Abstract
Background
The recombination-activating gene (RAG) 1/2 proteins play a critical role in the development of T and B cells by initiating the VDJ recombination process that leads to generation of a broad T-cell receptor (TCR) and B-cell receptor repertoire. Pathogenic mutations in the RAG1/2 genes result in various forms of primary immunodeficiency, ranging from T−B− severe combined immune deficiency to delayed-onset disease with granuloma formation, autoimmunity, or both. It is not clear what contributes to such heterogeneity of phenotypes.
Objective
We sought to investigate the molecular basis for phenotypic diversity presented in patients with various RAG1 mutations.
Methods
We have developed a flow cytometry–based assay that allows analysis of RAG recombination activity based on green fluorescent protein expression and have assessed the induction of the Ighc locus rearrangements in mouse Rag1−/− pro-B cells reconstituted with wild-type or mutant human RAG1 (hRAG1) using deep sequencing technology.
Results
Here we demonstrate correlation between defective recombination activity of hRAG1 mutant proteins and severity of the clinical and immunologic phenotype and provide insights on the molecular mechanisms accounting for such phenotypic diversity.
Conclusions
Using a sensitive assay to measure the RAG1 activity level of 79 mutations in a physiologic setting, we demonstrate correlation between recombination activity of RAG1 mutants and the severity of clinical presentation and show that RAG1 mutants can induce specific abnormalities of the VDJ recombination process.
Keywords: Recombination-activating gene 1, V(D)J recombination, severe combined immune deficiency, Omenn syndrome, autoimmunity, genotype-phenotype correlation, immune repertoire
The recombination-activating gene (RAG) 1 and RAG2 proteins initiate the VDJ recombination process by generating DNA double-strand breaks at the recombination signal sequences (RSSs) that flank the variable (V), diversity (D), and joining (J) gene segments of the immunoglobulin and T-cell receptor (TCR) genes.1 These DNA double-strand breaks are then joined by the ubiquitous mechanism of nonhomologous end-joining machinery.2 As a result of VDJ recombination, a polyclonal pool of functional T and B lymphocytes is generated, expressing a diverse repertoire of productive TCR and B-cell receptor rearrangements.
Null mutations of RAG1 and RAG2 genes result in the T−B− severe combined immune deficiency (SCID) phenotype.3 However, hypomorphic RAG mutations have been associated with a spectrum of clinical and immunologic phenotypes that include Omenn syndrome (OS),4–10 with erythroderma, lymphadenopathy, eosinophilia, increased serum IgE levels, and the presence of autologous, oligoclonal, and activated T lymphocytes; leaky/atypical SCID,10 with varying numbers of T and B cells but without the typical features of OS; SCID with expansion of γδ T lymphocytes (γδ-T),11,12 which us often associated with cytomegalovirus infection; delayed-onset combined immune deficiency with granuloma and/or autoimmunity (CID-G/A)13–15; and in a single case of idiopathic CD4+ T cell lymphopenia (ICL), presenting with extensive chickenpox and recurrent pneumonia.16
Attempts to correlate the phenotypic diversity of RAG-related disorders in human subjects with functional activity of the mutant proteins were largely based on a transient transfection assay in nonlymphoid adherent cells.17 In this assay cells are cotransfected with plasmids encoding for wild-type (or mutant) human RAG1 (hRAG1) and hRAG2 and a third plasmid containing a suitable recombination substrate that would allow expression of an antibiotic resistance gene upon recognition and cleavage by hRAG1 and hRAG2 and nonhomologous end joining–mediated ligation. However, with this assay, the recombination activity of RAG proteins is analyzed on an extrachromosomal substrate (ie, a nonphysiologic setting), and functional impairment of mutants that specifically affect nuclear translocation of the hRAG proteins might be missed. Furthermore, it has been shown that stability and posttranslational modifications of the RAG proteins differ significantly in lymphoid versus nonlymphoid cells.18
Recently, Abelson murine leukemia virus (A-MuLV)– transformed pro-B cells containing an inverted green fluorescent protein (GFP) cassette flanked by RSS (pMX-INV) have been used to measure VDJ recombination activity on an intrachromosomal substrate by using flow cytometry with GFP expression as a read-out.19 On the basis of this platform, we have analyzed the expression and recombination activity of 79 naturally occurring hRAG1 mutant proteins and thereby performed the largest comprehensive analysis of genotype-proteotype-phenotype correlation for hRAG1 deficiency. Our results provide novel insights into the molecular mechanisms underlying phenotypic diversity in patients with this disease.
