Abstract
Extensive use of β-lactam antibiotics has led to the selection of pathogenic streptococci resistant to β-lactams due to modifications of the penicillin-binding proteins (PBPs). PBP2b from Streptococcus pneumoniae is a monofunctional (class B) high-molecular-weight PBP catalyzing the transpeptidation between adjacent stem peptides of peptidoglycan. The transpeptidase domain of PBP2b isolated from seven clinical resistant (CR) strains contains 7 to 44 amino acid changes over the sequence of PBP2b from the R6 β-lactam-sensitive strain. We show that the extracellular soluble domains of recombinant PBP2b proteins (PBP2b*) originating from these CR strains have an in vitro affinity for penicillin G that is reduced by up to 99% from that of the R6 strain. The Thr446Ala mutation is always observed in CR strains and is close to the key conserved motif (S443SN). The Thr446Ala mutation in R6 PBP2b* displays a 60% reduction in penicillin G affinity in vitro compared to that for the wild-type protein. A recombinant R6 strain expressing the R6 PBP2b Thr446Ala mutation is twofold less sensitive to piperacillin than the parental S. pneumoniae strain. Analysis of the Thr446Ala mutation in the context of the PBP2b CR sequences revealed that its influence depends upon the presence of other unidentified mutations.
The high-molecular-weight (high-Mr) penicillin-binding proteins (PBPs) carry out the extracellular steps of peptidoglycan synthesis. These enzymes catalyze the glycosyltransferase reaction leading to the polymerization of the glycan chains and the transpeptidation (TP) activity establishing a covalent bond between two juxtaposed peptides (3, 15, 32). Streptococcus pneumoniae class A high-Mr PBPs (PBP1a, PBP1b, and PBP2a) are bifunctional enzymes bearing both activities, whereas class B high-Mr PBPs (PBP2b and PBP2x) have so far only been associated with the TP activity (15). Due to their structural similarity to the natural substrates, the d-Ala-d-Ala stem peptides, β-lactam molecules inhibit the TP reaction leading to bacterial cell lysis (35).
S. pneumoniae is a major human pathogen and the causative agent of ear infections in children, as well as meningitis and pneumonia. Extensive use of β-lactam antibiotics over the last 4 decades has led to the selection of pathogenic streptococci resistant to their action. The resistance to β-lactams in S. pneumoniae results from a decreased affinity of PBPs for the antibiotics. This phenotype results from genetic reshuffling. Mosaic genes encoding PBPs with lower affinity for β-lactams have been generated by homologous recombination events with other streptococcal species (18). The mosaicity of the pneumococcal pbp2b gene has been well established in penicillin clinical resistant (CR) isolates as resulting from recombination events between S. pneumoniae and Streptococcus mitis (8-10). DNA sequence analysis has produced evidence that, under clinical and laboratory conditions, mutants of the pbp2x and pbp2b genes are the first to be selected upon treatment with cefotaxime and piperacillin, respectively, indicating that PBP2x and PBP2b are primary resistance determinants for these antibiotics (22, 29).
To characterize the biochemical properties of PBP2b, we have expressed the periplasmic domain of PBP2b from the S. pneumoniae R6 sensitive strain (PBP2b*R6) and from seven CR isolates (PBP2b*CR). These latter proteins show a marked reduction in affinity for penicillin G and, when recombined in the R6 strain, significantly increase the MIC of piperacillin for the host strain. The preponderance of the Thr446Ala mutation in clinical strains, as well as its juxtaposition to the key motif (S443SN) of the PBP TP function, led us to investigate the role of this mutation in more detail. The Thr446Ala mutation lowers the affinity for β-lactams and may thus play a role in the development of antibiotic resistance.
MATERIALS AND METHODS
Pneumococcal strains.
Seven clinical pneumococcal strains were isolated in the Bacteriology Laboratory of the Centre Hospitalier Universitaire de Grenoble (Grenoble, France): one penicillin-sensitive strain for which the penicillin G MIC was ≤0.06 μg/ml, two intermediate sensitive strains for which the penicillin G MICs were between 0.12 and 1 μg/ml, and four resistant strains for which the penicillin G MICs were >1 μg/ml. The R6 strain of S. pneumoniae (MIC of penicillin G, <0.016 μg/ml) was used as the penicillin-susceptible reference strain. S. pneumoniae strains were grown at 37°C in an atmosphere of 95% air-5% CO2 on Columbia blood agar plates (bioMérieux). Clones were isolated, and overnight cultures were stored at −80°C.
Genomic DNA isolation from S. pneumoniae strains.
The same protocol was used for the unencapsulated R6 strain of S. pneumoniae and the pneumococcal clinical isolates. The various strains were anaerobically propagated in glucose-buffered broth (Bio-Rad) for 16 h at 30°C. Genomic DNA was extracted from 5 ml of culture with the a High Pure PCR template preparation kit (Roche) following the manufacturer's instructions; 100 ng of genomic DNA was recovered in 200 μl of water and was used as a template for PCR amplification of the pbp2b genes.
Sequencing of pbp2b genes.
A 1,360-kb segment of pbp2b, corresponding to the TP domain, was amplified using Vent polymerase (New England Biolabs). The upstream primer was 5′-823ACCTTACAAGGAAAACGCTCGG844-3′, and the downstream primer, which annealed to the 3′ noncoding regions of pbp2b genes, was 5′-CATCCCAATCGTATAAAAGGCC-3′. The PCR products were directly sequenced by Genome Express (Grenoble, France).
Cloning of the pbp2b* gene from the S. pneumoniae R6 strain.
