [Google Scholar]
- 4.Carson, M. 1991. Ribbons 2.0. J. Appl. Crystallogr. 24:958-961. [Google Scholar]
- 5.Chambers, S. P. 2002. High-throughput protein expression for the post-genomic era. Drug Discov. Today 7:759-765. [DOI] [PubMed] [Google Scholar]
- 6.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 7.Contreras, A., and A. Maxwell. 1992. GyrB mutations which confer coumarin resistance also affect DNA supercoiling and ATP hydrolysis by Escherichia coli DNA gyrase. Mol. Microbiol. 6:1617-1624. [DOI] [PubMed] [Google Scholar]
- 8.Crisona, N. J., T. R. Strick, D. Bensimon, V. Croquette, and N. R. Cozzarelli. 2000. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 14:2881-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drlica, K. 2001. Antibiotic resistance: can we beat the bugs? Drug Discov. Today 6:714-715. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, R., and M. Inouye. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25:24-28. [DOI] [PubMed] [Google Scholar]
- 11.Fersht, A. 1985. Enzyme structure and mechanism, 2nd ed., p. 99. W. H. Freeman and Company, New York, N.Y.
- 12.Gellert, M., M. H. O'Dea, T. Itoh, and J. Tomizawa. 1976. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. USA 73:4474-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, C. H., J. D. Parsons, T. H. Grossman, P. S. Charifson, S. Bellon, J. Jernee, M. Dwyer, S. P. Chambers, W. Markland, M. Botfield, and S. A. Raybuck. 2003. Active-site residues of Escherichia coli DNA gyrase required in coupling ATP hydrolysis to DNA supercoiling and amino acid substitutions leading to novobiocin resistance. Antimicrob. Agents Chemother. 47:1037-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins, N. P., C. L. Peebles, A. Sugino, and N. R. Cozzarelli. 1978. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc. Natl. Acad. Sci. USA 75:1773-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, X., M. Machius, and W. Yang. 2003. Monovalent cation dependence and preference of GHKL ATPases and kinases. FEBS Lett. 544:268-273. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, A. P., and A. Maxwell. 1993. Identifying the catalytic residue of the ATPase reaction of DNA gyrase. Proc. Natl. Acad. Sci. USA 90:11232-11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khodursky, A. B., E. L. Zechiedrich, and N. R. Cozzarelli. 1995. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:11801-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamour, V., L. Hoermann, J. M. Jeltsch, P. Oudet, and D. Moras. 2002. An open conformation of the Thermus thermophilus gyrase B ATP-binding domain. J. Biol. Chem. 277:18947-18953. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. Procheck: a program to check the stereo chemical quality of protein structure. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 20.Lee, M. P., M. Sander, and T. Hsieh. 1989. Nuclease protection by Drosophila DNA topoisomerase II. J. Biol. Chem. 264:21779-21787. [PubMed] [Google Scholar]
- 21.Leslie, A. G. W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. News Lett. Protein Crystallogr. 26:73-80. [Google Scholar]
- 22.Lewis, R. J., O. M. P. Singh, C. V. Smith, T. Skarzynski, A. Maxwell, A. J. Wonacott, and D. B. Wigley. 1996. The nature of inhibition of DNA gyrase by the coumarins and cyclothialidine revealed by X-ray crystallography. EMBO J. 15:1412-1420. [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, L. F., and J. C. Wang. 1978. Micrococcus luteus DNA gyrase: active components and a model for its supercoiling of DNA. Proc. Natl. Acad. Sci. USA 75:2098-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcu, M. G., T. W. Schulte, and L. Neckers. 2000. Novobiocin and related coumarins and depletion of heat shock protein 90-dependent signaling proteins. J. Natl. Cancer Inst. 92:242-248. [DOI] [PubMed] [Google Scholar]
- 25.Marcu, M. G., A. Chadli, I. Bouhouche, M. Catelli, and L. M. Neckers. 2000. The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J. Biol. Chem. 275:37181-37186. [DOI] [PubMed] [Google Scholar]
- 26.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33:491-497. [DOI] [PubMed] [Google Scholar]
- 27.Maxwell, A., and D. M. Lawson. 2003. The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr. Top. Med. Chem. 3:283-303. [DOI] [PubMed] [Google Scholar]
- 28.McLaughlin, S. H., H. W. Smith, and S. E. Jackson. 2002. Stimulation of the weak ATPase activity of human Hsp90 by a client protein. J. Mol. Biol. 315:787-798. [DOI] [PubMed] [Google Scholar]
- 29.Navaza, J. 1994. AmoRe: an automated package for molecular replacement Acta Crystallogr. A 50:157-163.
- 30.O'Dea, M. H., J. K. Tamura, and M. Gellert. 1996. Mutations in the B subunit of Escherichia coli DNA gyrase that affect ATP-dependent reactions. J. Biol. Chem. 271:9723-9729. [DOI] [PubMed] [Google Scholar]
- 31.Orphanides, G., and A. Maxwell. 1994. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 22:1567-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 33.Pan X. S., and L. M. Fisher. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng, H., and K. J. Marians. 1993. Escherichia coli topoisomerase IV purification, characterization, subunit structure and subunit interactions. J. Biol. Chem. 268:24481-24490. [PubMed] [Google Scholar]
- 35.Peng, H., and K. J. Marians. 1995. The interactions of Escherichia coli topoisomerase IV with DNA. J. Biol. Chem. 270:25286-25290. [DOI] [PubMed] [Google Scholar]
- 36.Prodromou, C., S. M. Roe, R. O'Brien, J. E. Ladbury, P. W. Piper, and L. H. Pearl. 1997. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90:65-75. [DOI] [PubMed] [Google Scholar]
- 37.Reece, R. J., and A. Maxwell. 1991. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 19:1399-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, C. E., and A. Maxwell. 1998. Identification of a residue involved in transition-state stabilization in the ATPase reaction of DNA gyrase. Biochemistry 37:9658-9667. [DOI] [PubMed] [Google Scholar]
- 39.Soti, C., A. Racz, and P. Csermely. 2002. A nucleotide-dependent molecular switch controls ATP binding at the C-terminal domain of Hsp90. J. Biol. Chem. 277:7066-7075. [DOI] [PubMed] [Google Scholar]
- 40.Sugino, A., P. Higgins, P. O. Brown, C. L. Peebles, and N. R. Cozzarelli. 1978. Energy coupling in DNA gyrase and mechanism of action of novobiocin. Proc. Natl. Acad. Sci. USA 75:4838-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takei, M., H. Fukuda, R. Kishii, and M. Hosaka. 2001. Target preference of 15 quinolones against Staphylococcus aureus, based on antibacterial activities and target inhibition. Antimicrob. Agents Chemother. 45:3544-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai, F. T. F., O. M. P. Singh, T. Skarzynski, A. J. Wonacott, S. Weston, A. Tucker, R. A. Pauptit, A. L. Breeze, J. P. Poyser, R. O'Brien, J. E. Ladbury, and D. B. Wigley. 1997. The high-resolution crystal structure of a 24-kDa gyrase B fragment from E. coli complexed with one of the most potent coumarin inhibitors, clorobiocin. Proteins 28:41-52. [PubMed] [Google Scholar]
- 43.Ullsperger, C., and N. R. Cozzarelli. 1996. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 271:31549-31555. [DOI] [PubMed] [Google Scholar]
- 44.Wigley, D. B., G. J. Davies, E. J. Dodson, A. Maxwell, and G. Dudson. 1991. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351:624-629. [DOI] [PubMed] [Google Scholar]