METHODS
Patient selection and assignment to phenotypic subgroups
Deidentified patients’ clinical, immunologic, and molecular data were provided by an international network of physicians in Europe, the Middle East, South America, and the United States, according to protocols approved by the local institutional review boards. On the basis of phenotype, each patient was assigned to one of the following subgroups: T−B− SCID, OS, γδ-T, atypical/leaky SCID-G/A, and ICL.
Determination of recombinase activity level of wild-type and mutant RAG1
Analysis of recombination activity of wild-type and mutant RAG1 constructs was performed, as previously described.14 For detailed methods, see the Methods section in this article’s Online Repository at www.jacionline.org.
Immune repertoire analysis
Analysis of rearrangements at the endogenous Ighc locus was performed by using as a template the mRNA extracted from A-MuLV pro-B cells transduced with retroviral vectors encoding for wild-type or mutant hRAG1. A set of nested primers specific for various VH and CH elements of the Ighc locus were used for the first amplification cycles, followed by amplification with communal primers according to the manufacturer’s protocol for MBCR (iRepertoire, Huntsville, Ala). By using this protocol, ampliconrescued multiplex PCR allows semiquantitative amplification of the immune repertoire.20 Purified PCR products were sequenced with the GS Junior 454 platform (Roche, Mannheim, Germany).
Raw sequences were filtered for PCR errors, and a tree map and 2-dimensional map were generated from the total complementarity-determining region (CDR)–H3 sequences to analyze VH-JH pairing and the relative distribution of distinct rearrangements (iRepertoire). The filtered sequences, excluding duplicates, were further analyzed for VH, DH, and JH gene usage; composition; CDR-H3 length; reading frame (RF) determination; and the diversity index of Shannon entropy by using IMGT HighV-QUEST output files21 and IgAT software.22 Rarefaction curves were generated by using the VDJ statistics file from IgAT analysis and the PAST program.23 To assess reproducibility, 2 runs on independent amplification libraries were performed for R699Q, and 2 to 3 repeats of the same amplification library were run for the R314W and M435V mutants.
Statistical analysis
Results of recombination activity of hRAG1 proteins were analyzed with the Mann-Whitney U test. The χ2 test was used to assess skewing in the use of VH, DH, and JH genes by various hRAG1 mutant proteins compared with wild-type hRAG1. The unpaired 2-tailed t test was used to analyze the average CDR-H3 length, the average index of hydrophobicity according to a normalized Kyte-Doolittle scale, differential use of 3 RFs, and the average Shannon entropy of the CDR-H3 loop. Nonparametric Spearman correlation was used to determine correlation between the recombination activity level and protein expression of various hRAG1 mutants by using the Spearman correlation coefficient (rs).
RESULTS
Recombination activity and protein expression of hRAG1 mutants
We have compiled a list of 79 distinct, naturally occurring hRAG1 genetic variants, including 3 known polymorphisms (G99S, H249R, and K820R) and 76 distinct mutations identified in 68 patients (among which were 34 novel cases, Table I).4–9,11–16,24–28 To investigate correlation between the expression and function of mutant hRAG1 and the patients’ clinical and immunologic phenotypes, we used A-MuLV– transformed Rag1−/− tg.Eμ-bcl2 pro-B cells (hereafter referred to as Rag1−/− Abl pro-B cells), containing a single integration of the pMX-INV GFP cassette flanked by RSSs (Fig 1, A, and see Fig E1 in this article’s Online Repository at www.jacionline.org),21 which were further transduced with retroviral vectors encoding wild-type or mutant hRAG1 or wild-type mouse Rag1 (mRag1) and hCD2 as a reporter. To control for expression of wild-type and mutant RAG1 proteins, Rag1−/− Abl pro-B cells expressing either wild-type or mutant hRAG1 were equally enriched to contain greater than 90% cells expressing the hCD2 reporter. Comparable levels of GFP expression were observed on induction of wild-type hRAG1 and mRag1 expression with STI-571 (imatinib; Fig 1, B). This treatment arrests cells in the G0/G1 phase of the cell cycle while RAG proteins are expressed.29 We then measured the VDJ recombination activity of 79 different naturally occurring hRAG1 variants by normalizing GFP expression to that observed in the presence of wild-type hRAG1 (Fig 1, C, and see Table E1 in this article’s Online Repository at www.jacionline.org). Wild-type levels of VDJ recombination activity were observed for 3 known hRAG1 polymorphisms (G99S, H249R, and K820R) and for N106K, R449K, R474C, H612R, A868V, and M1006V mutants, suggesting that the latter represent neutral variants and not disease-causing mutations.
TABLE I.