Genomic DNA from the R6 strain of S. pneumoniae was used as a template to amplify the complete pbp2b gene, i.e., containing the cytoplasmic portion, the transmembrane anchor, and the periplasmic domain. Primers were designed for the PCR amplification: 5′-3CGCATATGAGAAAATTTAACAGCCATTC23-3′ and 5′-2041CGCTCGAGGTTCATTGGATGGTATTTTTG2021-3′, which contained the restriction sites NdeI and XhoI (underlined), respectively. Amplification by PCR was performed using the following reaction mix:1 μM each oligonucleotide, 1 μM dNTP, 60 μg of DNA template/ml, and 6 mM MgCl2. Reactions used 4 U of Vent DNA polymerase (New England Biolabs) for 25 cycles as follows: 1 min at 94°C, 1 min at 60°C, and 2.5 min at 72°C. A PCR product of 2 kb was produced, corresponding to the expected size of the pbp2b gene (2,044 bp). A step subcloning the pbp2b gene into the pCR-Script vector (Stratagene) was performed by following the manufacturer's instructions. The pbp2b gene was subcloned into a pET30b+ vector (Novagen) to give the construct pET30b/pbp2bR6. This construct encodes the complete membrane-associated form of PBP2b with a C-terminally linked His6 tag. This construct was entirely sequenced (Genome Express) to confirm that no mutations had been introduced during PCR.
From the construct pET30b/pbp2bR6, site-directed mutagenesis was performed to remove the first 34 amino acids, which correspond to the N-terminal membrane anchor of PBP2b and the short cytoplasmic region. The resulting construct, pET30b/pbp2b*R6, allows the expression of the periplasmic form of the PBP2b protein (PBP2b*) C-terminally fused to a His6 tag.
For the construct pET30b/pbp2b*R6, site-directed mutagenesis at position 446 was performed to mutate the codon Thr into Ala with a Quick Change site-directed mutagenesis kit (Stratagene), leading to the construct pET30b/pbp2b*R6Thr446Ala.
Cloning of pbp2b* genes from S. pneumoniae clinical isolates.
Genomic DNA from each clinical isolate of S. pneumoniae was used as a template to amplify the periplasmic form of the pbp2b gene (pbp2b*), i.e., deleted from the cytoplasmic portion and the transmembrane anchor. The upstream (5′-103CGCGGATCCCAGGTTTTGAACAAGGATTTTTACGAAAAAAAGCTA138-3′) and downstream (5′-2041CCGCTCGAGAGCATAATTTCCTTTCTAATTCATTGGATGGTATTTTTG2080-3′) primers had BamHI and XhoI sites (underlined), respectively. PCR products, digested with BamHI and XhoI, were cloned into pGEX-4T1 (Amersham Biosciences). All cloned pbp2b* genes were sequenced by Genome Express, and the constructs were called pGEX-4T1/pbp2b*5023, pGEX-4T1/pbp2b*5259, pGEX-4T1/pbp2b*4935, pGEX-4T1/pbp2b*5245, pGEX-4T1/pbp2b*4790, pGEX-4T1/pbp2b*5268, pGEX-4T1/pbp2b*5204, and in a more general way, pGEX-4T1/pbp2b*CR.
Homologous recombination of pbp2b* genes in the S. pneumoniae R6 strain.
R6 strain cells were made competent with a C-medium culture supplemented with 0.18% albumin at 8%. Upon achieving a culture with an optical density at 620 nm (OD620) of 0.15, 1-ml aliquots were added to 20% glycerol and frozen at −80°C. Competent cells were diluted 1:10 in C-medium supplemented with 0.18% albumin at 8% for a final volume of 1 ml. Fifty nanograms of plasmid DNA carrying native or mutated pbp2b* genes was added for 30 min at 30°C. After a subsequent incubation for 2 h at 37°C, 100 μl of cell suspension aliquots were deposited on an empty petri dish overlaid with 20 ml of blood agar gelose (Columbia blood agar base EH; DIFCO) containing 4% horse blood (Eurobio) and the antibiotics at various concentrations. After incubation at 37°C for 18 h, three clones were isolated from each transformation, and the MICs of penicillin G and piperacillin were measured. Homologous recombination was verified for one selected clone from each transformation by sequencing the periplasmic region of the pbp2b gene amplified by PCR.
MIC testing.
For each recombinant strain, the MIC of penicillin G and piperacillin was determined using the Etest method. After overnight growth, Mueller-Hinton plates were inoculated, and penicillin G or piperacillin (0.016 to 256 μg/ml) Etest strips (AB Biodisk) were laid. Plates were incubated for 18 to 24 h before the MICs were read where the ellipse of growth inhibition intersects the strip. MICs were measured independently between three and six times. When different discrete values were measured, the results were expressed as a range of the maximal and minimal values.
Expression and purification of the different forms of PBP2b* proteins. (i) Purification of PBP2b*R6 and PBP2b*R6Thr446Ala.
An overnight culture of an Escherichia coli BL21(DE3) expression strain transformed with plasmid pET30b/pbp2b*R6 was used to inoculate (1:50) 1 liter of Lennox-LB broth (LB) medium supplemented with 30 μg of kanamycin/ml. Upon achieving a culture with an OD600 of 1 in LB medium at 37°C, protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) while incubating for 16 h at 15°C. Purification steps were all performed either at 4°C or on ice. Cells were spun at 6,000 × g for 15 min, and the pellet was resuspended in 50 mM Tris-HCl (pH 8.0)-200 mM NaCL-50 mM imidazole containing 1 μg of aprotinin/ml, 1 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride. Cell lysis was achieved by sonication, and the soluble fraction was obtained by centrifugation at 39,200 × g for 20 min. The supernatant was loaded at a flow rate of 1 ml/min onto a 10-ml chelating Sepharose column (Amersham Biosciences) equilibrated in buffer A (50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 50 mM imidazole). The resin was washed with 10 column volumes of buffer A. Steps of 30 ml of increasing concentrations of imidazole (115 mM, 130 mM, and 175 mM) were used to elute PBP2b*. Fractions containing PBP2b* (115 mM imidazole elution step) were pooled and loaded onto a Sephadex G75 gel filtration column (Amersham Biosciences) previously equilibrated in 50 mM Tris-HCl (pH 8.0)-200 mM NaCl at a flow rate of 0.7 ml/min.
(ii) Purification of PBP2b*CR.