List of patients subdivided into 6 clinical groups
| PID type | Patient no. | Mutation | Age | T cells/µL | B cells/µL | Eosinophils/µL | IgE (IU/µL) | Autoimmunity | Reference |
|---|---|---|---|---|---|---|---|---|---|
| ICL | 1* | a. R474C; b. L506F | 8 y | 810 | 480 | 670 | <5 | No | 16 |
| CID-G/A | 2* | a. R975W; b. R778Q | 7.5 y | 606 | 132 | 200 | ND | No | 13 |
| 3* | a. R314W; b. R507W/R737H | 2.6 y | 315 | 30 | 21 | <4.4 | No | 13 | |
| 4* | a. R975Q; b. R396C | 4 y | 865 | 51 | NA | <30 | Yes | 24 | |
| 5* | a. W522C; b. L541CfsX30 | 14 y | 1404 | 552 | NA | <5 | Yes | 14 | |
| 6 | a. F974L; b. R841Q | 0 | 34 | 256 | ND | 38 | Yes | 15 | |
| 7* | a. G516A; b. S601P | 2 y | 50 | 94 | 2700 | ND | |||
| 8 | a. R699Q; b. C730F | 15 mo | 187 | 51 | 2100 | 3.88 | Yes | ||
| 9† | a. W522C; b. R973C | 18 mo | 3973 | 0 | 100 | 126 | Yes | ||
| 10 | a. M435V; b. R699W | 5 y | 180 | 90 | ND | <5 | Yes | ||
| 11 | a. R474C; b. K86VfsX33 | 3 y | 327 | 266 | ND | <2 | Yes | ||
| 12 | a. R841W; b. R841W | 18 mo | 407 | 44 | 0 | 1 | Yes | ||
| AS/LS | 13 | a. R975Q; b. R396L | 4 mo | 1350 | 432 | 1150 | 1690 | 4 | |
| 14 | a. R841W; b. R410Q | 1 mo | 202 | 152 | 1200 | >1000 | 4 | ||
| 15 | a. A444V; b. A444V | 1 mo | 822 | 9 | 1680 | 45 | 4 | ||
| 16 | a. V433M; b. A444V | 2 mo | 51 | 8 | 140 | ND | 4 | ||
| 17 | a. W896R; b. W896R | 4.5 mo | 1660 | 0 | ND | <3 | 25 | ||
| 18‡ | a. E770K; b E770K | 0 | 36 | <5 | ND | ND | 26 | ||
| 19 | a. H612R; b. H612R | 7.5 y | 245 | 0 | 33 | <2 | 27 | ||
| 20 | a. R142X; b. unknown | 12 mo | 778 | 301 | 1045 | >5000 | |||
| γδ-T | 21 | a. R841W; b. R841W | 13 mo | 4050 | 200 | 100 | 3.6 | Yes | 11 |
| 22 | a. Q981P; b. Q981P | 12 mo | 3900 | 2550 | 100 | 11 | Yes | 11 | |
| 23§3 | a. K86VfsX33; b. K86VfsX33 | 19 mo | 1925 | 420 | ND | <4 | Yes | 11 | |
| 24 | a. E174SfsX27; b. E174SfsX27 | 3 mo | 420 | 0 | 800 | ND | 11 | ||
| 25 | a. R561H; b. R561H | 11 mo | 350 | 24 | 420 | 20 | Yes | 12 | |
| 26 | a. R561H; b. R561H | 11 mo | 135 | 10 | 0 | ND | Yes | 12 | |
| 27§1 | a. K86VfsX33; b. K86VfsX33 | 13 mo | 3400 | 320 | ND | <10 | Yes | 7 | |
| 28 | a. R474C; b. L732P | 8 mo | 294 | 63 | ND | ND | Yes | 26 | |
| 29 | a. K86VfsX33; b. K86VfsX33 | 4 y | 2175 | 1535 | ND | ND | Yes | ||
| 30§3 | a. K86VfsX33; b. K86VfsX33 | 0 | 135 | 44 | 600 | ND | Yes | ||
| 31 | a. K86VfsX33; b. K86VfsX33 | 11 mo | 1010 | 310 | 590 | 5.6 | Yes | ||
| 32 | a. R108X; b. R108X | 2 mo | 2550 | 193 | 1400 | Yes | |||
| 33 | a. E174SfsX27; b. E174SfsX27 | 2 mo | 399 | 3 | 0 | 113 | Yes | ||
| OS | 34 | a. M435V; b. R559S | 1 mo | 5461 | <5 | 59 | ND | 4 | |
| 35 | a. Y912C; b. R396C | 3 mo | 21580 | <5 | 2250 | >3000 | 4 | ||
| 36 | a. C328Y; b. C328Y | 3 mo | 371 | <5 | 10 | 19.9 | 4 | ||
| 37 | a. K86VfsX33; b. D429G | 1.5 mo | 96 | <6 | 8 | 500 | 4 | ||
| 38 | a. R396C; b. R396C | 4 mo | 7544 | <5 | 5150 | 182 | 4 | ||
| 39 | a. S401P; b. S401P | 1.5 mo | 2455 | <5 | 1350 | 166 | 4 | ||
| 40 | a. A444V; b. A444V | 3 mo | 310 | <5 | 1700 | ND | 4 | ||
| 41 | a. R624H; b. K992E | 5 mo | 1000 | 0 | 500 | >1000 | 5 | ||
| 42 | a. R561H; b. R561H | 2 mo | 245 | 14 | 4550 | 190 | 12 | ||
| 43 | a. I956T; b. I956T | 1 mo | 12200 | 0 | NA | NA | 6 | ||
| 44§1 | a. K86VfsX33; b. K86VfsX33 | 3 mo | 820 | 0 | ND | ND | 7 | ||
| 45§1 | a. K86VfsX33; b. K86VfsX33 | 18 mo | 428 | 0 | ND | <10 | 7 | ||
| 46 | a. R142X; b. M458SfsX34 | 1 wk | 10491 | 2 | ND | ND | 9 | ||
| 47 | a. L454Q; b. L454Q | NA | 1584 | 26 | 1970 | 260 | 8 | ||
| 48 | a. R559S; b. M435V | 3 wk | 10755 | 0 | 5400 | 95 | |||
| 49 | a. M435V/M1006V; b. M435V/M1006V | 1 wk | 18315 | 0 | 2600 | ND | |||
| 50 | a. K992E; b. K992E | 6 mo | 168 | 0 | 203 | 1 | |||
| 51 | a. G99S/K391E; b. G99S/K391E | 6 mo | 629 | 2 | 150 | 12 | |||
| 52 | a. R778W; b. R410W | 2 wk | 2138 | 0 | ND | >2000 | Yes | ||
| 53 | a. D539V; b. A246TfsX17 | 1 mo | 2025 | 0 | 5020 | 501 | |||
| 54 | a. Q248X; b. Q248X | NA | 4792 | 0 | ND | ND | |||
| 55 | a. R404Q; b. R396C | 2 mo | 36670 | 0 | 8640 | Increased | |||
| 56 | a. R559S; b. P786L | 9 mo | 391 | 4 | ND | 310 | Yes | ||
| 57 | a. R396H; b. R396H | 2 mo | NA | NA | NA | NA | |||
| 58 | a. K383RfsX7; b. K383RfsX7 | 6 mo | 205 | 31 | 390 | ND | |||
| 59 | a. W959X; b. W959X | 3 wk | 3424 | 0 | 11700 | <2 | No | ||
| T−/B− | 60 | a. M1006V; b. M1006V | NA | NA | NA | NA | NA | 28 | |
| 61 | a. A444V; b. A444V | 3 mo | 64 | 0 | ND | ND | |||
| 62 | a. A444V; b. A444V | 0 | 179 | 30 | ND | ND | |||
| T−/B− | 63 | a. R404Q/R449K; b. R764P | 1 mo | 28 | <20 | 30 | ND | No | |
| 64 | a. R394Q; b. R394Q | NA | 20 | 0 | ND | ND | |||
| 65 | a. T403P; b. T403P | 3 wk | 0 | 0 | 350 | ND | |||
| 66 | a. V475AfsX17; b. V475AfsX17 | 2 mo | 2 | 1 | 26 | <2 | |||
| 67 | a. L411P; b. L411P | 1.5 y | 25930 | 0 | 900 | 29464 | No | ||
| 68‡ | a. S626X; b. S626X | 2 mo | 1836 | 0 | ND | ND |
a, One allele; AS/LS, atypical/leaky SCID; b, the other allele; NA, not available; ND, not determined; PID, primary immunodeficiency.
Patients with granuloma.
Patients treated with rituximab.
Maternal engraftment.
Siblings.
FIG 1.
Schematic representation of the experimental outline and readout. A, A mouse Abelson (Abl) virus–transformed pro–B-cell line deficient for mRag1 was infected with a retrovirus containing an inverted GFP cassette. Subclones with single-copy stable integrants were transduced with vectors expressing wild-type RAG1 or various hRAG1 mutations (see Table E1) and then treated with imatinib to promote cell differentiation and induction of RAG1 activity.21 B and C, The level of GFP expression indicated the recombinase activity level on imatinib stimulation of Rag1−/− Abl pro-B cells transduced with an empty vector, or with vectors encoding either for wild-type hRAG1 (hWT) or wild-type mRag1 (mWT; Fig 1, B) or for one representative hRAG1 mutation for each of the 5 different phenotypic subgroups of the disease (Fig 1, C). LTR, Long terminal repeats.