An overnight culture of an E. coli DH5α strain transformed with the different pGEX-4T1/pbp2b*CR plasmids was used to inoculate (1:50) 1 liter of LB medium supplemented with 100 μg of ampicillin/ml. Upon achieving a culture with an OD600 of 1 in LB medium at 37°C, protein expression was induced with 1 mM IPTG while incubating for 16 h at 15°C. Cells lysis was performed by sonication in buffer B (Tris-HCl [pH 8.0], 200 mM NaCl, 1 mM EDTA). The supernatant was loaded at a flow rate of 1 ml/min onto a 7-ml glutathione Sepharose column (Amersham Biosciences). The fusion proteins glutathione S-transferase (GST)-PBP2b* were eluted with 10 mM reduced glutathione (Roche) in buffer B and extensively dialyzed against buffer B. The fusion proteins were cleaved in the presence of 100 U of thrombin (Sigma) for 1 h 30 min at room temperature. Thrombin was inhibited by 1 mM PMSF, the protein solution was loaded onto the glutathione Sepharose column in order to retain GST, and the PBP2b*CR proteins were collected in the subsequent flow.
β-lactam binding assays.
The titration of PBP was performed using [3H]benzylpenicillin (20 Ci/mmol, 1 mCi/ml; Amersham Biosciences). Purified PBP2b*R6 (1 μM) in 50 mM Tris-HCl (pH 8.0)-200 mM NaCl was incubated at 37°C with various concentrations of [3H]benzylpenicillin for 15 min. The samples were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and [3H]benzylpenicillin bound to proteins was monitored. The gel was stained with Coomassie blue, destained, incubated with Amplifya (Amersham Biosciences), dried, and either exposed to film for 16 h or cut around the protein bands. The radioactivity of the gel slice was measured using a liquid scintillation analyzer (Packard model 2100TR) after mixing with 5 ml of liquid scintillation counting cocktail (Picofluor 15; Packard).
The relative affinity of PBP2b*R6 for penicillin G and piperacillin was determined by measuring the quantity of β-lactam necessary to inhibit 50% of [3H]benzylpenicillin binding (IC50). PBP2b*R6 (1 μM) was incubated with 1 μM [3H]benzylpenicillin and variable concentrations of antibiotics. The mixture was incubated at 37°C for 15 min, denaturing loading buffer was added, and the samples were analyzed by SDS-12.5% PAGE. The procedure to quantify the radioactivity is described above.
The relative affinity of PBP2b*R6, PBP2b*R6Thr446Ala, and PBP2b*CR for penicillin G was determined as follows. The various purified PBP2b* proteins (0.5 μM) were incubated at 37°C for 30 min in buffer B with different concentrations of [3H]benzylpenicillin ranging between 0.5 μM and 30 μM. The reaction was stopped by adding denaturing loading buffer and boiling at 100°C for 5 min. The samples were then subjected to SDS-PAGE, and the procedure to quantify the radioactivity is described above.
The deacylation reaction follows the equation −k3t = ln [EI*]t/[EI*]0, where [EI*]0 is the initial concentration of acyl-enzyme and [EI*]t is its concentration at time t. Determination of k3 was performed as follows. Ten micromolar purified PBP2b*R6 was labeled with 10 μM [3H]benzylpenicillin during 15 min at 37°C. A 15 mM excess of cold penicillin G was added, and the incubation was continued at 37°C. Samples were removed at various times, analyzed by SDS-PAGE, and the amount of radioactivity was measured in the protein bands as mentioned above.
The activity of the TP domain of PBP2b*R6 was measured by its ability to hydrolyze the N-benzoyl-d-alanylmercaptoacetic acid (S2d) molecule, a synthetic thiolester analog of the cell wall stem peptide, according to the method described by Zhao et al. (36). The thiol group produced by the hydrolysis reaction is coupled to 4,4′-dithiodipyridine, and the formation of this complex is followed by the increase of absorbance at 325 nm, leading to the determination of the kcat/Km value.
RESULTS
β-lactam susceptibility of pneumococcal strains.
Seven clinical pneumococcal strains were isolated in the Bacteriology Laboratory of the Centre Hospitalier Universitaire de Grenoble at different periods and from different patients. The origins of these strains are shown in Table 1. The MICs of penicillin G and piperacillin for these pneumococcal strains are clearly correlated (Table 2) and span a wide range of values. One isolate, 5023, is penicillin susceptible with a penicillin G MIC of 0.064 μg/ml. Two strains (5259 and 4935) present an intermediate susceptibility to penicillin G, as the MICs range between 0.12 and 1 μg/ml. The last four strains (4790, 5245, 5268, and 5204) are penicillin-resistant strains, as the MICs are >1 μg/ml. The strains 5268 and 5204 are highly resistant to penicillin G, as the MICs are 3 and 6 μg/ml, respectively. The pattern observed with piperacillin is slightly different. Strains 5023 and 5259 are considered susceptible to piperacillin, as the MICs range between 0.05 and 0.1 μg/ml, while strains 4935, 4790, 5245, and 5268 display intermediate piperacillin resistance, since the MICs range between 1 and 2 μg/ml. Only strain 5204 is highly resistant to piperacillin, with a MIC equal to 4 μg/ml.
TABLE 1.
Serotypes and origins of the pneumococcal isolates tested in this study
| Strain | Serotype | Site of isolation | Yr of isolation | Geographic origin |
|---|---|---|---|---|
| R6 | Unencapsulated | 1930s | United States | |
| 5023 | 3 | Blood culture | 1999 | France |
| 5259 | 15 | Respiratory tract | 2000 | France |
| 4935 | 3 | Blood culture | 1998 | France |
| 4790 | 23 | Blood culture | 1996 | France |
| 5245 | 6 | Sputum | 2000 | France |
| 5268 | 14 | Superior respiratory tract | 2000 | France |
| 5204 | 14 | Sputum | 1999 | France |
TABLE 2.
MICs and distribution of nucleotide and amino acid substitutions in the TP domains of PBP2b of the seven pneumococcal isolates tested in this studya
| Strain | Penicillin
|
Piperacillin
|
Nucleotides alteredb | Amino acids alteredb | ||
|---|---|---|---|---|---|---|
| Classification | MIC (μg/ml) | Classification | MIC (μg/ml) | |||
| R6 | S | <0.016 | S | 0.016 | 0 | 0 |
| 5023 | S | 0.064 | S | 0.047 | 39 | 7 |
| 5259 | I | 0.19 | S | 0.094 | 85 | 15 |
| 4935 | I | 0.5 | I | 1 | 75 | 13 |
| 4790 | R | 1.5 | I | 1 | 76 | 13 |
| 5245 | R | 1.5 | I | 1.5 | 138 | 29 |
| 5268 | R | 3 | I | 2 | 186 | 43 |
| 5204 | R | 6 | R | 4 | 182 | 44 |
S, susceptible (penicillin G MICs of ≤0.06 μg/ml); I, intermediate (penicillin G MICs from 0.12 to 1 μg/ml); R, resistant (penicillin G MICs of >1 μg/ml). The region of the pbp2b gene coding for the catalytic region of PBP2b, i.e., the TP domain, is comprised from nucleotide 937 to 2,044, which corresponds to amino acids 313 to 680 (the numbering takes into account the Met1).