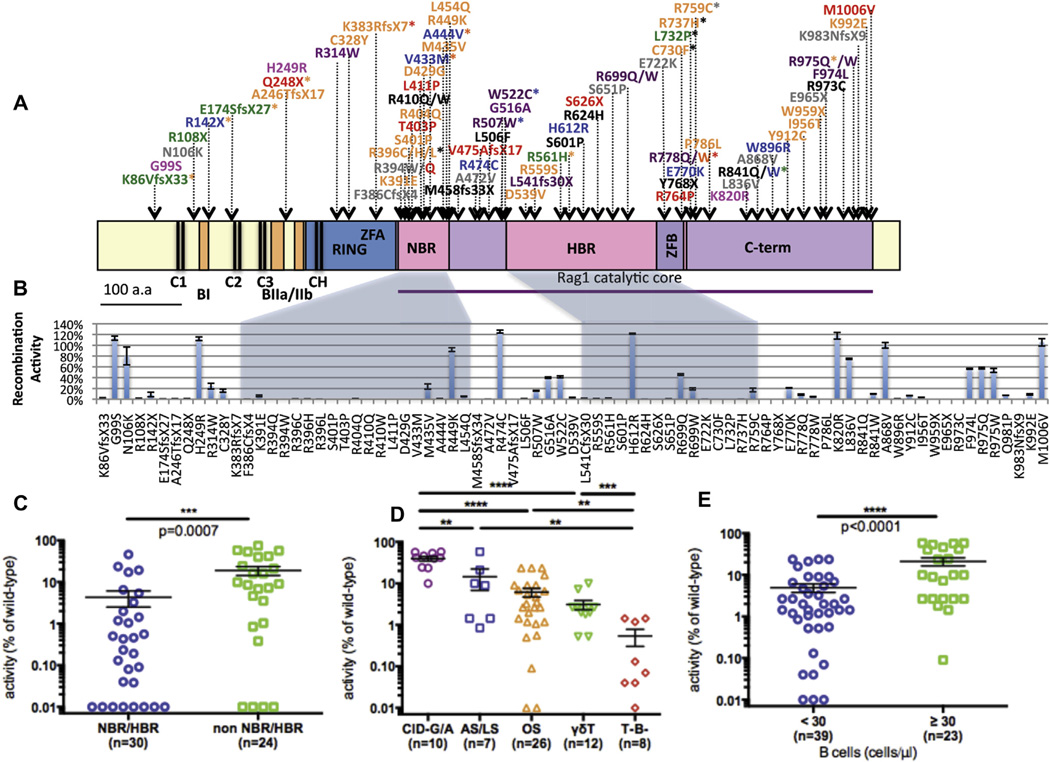
We investigated whether the specific location of RAG1 mutations might impinge on recombination activity. Fig 2, A, shows the location of the mutations analyzed in this study along the length of the hRAG1 protein; different colors were used to indicate the diverse clinical and immunologic phenotypes. For mutations associated with more than 1 phenotype, the color code corresponds to the least severe phenotype under the assumption that the mutation is potentially permissive for some V(D)J recombination activity. We observed a relative overrepresentation of mutations affecting the nonamer-binding region (NBR) of hRAG1 (Fig 2, B). Moreover, missense mutations in the NBR and in the heptamer-binding region (HBR) had a significantly lower activity level than mutations in other regions of the RAG1 protein (Fig 2, C), which is consistent with the critical roles played by the NBR and HBR domains in DNA recognition and binding.1 In contrast, mutations at the C-terminus of RAG1 and some missense mutations in the central region of the molecule showed significant residual VDJ recombination activity (Fig 2, B). These mutations have been identified mostly in patients with CID-G/A (ie, a less severe and often delayed-onset form of the disease), suggesting that mutations in the C-terminus of RAG1 are more tolerable than mutations in the NBR/HBR.
FIG 2.
Activity levels of 79 genetic variants of hRAG1. A, Schematic representation of 79 genetic variants of hRAG1 affecting the various domains: RING, zinc finger RING type domain (amino acids 168–283); ZFA, zinc finger A; NBR (amino acids 387–461); HBR (amino acids 531–763); ZFB, zinc finger B; and the core domain from amino acids 385 to 1011. The conserved cysteine (C) and histidine (H) residues are marked with black lines, and the basic domains are marked with bars. They are as follows: C1 (103/115); C2 (169/181); C3 (204/214) CH (269/275); CC (905/910); HH (940/945); BI (142–147); BIIa (219–225); BIIb (234–237); and BIII (244–257). The RAG1 variants are color coded, corresponding to the clinical phenotype of patients in which they were identified (red = T−/B− SCID, orange = OS, green = γδ-T, blue = atypical/leaky SCID, and purple = CID-G/A). The asterisk marks mutations associated with other phenotypes. Mutations in black correspond to alleles with the lower recombination activity that had been identified in patients who were compound heterozygous for RAG1 mutations. Known polymorphisms are indicated in pink, and gray is used to identify variants detected in patients for whom incomplete clinical and immunologic information was available. B, Bar diagram representing the activity level of various hRAG1 mutants relative to wild-type hRAG1. Values are expressed as percentages ± SEMs. For each mutant, 3 to 5 independent experiments were performed. Mutations falling in the NBR and HBR are contained in shaded areas. C, Recombination activity of missense mutations falling in the NBR/HBR versus other regions of hRAG1. D, Recombination activity of hRAG1 mutants identified in patients with a distinct clinical and immunologic phenotype. E, Recombination activity of hRAG1 mutants identified in patients with virtual lack of circulating B cells (<30 cells/µL) and in those with residual B cells (≥30 cells/µL). The Mann-Whitney U test was performed to demonstrate statistical significance for all the 1-tailed P values in the graphs: **P < .01, ***P < .001, and ****P < .0001.
Next, we analyzed correlation between recombination activity of hRAG1 mutant proteins and the patients’ clinical and immunologic phenotype (Fig 2, D). Recombination activity of those hRAG1 mutants that had been identified in patients belonging to various clinical subgroups were reported for each of the subgroups to avoid a bias in the statistical analysis of the data. VDJ recombination activity levels were greater for RAG1 mutations identified in patients with CID-G/A and were progressively lower in patients with more severe phenotypes, being extremely low for patients belonging to the T−B− SCID subgroup (Fig 2, D). Furthermore, significantly lower VDJ recombination activities of hRAG1 mutants were observed in patients with very low to absent circulating B cells (Fig 2, E). Altogether, these data indicate that there is a strong, although imperfect, correlation between the VDJ recombination activity of mutant hRAG1 proteins and the associated clinical and immunologic phenotype.
To determine whether differences in recombination activity of hRAG1 mutants might reflect different levels of protein expression, we performed Western blotting analysis of 41 RAG1 mutants introduced into Rag1−/− Abl pro-B cells. The same amount of cells (400,000 cells), of which greater than 90% expressed hCD2, were loaded in each lane, and adjusted RAG1 expression was calculated by using the loading control as a reference. Of 27 hRAG1 mutants in the NBR/HBR that were studied, 13 showed very low levels of protein expression (adjusted expression, <0.2; Fig 3, A and B). Variable levels of protein expression were observed for NBR mutants with different amino acid substitutions at the same codon (R394 and R396; Fig 3, A), yet all of them showed very low to undetectable VDJ recombination activity (see Table E1). Among 15 missense mutations located outside of NBR/HBR domains, 4 had very low levels of protein expression, and 7 had expression levels comparable with those of wild-type hRAG1 (Fig 3, C). Overall, levels of protein expression of hRAG1 mutants affecting the NBR/HBR regions were not significantly different than those of mutants affecting other domains (Fig 3, D). Furthermore, we observed only a modest correlation between RAG1 mutant protein expression and recombination activity (Fig 3, E).
FIG 3.
Protein expression of hRAG1 mutants. A–C, Protein expression of hRAG1 mutants affecting NBR (Fig 3, A), HBR (Fig 3, B), and non-NBR/HBR (Fig 3, C) domains. Expression of β-actin was used to normalize the density for each of the hRAG1 mutants. Results are shown as adjusted density (ImageJ). One representative of 2 immunoblots is shown. D, Adjusted density of hRAG1 protein expression of mutants affecting the NBR/HBR or non-NBR/HBR domains of the molecule. ns, Not significant. Statistical analysis was performed with the Mann-Whitney U test. E, Correlation between adjusted density of protein expression and recombination activity of RAG1 mutants (Spearman rs = 0.351, P = .023).