The published sequence of penicillin-susceptible S. pneumoniae R6 was used for comparison.
DNA sequencing and amino acid alterations in PBP2b from clinical isolates.
Each PBP2b TP domain from CR strains includes numerous nucleotide changes from the corresponding region of strain R6. Comparison of the nucleotide and amino acid changes of each sequence indicates that many mutations are silent (Table 2). Most of these amino acid alterations are located downstream from the TP catalytic motif S386VVK (Fig. 1). Amino acid sequences from strains 5268 and 5204 are identical, with the exception of position 351. In addition, a cluster of mutations around the K615TG motif are common to the PBP2b of strains 5245, 5268, and 5204 (Fig. 1).
FIG. 1.
Alignment of amino acid sequences of the TP domains from residues 313 to 680 of PBP2b from clinical isolates and from the R6 strain. The catalytic motifs of the TP domain are enclosed in dark boxes, and all the mutations present in clinical isolate sequences, compared to the sequence of PBP2b from the R6 strain, are underlined. The mutations present in at least three of the six penicillin-intermediate and penicillin-resistant clinical isolates are underlined and highlighted by shaded boxes. The stars highlight amino acid substitutions present in all clinical isolates: Thr446Ala, Glu476Gly, and Thr489Ser/Ala.
The Thr446Ala mutation, which lies downstream from the conserved S443SN catalytic motif, has been identified in all resistant isolates analyzed to date (8, 9, 12-14, 26, 27, 30, 31, 33, 34). This high mutation frequency prompted us to investigate the role of position 446 in the biochemical properties of PBP2b and resistance to β-lactams.
Purification and characterization of PBP2b*R6.
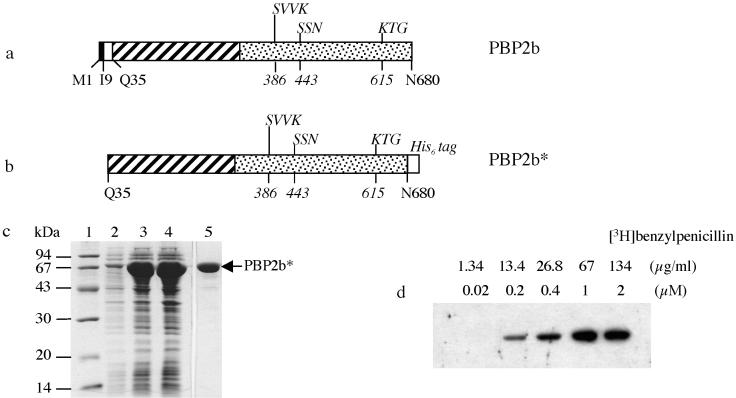
The 2,043-bp pbp2b gene was amplified by PCR from genomic DNA of the penicillin-sensitive S. pneumoniae R6 strain, and its sequence corresponds to the one previously reported (11, 18). The encoded PBP2b amino acid sequence reveals the classical topology shared by all high-Mr PBPs with a very small cytoplasmic domain (M1 to S8), a predicted transmembrane region (I9 to M34), and a large periplasmic region (Q35 to N680) (Fig. 2a). This extracellular region comprises an N-terminal region for which no function has yet been assigned and the TP domain with the conserved amino acid motifs S386VVK, S443SN, and K615TG (Fig. 2a).
FIG. 2.
Expression and purification of PBP2b*R6. (a) Schematic representation of the native PBP2b protein. The solid and empty boxes indicate the cytoplasmic domain and the membrane anchor, respectively. In the periplasmic region, the N-terminal and the TP domains are represented by hatched and dotted boxes, respectively. The conserved motifs of the TP domain are indicated above the boxes (with the first position numbered in italics below). (b) Schematic representation of the construct yielding the soluble form of PBP2b*R6. The His6 tag added in the C terminus of PBP2b* is shown (shadowed box). (c) Expression and purification of PBP2b*R6. Proteins were separated by SDS-12.5% PAGE and stained with Coomassie blue. Lanes: 1, standard molecular mass markers; 2, flowthrough from nickel column; 3, total cell lysate; 4, soluble supernatant following cell lysis; 5, protein eluted at 115 mM imidazole. (d) Saturation binding of PBP2b* by [3H]benzylpenicillin. Fluorogram of a SDS-12.5% PAGE dried gel of PBP2b* (1 μM) labeled with different concentrations of [3H]benzylpenicillin indicated above the gel.
The pbp2b gene was cloned under the control of an inducible promoter (construct pET30b/pbp2bR6). An abundant protein of 74 kDa was observed in E. coli extracts upon induction of a bacterial culture harboring pET30b/pbp2bR6 (data not shown). The protein was insoluble, probably due to the presence of the transmembrane region.
In previous work, the soluble GST fusion forms of various PBPs from S. pneumoniae have been expressed and purified following deletion of their cytoplasmic and transmembrane regions (4, 5, 23, 24). The soluble form of PBP2b of the R6 strain (PBP2b*R6) was thus constructed by deleting the cytoplasmic and transmembrane regions and by adding a C-terminal His6 tag (Fig. 2b). Induction of the E. coli BL21(DE3) strain bearing pET30b/pbp2b*R6 resulted in the production of a protein migrating on SDS-PAGE with an apparent molecular mass of 70 kDa (Fig. 2c, lane 3), corresponding to the expected molecular mass of PBP2b*R6. Most of the protein was present in the soluble fraction (Fig. 2c, compare lanes 3 and 4) and was retained efficiently on a nickel-Sepharose resin (Fig. 2c, lane 2). The presence of 50 mM imidazole during protein loading on the column allowed efficient removal of contaminating proteins. PBP2b*R6 was eluted from the column with 115 mM imidazole and was estimated by SDS-PAGE to be over 95% pure (Fig. 2c, lane 5). This fraction contained a fraction of oligomerized PBP2b* that was resolved on a gel Superdex G75 as two peaks (data not shown). The second peak corresponds to a protein with an apparent molecular mass of 70 kDa, confirming the monomeric state of PBP2b*R6. The purification of monomeric PBP2b*R6 yielded approximately 50 mg of E. coli culture/liter.