Comparison of recombination activity and predicted pathogenicity of hRAG1 mutants
Bioinformatics tools might help predict the pathogenicity of DNA variants and missense mutations in particular. To predict the pathogenicity of 63 hRAG1 missense variants, we used the Pathogenic Or Not Pipeline (PON-P), which integrates information provided by Phd-SNP, SNAP, PolyPhen-2, SIFT, and I-Mutant predictors into a single consensus prediction with a reliability score.30 PON-P assigned each hRAG1 variant to a specific class according to probability and reliability scores (P, pathogenic; N, neutral; and UV, unclassified variant). Most variants in class P had absent or negligible recombination activity, whereas most variants in class N had wild-type recombination activity (see Table E1). Overall, correlation between predicted pathogenicity and in vitro recombination activity was not perfect. However, among missense mutations located in the NBR, those affecting residues that are directly involved in DNA binding31 were assigned to class P by using PON-P, and they all showed absent or markedly decreased recombination activity (see Fig E2 in this article’s Online Repository at www.jacionline.org). In contrast, missense mutations in the more distal α-helix of the NBR, affecting residues that are not directly involved in DNA binding, were mostly assigned to class UV and had low but detectable recombination activity. Therefore PON-P can be informative in validating experimental results and predicting mutations in regions with distinct and imperative activities.
Analysis of rearrangements at the endogenous Ighc locus
Introduction of hRAG1 mutants into Rag1−/− Abl pro-B cells offers the opportunity to analyze VDJ recombination activity in a physiologic setting through the study of rearrangements at the endogenous Ighc locus. For this purpose, we used a semiquantitative deep-sequencing approach based on amplification of mRNA transcripts by using VH forward primers and CH reverse primers with a communal barcode (provided by iRepertoire) and use of the Roche 454 pyrosequencing platform. In particular, we tested the frequency, quality, and diversity of Ighc rearrangements induced by transduction of Rag1−/− Abl pro-B cells with retroviral vectors encoding either wild-type hRAG1 or mRag1 or 10 distinct hRAG1 mutants (K86Vfs33X, R142X, R314W, C328Y, M435V, G516A, R699Q, R841W, Y912C, and K992E). These mutants were selected because they occur in different domains of the protein, support different levels of VDJ recombination activity (as assessed based on GFP expression), and were identified in patients representative of the phenotypic spectrum of the disease. The total number of reads, as well as total and unique CDR-H3 sequences, were comparable between mRag1, hRAG1, and R699Q, whereas a reduced number of reads or no reads were obtained for all other mutants with lower activity levels than R699Q (see Table E2 in this article’s Online Repository at www.jacionline.org).
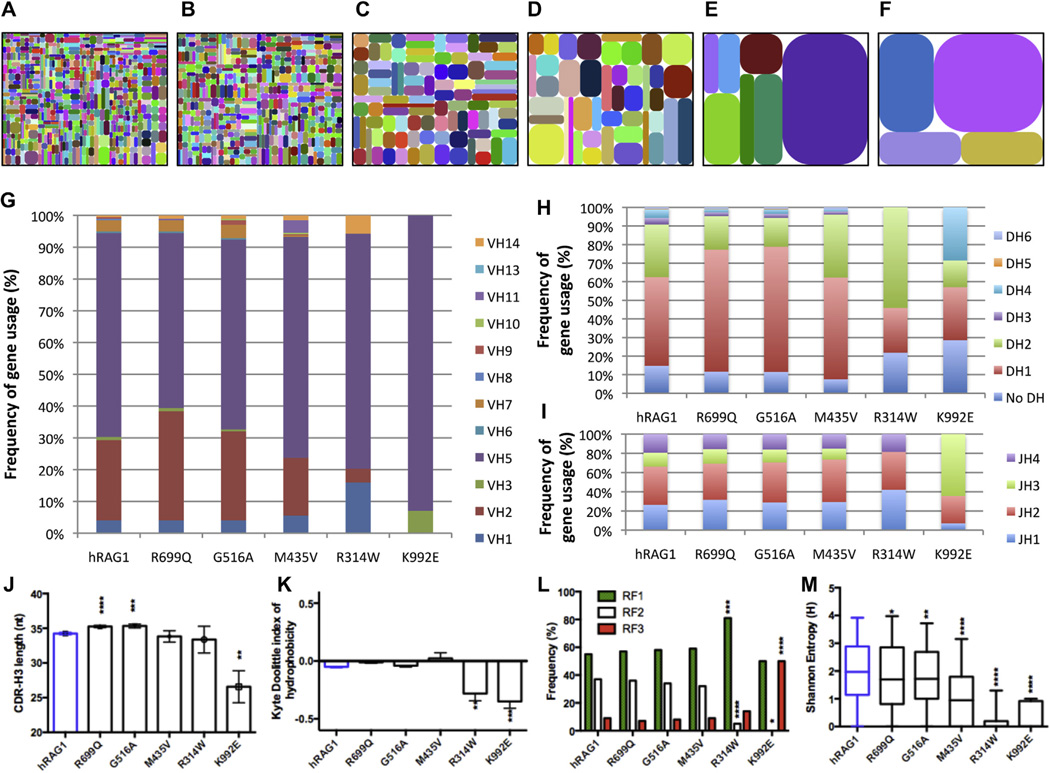
To assess whether we had collected a sufficient number of reads to cover the diversity of rearrangements induced by each mutant, we analyzed individual rarefaction curves in which the number of species (VDJ rearrangement products) is plotted as a function of the number of sequences obtained, thereby informing on the richness of the reads (see Fig E3, A, in this article’s Online Repository at www.jacionline.org). Although a very low number of reads was obtained for both the R314W and K992E mutants, the rarefaction curves for these mutants were flatter than for wild-type hRAG and for other mutations of the same sample size, indicating that an increase in sequence reads for R314W and K992E samples would not result in a significant increase in the number of diverse VDJ recombination products (see Fig E3, B). Furthermore, analysis of individual VH to JH pairing demonstrated progressive restriction of the Ighc rearrangement repertoire when moving from hRAG1 mutants with high to those with low VDJ recombination activity (Fig 4, A–F).