The absence of posttranslational modifications on the protein was verified by electrospray ionization mass spectrometry. Indeed, the measured mass of PBP2b*R6 of 71,024 Da is within the limits of error (± 8 Da) of the theoretical mass of 71,022 Da (data not shown). An isoelectric point of 4.95 was determined for PBP2b*R6 by isoelectric focusing, and this value is close to the calculated one of 5.08 (data not shown).
Purified PBP2b*R6 was functional with respect to [3H]benzylpenicillin binding as shown in Fig. 2d. A 1 μM solution of PBP2b*R6 was mixed with increasing concentrations of labeled antibiotic, and the intensity of binding increased until a plateau was reached at 1 μM benzylpenicillin. This stoichiometric binding demonstrated the functional homogeneity of the PBP2b*R6 preparation.
β-lactam binding on PBP2b*R6.
The β-lactam acylation efficiency of PBPs is characterized by the constant k2/K, which is usually determined by monitoring the decrease in the intrinsic fluorescence of the protein in the presence of β-lactam (21, 23, 24). Unfortunately, we could not rely upon this approach, since PBP2b*R6 intrinsic fluorescence remains constant following acylation by either piperacillin or penicillin G. Therefore, we used an alternative method based on competitive binding to analyze the affinity of PBP2b*R6 for β-lactam molecules. The IC50 values were measured (for [3H]benzylpenicillin binding), and the data were expressed as relative values compared to those for penicillin G (Table 3). We performed IC50 measurements with PBP2b*R6 and PBP2x*R6 in order to compare the affinity patterns of both class B PBPs from S. pneumoniae for β-lactams (23, 24). The patterns of reactivity for both PBPs towards piperacillin and cefotaxime are clearly different. The affinity of PBP2b*R6 for piperacillin is nearly sevenfold higher than that of PBP2x*R6, whereas affinity of PBP2b*R6 for cefotaxime is at least 10-fold lower than that of PBP2x*R6.
TABLE 3.
Enzymatic parameters of PBP2b*R6a
| Variant | Relative value compared to penicillin IC50 (%)
|
k3 (s−1) for penicillin G | kcat/Km (M−1 · s−1) for S2d | |
|---|---|---|---|---|
| Piperacillin | Cefotaxime | |||
| PBP2b* | 44 | >1,300 | 3.8 × 10−5 | 80 |
| PBP2x* | 300 | 130 | ||
Data were obtained from two to three independent experiments.
The efficiency of hydrolysis, kcat/Km value, of PBP2b*R6 was determined using the substrate analogue S2d. The efficiency of hydrolysis of S2d thiolester by PBP2b*R6 is 10- to 100-fold lower than the efficiency obtained when using other S. pneumoniae PBPs from the R6 strain (5, 6). The value obtained (80 M−1 · s−1) is three- to fourfold higher than the value of PBP2x* from a CR isolate (20 M−1 · s−1) but 30-fold lower than that for the sensitive PBP2x* (2,500 M−1 · s−1) (23, 24).
The k3 deacylation rate (3.8 × 10−5 · s−1) of PBP2b*R6 for penicillin G is in the same range as those reported for other S. pneumoniae PBPs originating either from sensitive or resistant strains (4-7, 23, 24).
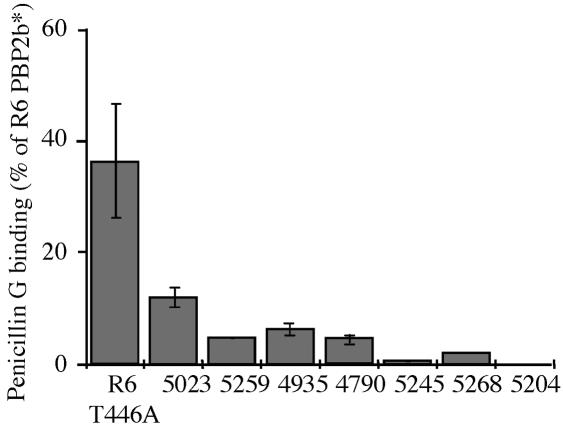
Affinity for penicillin G of PBP2b* variants.
PBP2b* variants from sensitive (PBP2b*R6) and CR strains (PBP2b*CR) were compared with respect to their affinity for penicillin G following purification of the various recombinant proteins (Fig. 3). PBP2b* from the penicillin-susceptible strain 5023 has a penicillin G affinity reduced by more than 85% compared to that of PBP2b*R6, whereas PBP2b* from intermediate and resistant penicillin strains 5259, 4935, and 4790 harbor a penicillin G affinity that is reduced by about 90%. The PBP2b* with the most reduced measurable penicillin G affinity came from strains 5245 and 5268, with decreases of 95% and 99%, respectively. In our experimental conditions, no binding was observed with PBP2b* from strain 5204, even with the highest [3H]benzylpenicillin concentration, indicating a major reduction of affinity for the antibiotic.
FIG. 3.
Relative affinity of PBP2b* proteins for penicillin G. The various purified PBP2b* proteins (0.5 μM) were incubated at 37°C for 30 min with 2 μM [3H]benzylpenicillin. All components were added at the same time, and the reaction was stopped by adding denaturing loading buffer. The affinity for penicillin G for each protein is expressed as a percentage of the PBP2b*R6 value.
In order to characterize the role of the Thr446Ala mutation in PBP2bCRs, this mutation was introduced into PBP2b*R6. The mutant (PBP2b*R6Thr446Ala) was expressed and purified in a similar manner as the wild-type protein (PBP2b*R6). The unique point mutation Thr446Ala in PBP2b*R6 decreases the affinity for penicillin G by 60% compared to that of the wild-type protein (Fig. 3).
Effect of PBP2b variants on MICs for S. pneumoniae R6.
Plasmids bearing genes encoding PBP2b*R6Thr446Ala or the various PBP2b*CRs were used to transform competent S. pneumoniae R6 cells. Recombinant strains were selected by using either penicillin G or piperacillin. Despite repeated attempts, no penicillin G-resistant clones were obtained following transformation with the PBP2b*R6Thr446Ala or PBP2b*CR genes, indicating that neither the point mutation nor the CR sequences reduce the sensitivity of R6 for penicillin G to allow selection, which indicates that other modified PBPs are probably involved in the increase of the MICs for the CR strains.
At the highest piperacillin concentration allowing recombinant strain growth, three to six independent clones were isolated, and the absence of contamination by other bacterial species was checked. The MIC of piperacillin for the S. pneumoniae R6 bearing the PBP2b*R6Thr446Ala gene is 0.032 μg/ml, an increase of twofold over that for the parental R6 strain (<0.016 μg/ml). As expected, the recombinant strains had a sensitivity to penicillin G as high as the one displayed by R6 (<0.016 μg/ml).
Except for transformants obtained with CR pbp2b originating from strain 5023, for which the piperacillin MIC was 0.023 to 0.047 μg/ml, all other strains produced similar MICs of 0.032 to 0.047 μg/ml. These values are at least double the one for R6 (0.016 μg/ml). For transformants originating from CR strains 5023, 5259, 4935, 4790, 5245, and 5204, the penicillin G MIC was 0.016 to 0.023 μg/ml. The MICs for clones transformed with PBP2b*CR from strain 5268 were 0.016 to 0.032 μg/ml. In all cases, the MICs were higher than the one for R6 (<0.016 μg/ml).
In order to further investigate the influence of PBP2b* on β-lactam affinity, the Ala446 was mutated back to Thr in all seven PBP2b*CRs and was used to transform the R6 strain. Piperacillin-resistant transformants could be selected following transformation with five reversed CR pbp2b* sequences but, despite repeated attempts, not with sequences originating from strains 5023 and 5259. The MICs for all five transformants did not differ significantly from those obtained with the nonmutated PBP2b sequences (data not shown).
Recombinant clones were selected with piperacillin following transformation of R6 with each of the seven PBP2b*CR genes. Sequencing of the pbp2b genes isolated from some piperacillin-resistant transformants revealed that part of the 3′ region of the pbp2b sequence encoding for a region of the TP domain (around amino acid 520 to 530 to the C terminus of PBP2b) did not recombine with the R6 pbp2b gene. Extending the end of the CR pbp2b genes used for the transformation by 660 nucleotides did not result in a significant modification of the recombination event. Therefore, the transformants express a hybrid PBP2b protein in which the C terminus originates from R6 PBP2b.
DISCUSSION
β-lactam-resistant S. pneumoniae strains isolated in hospitals harbor high-Mr pbp genes with mosaic structures. The mosaicity of S. pneumoniae pbp genes results from recombination events between S. pneumoniae and S. mitis (8-10). Consequently, sequences of PBP2x, PBP2b, and PBP1a isolated from CR strains carry many amino acid changes compared to those of PBPs from susceptible strains, with only a fraction likely being related to the reduction of PBP affinity for β-lactam antibiotics. An understanding of the consequences of such mutations at a biochemical level requires the in vitro analysis of purified PBPs. For instance, a subset of mutations within the TP domain of PBP2x has been implicated in its reduction of inhibition efficiency by cefotaxime and increased S. pneumoniae resistance to β-lactams (1, 23, 24). PBP2b is the primary mutated PBP when piperacillin selection is applied to S. pneumoniae cultures in the laboratory (22, 29). We present here the enzymatic characteristics of PBP2b and show that PBP2b proteins from CR strains have a reduced in vitro affinity for β-lactams. Moreover, we have identified, among tens of mutations, Thr446Ala of the TP domain as a molecular determinant for resistance to piperacillin.
We have cloned, expressed, and purified the periplasmic region of the S. pneumoniae PBP2b from the R6 β-lactam-sensitive strain (PBP2b*R6) and from seven CR strains isolated in Grenoble, France. The in vitro characterization of PBP2b*R6 provides a rationale for the PBP2b reactivity pattern towards β-lactams (20). Since PBP2b is the sole mutated high-Mr PBP under piperacillin selection (22, 29), it is understandable that PBP2b should harbor the highest piperacillin affinity among the five S. pneumoniae high-Mr PBPs. The relatively low affinity of PBP2b*R6 for cefotaxime appears to be in the same range as the values obtained previously with PBP1a*R6 and PBP2a*R6 (5, 6). This is in accordance with the low mutational susceptibility of the pbp2b gene under cefotaxime selection (17-19).
PBP2b*CRs which display the highest in vitro reduction in affinity (95 to 99%) for penicillin G are from strains 5245, 5268, and 5204. These proteins harbor a common set of point mutations localized around the K615TG motif (Fig. 1), which are also found in PBP2b from highly penicillin-resistant strains (penicillin G MICs of 2 to 8 μg/ml) (2, 12, 33). The role of some of these substitutions may be proposed by the light of tridimensional structure of S. pneumoniae PBP2x (16, 28). In PBP2b, Ala619, downstream of the K615TG motif, is proposed to be part of the strand β3 bordering the active site cleft. Thus, mutation Ala619Gly might increase the flexibility of the β3 strand and, in consequence, modify the kinetics of β-lactam binding. On the other hand, amino acids Asn659, Gly660, and Ser664 are most probably located at the N terminus of helix α11; thus, their mutation into Lys, Asn, and Ala, respectively, might induce structural rearrangements near the active site and again interfere with antibiotic binding. Unfortunately, the effect of these mutations in PBP2b of strains 5245, 5268, and 5204 could not be investigated in vivo. We observed that, once recombined into the R6 genome, these CR pbp2b* genes lacked a fragment encoding a region around positions 525 to 680, which includes the K615TG motif. Extension of the DNA fragment used for recombination by 660 bp downstream of the 3′ end of the pbp2b genes did not allow insertion of the full gene. This problem is specific to the PBP2b gene, as PBP2x could be fully recombined into the R6 genome (1). It might be that the C-terminal region of PBP2bCR sequences is incompatible with the expression in R6. Indeed, a complex relationship between PBPs had been inferred previously (1).
The R6 strain transformed with sequences originating from the seven CR strains provided recombinant S. pneumoniae for which the MIC of penicillin G was within a narrow range (0.016 to 0.032 μg/ml), contrary to the MIC for the donor CR strains (0.064 to 6 μg/ml). This threshold effect is likely due to the high penicillin G sensitivity of the host strain PBP1a (1, 2, 25). Similarly, the MICs of piperacillin for the recombinant strains do not exceed 0.023 to 0.047 μg/ml, whereas the corresponding values for the CR strains range from 0.047 to 4 μg/ml. The MIC for the recombinant strain obtained with the PBP2b sequence from strain 5023 is similar to that for the donor CR strain. Indeed, this PBP2b TP sequence is close to the one from R6 PBP2b, with only seven amino acid changes. In strain 5023, the PBP1a sequence is identical to the one from R6, contrary to the six other CR strains studied here. Therefore, it is likely that the threshold effect for piperacillin observed in our recombined strains is also due to the R6 PBP1a sensitivity for the antibiotic. Indeed, it has recently been shown that the threshold effect for penicillin G resistance observed in our recombined strains is due to the R6 PBP1a and PBP2x proteins (Chesnel et al., unpublished results).
The analysis of the amino acid sequences of the TP domain of PBP2b from the seven CR strains revealed that the most predominant mutations are Thr446Ala, Glu476Gly, and Thr489Ser/Ala, which have been identified in other CR strains around the world (8, 9, 12-14, 26, 27, 30, 31, 33, 34). The amino acid sequences of the TP domain of PBP2b from strains 5023, 5259, 4935, and 4790 are already represented in databases, contrary to sequences from our most highly resistant strains 5245, 5268, and 5204, which are new. The preponderance of the Thr446Ala mutation in CR isolates and in laboratory resistant strains (17), as well as its juxtaposition to a key motif of PBP function (S443SN), led us to investigate the role of this mutation in detail. To achieve this purpose, the Thr446Ala mutation was introduced into PBP2b*R6, leading to the protein PBP2b*R6Thr446Ala. The measurements of relative affinity for penicillin G showed that the point mutation Thr446Ala in PBP2b*R6 decreases the affinity for penicillin G by 60%, compared to that of the wild-type protein. When this mutation was introduced into S. pneumoniae strain R6, the MIC of piperacillin increased twofold. These results show that the Thr446Ala mutation in PBP2b from the R6 strain directs the affinity for β-lactams. Surprisingly, when the reverse experiment was attempted, that is, the insertion in the R6 genome of PBP2b*CRs with the Ala446Thr mutation, the piperacillin MICs for the resulting five strains that could be selected were similar to those for the nonmutated strains. The absence of the C-terminal region of the TP domain might be responsible for this effect. Moreover, the inability to select R6 transformants expressing PBP2bCRAla446Thr from strains 5023 and 5259 might indicate that the mutation has lowered the MIC to a level comparable to that for the R6 strain. The MICs for strains 5023 and 5259 were the lowest among our selection of strains. Taken together, these results illustrate the complex relationship between mutation at position 446 and other mutations present in PBP2bCR sequences.
Acknowledgments
This work was supported by a fellowship to L.C., a grant from the Rhône-Alpes region, and European Commission grant LSMH-CT-2003-503335. E.P. is a recipient of a CEA CFR fellowship.
We are much indebted to Nicolas Mouz (Protein'eXpert, Grenoble, France) for excellent advice on recombinant protein expression. We thank Genome Express for providing DNA sequencing services. We are very grateful to Andréa Dessen and André Zapun (Institut de Biologie Structurale Jean-Pierre Ebel, Grenoble, France) for constructive comments and critical review of the manuscript.
REFERENCES
- 1.Chesnel, L., L. Pernot, D. Lemaire, D. Champelovier, J. Croize, O. Dideberg, T. Vernet, and A. Zapun. 2003. The structural modifications induced by the M339F substitution in PBP2x from Streptococcus pneumoniae further decreases the susceptibility to beta-lactams of resistant strains. J. Biol. Chem. 278:44448-44456. [DOI] [PubMed] [Google Scholar]
- 2.Coffey, T. J., M. Daniels, L. K. McDougal, C. G. Dowson, F. C. Tenover, and B. G. Spratt. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 39:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Guilmi, A. M., A. Dessen, O. Dideberg, and T. Vernet. 2003. The glycosyltransferase domain of penicillin-binding protein 2a from Streptococcus pneumoniae catalyzes the polymerization of murein glycan chains. J. Bacteriol. 185:4418-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Guilmi, A. M., A. Dessen, O. Dideberg, and T. Vernet. 2003. Functional characterization of penicillin-binding protein 1b from Streptococcus pneumoniae. J. Bacteriol. 185:1650-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Guilmi, A. M., N. Mouz, J. P. Andrieu, J. Hoskins, S. R. Jaskunas, J. Gagnon, O. Dideberg, and T. Vernet. 1998. Identification, purification, and characterization of transpeptidase and glycosyltransferase domains of Streptococcus pneumoniae penicillin-binding protein 1a. J. Bacteriol. 180:5652-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Guilmi, A. M., N. Mouz, L. Martin, J. Hoskins, S. R. Jaskunas, O. Dideberg, and T. Vernet. 1999. Glycosyltransferase domain of penicillin-binding protein 2a from Streptococcus pneumoniae is membrane associated. J. Bacteriol. 181:2773-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Guilmi, A. M., N. Mouz, Y. Petillot, E. Forest, O. Dideberg, and T. Vernet. 2000. Deacylation kinetics analysis of Streptococcus pneumoniae penicillin-binding protein 2x mutants resistant to beta-lactam antibiotics using electrospray ionization-mass spectrometry. Anal. Biochem. 284:240-246. [DOI] [PubMed] [Google Scholar]
- 8.Dowson, C. G., T. J. Coffey, C. Kell, and R. A. Whiley. 1993. Evolution of penicillin resistance in Streptococcus pneumoniae; the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol. Microbiol. 9:635-643. [DOI] [PubMed] [Google Scholar]
- 9.Dowson, C. G., A. Hutchison, J. A. Brannigan, R. C. George, D. Hansman, J. Linares, A. Tomasz, J. M. Smith, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowson, C. G., A. Hutchison, and B. G. Spratt. 1989. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol. Microbiol. 3:95-102. [DOI] [PubMed] [Google Scholar]
- 11.Dowson, C. G., A. Hutchison, and B. G. Spratt. 1989. Nucleotide sequence of the penicillin-binding protein 2B gene of Streptococcus pneumoniae strain R6. Nucleic Acids Res. 17:7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.du Plessis, M., E. Bingen, and K. P. Klugman. 2002. Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob. Agents Chemother. 46:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright, M. C., and B. G. Spratt. 1999. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol. Biol. Evol. 16:1687-1695. [DOI] [PubMed] [Google Scholar]
- 14.Ferroni, A., and P. Berche. 2001. Alterations to penicillin-binding proteins 1A, 2B and 2X amongst penicillin-resistant clinical isolates of Streptococcus pneumoniae serotype 23F from the nasopharyngeal flora of children. J. Med. Microbiol. 50:828-832. [DOI] [PubMed] [Google Scholar]
- 15.Goffin, C., and J. M. Ghuysen. 2002. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 66:702-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, E., N. Mouz, E. Duee, and O. Dideberg. 2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477-485. [DOI] [PubMed] [Google Scholar]
- 17.Grebe, T., and R. Hakenbeck. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob. Agents Chemother. 40:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakenbeck, R., A. Konig, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakenbeck, R., C. Martin, C. Dowson, and T. Grebe. 1994. Penicillin-binding protein 2b of Streptococcus pneumoniae in piperacillin-resistant laboratory mutants. J. Bacteriol. 176:5574-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hakenbeck, R., S. Tornette, and N. F. Adkinson. 1987. Interaction of non-lytic beta-lactams with penicillin-binding proteins in Streptococcus pneumoniae. J. Gen. Microbiol. 133:755-760. [DOI] [PubMed] [Google Scholar]
- 21.Jamin, M., C. Damblon, S. Millier, R. Hakenbeck, and J. M. Frère. 1993. Penicillin-binding protein 2x of Streptococcus pneumoniae: enzymic activities and interactions with beta-lactams. Biochem. J. 292:735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krauss, J., M. van der Linden, T. Grebe, and R. Hakenbeck. 1996. Penicillin-binding proteins 2x and 2b as primary PBP targets in Streptococcus pneumoniae. Microb. Drug Resist. 2:183-186. [DOI] [PubMed] [Google Scholar]
- 23.Mouz, N., A. M. Di Guilmi, E. Gordon, R. Hakenbeck, O. Dideberg, and T. Vernet. 1999. Mutations in the active site of penicillin-binding protein PBP2x from Streptococcus pneumoniae. Role in the specificity for beta-lactam antibiotics. J. Biol. Chem. 274:19175-19180. [DOI] [PubMed] [Google Scholar]
- 24.Mouz, N., E. Gordon, A. M. Di Guilmi, I. Petit, Y. Pétillot, Y. Dupont, R. Hakenbeck, T. Vernet, and O. Dideberg. 1998. Identification of a structural determinant for resistance to beta-lactam antibiotics in gram-positive bacteria. Proc. Natl. Acad. Sci. USA 95:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz, R., C. G. Dowson, M. Daniels, T. J. Coffey, C. Martin, R. Hakenbeck, and B. G. Spratt. 1992. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 6:2461-2465. [DOI] [PubMed] [Google Scholar]
- 26.Nagai, K., T. A. Davies, M. R. Jacobs, and P. C. Appelbaum. 2002. Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob. Agents Chemother. 46:1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichol, K. A., G. G. Zhanel, and D. J. Hoban. 2002. Penicillin-binding protein 1A, 2B, and 2X alterations in Canadian isolates of penicillin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:3261-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pares, S., N. Mouz, Y. Petillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3:284-289. [DOI] [PubMed] [Google Scholar]
- 29.Reichmann, P., A. Konig, A. Marton, and R. Hakenbeck. 1996. Penicillin-binding proteins as resistance determinants in clinical isolates of Streptococcus pneumoniae. Microb. Drug Resist. 2:177-181. [DOI] [PubMed] [Google Scholar]
- 30.Rieux, V., C. Carbon, and E. Azoulay-Dupuis. 2001. Complex relationship between acquisition of beta-lactam resistance and loss of virulence in Streptococcus pneumoniae. J. Infect. Dis. 184:66-72. [DOI] [PubMed] [Google Scholar]
- 31.Sa-Leao, R., S. E. Vilhelmsson, H. de Lencastre, K. G. Kristinsson, and A. Tomasz. 2002. Diversity of penicillin-nonsusceptible Streptococcus pneumoniae circulating in Iceland after the introduction of penicillin-resistant clone Spain(6B)-2. J. Infect. Dis. 186:966-975. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz, B., J. A. Markwalder, S. P. Seitz, Y. Wang, and R. L. Stein. 2002. A kinetic characterization of the glycosyltransferase activity of Escherichia coli PBP1b and development of a continuous fluorescence assay. Biochemistry 41:12552-12561. [DOI] [PubMed] [Google Scholar]
- 33.Smith, A. M., and K. P. Klugman. 1995. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 39:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song, J. H., J. W. Yang, J. H. Jin, S. W. Kim, C. K. Kim, H. Lee, K. R. Peck, S. Kim, N. Y. Lee, M. R. Jacobs, P. C. Appelbaum, et al. 2000. Molecular characterization of multidrug-resistant Streptococcus pneumoniae isolates in Korea. J. Clin. Microbiol. 38:1641-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tipper, D. J., and J. L. Strominger. 1965. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc. Natl. Acad. Sci. USA 54:1133-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, G., W. K. Yeh, R. H. Carnahan, J. Flokowitsch, T. I. Meier, W. E. Alborn, Jr., G. W. Becker, and S. R. Jaskunas. 1997. Biochemical characterization of penicillin-resistant and -sensitive penicillin-binding protein 2x transpeptidase activities of Streptococcus pneumoniae and mechanistic implications in bacterial resistance to β-lactam antibiotics. J. Bacteriol. 179:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]