FIG 4.
Diversity and CDR-H3 characteristics and composition of the rearranged Ighc repertoire of hRAG1 mutations. A–F, Tree maps (iRepertoire) were generated to depict graphically the diversity and frequency of different V–J pairings induced by various hRAG1 mutants: wild-type hRAG1 (Fig 4, A); R699Q (Fig 4, B); G516A (Fig 4, C); M435V (Fig 4, D); R314W (Fig 4, E); and K992E (Fig 4, F). Each dot represents a unique VH-JH recombination, and the size of the dot indicates the relative frequency of that specific V-JH rearrangement. G–I, Bar diagrams representing the uses of VH (Fig 4, G), DH (Fig 4, H), and JH (Fig 4, I) genes for wild-type hRAG1 and various hRAG1 mutants. For the VH genes, only the genes that had positive values are included. J–M, Characterization of CDR-H3 sequences: average CDR-H3 length (± SEM; Fig 4, J); average index of hydrophobicity (± SEM) according to a normalized Kyte-Doolittle scale (Fig 4, K); differential use of 3 RFs (Fig 4, L); and average Shannon entropy index (± SEM) of the CDR-H3 loop (Fig 4, M). *P < .05, **P < .01, ***P < .001, and ****P < .0001 unpaired 2-tailed t test.
As expected,18,32 introduction of wild-type hRAG1 or mRag1 into Rag1−/− Abl pro-B cells induced Ighc rearrangements that often use the most proximal VH gene segment Ighv5-2 (see Fig E4 in this article’s Online Repository at www.jacionline. org). In addition to preferential use of this VH gene segment, skewing in the use of individual VH, DH, and JH genes was observed for all mutants tested and especially for the R314W and K992E mutants (Fig 4, G–I, and see Fig E5 in this article’s Online Repository at www.jacionline.org) and for DH use (Fig 4, H, and see Fig E5, C).
Differences in the composition and length of the CDR-H3 region might affect the ability of immunoglobulins to bind to antigens. Dynamic changes in CDR-H3 composition occur during differentiation from pro-B to mature B cells.33 In particular, immature B cells enriched for self-reactive specificities tend to have longer CDR-H3 sequences with presence of positively charged amino acid residues.34,35 Because immune dysregulation is a common feature in patients with hypomorphic RAG mutations, we sought to determine whether introduction of these mutants into Rag1−/− Abl pro-B cells induces expression of CDR-H3 sequences that differ in length and composition from those induced by wild-type hRAG1. There was a slight but significant increase in the length of CDR-H3 for both the R699Q (35.24 ± 8.76 nt) and G516A (35.34 ± 8.54 nt) mutants compared with hRAG1 (34.24 ± 8.76 nt; P < .0001 and P = .0007, respectively; Fig 4, J), mainly because of a longer DH gene segment (see Fig E6, A and B, in this article’s Online Repository at www. jacionline.org). However, the more severe K992E mutant, which is associated with OS, supported generation of Ighc rearrangements characterized by markedly shorter CDR-H3 sequence (26.57 ± 8.64 nt, P = .0055; Fig 4, J). A reduced number of N nucleotide additions at the VDJ recombination junctions (see Fig E6, A and C), a higher frequency of direct VH-to-JH rearrangements (see Fig E6, D), a significant decrease in the average DH length (see Fig E6, B), and a higher frequency of use of the shorter JH3 gene segment (Fig 4, I, and see Fig E5, D) all contributed to the shorter CDR-H3 sequences, likely reflecting the impaired function and abnormal VDJ recombination activity of the K992E mutant.
For the R699Q, G516A, and M435V mutants with higher levels of recombination activity, the average index of CDR-H3 hydrophobicity according to Kyte-Doolittle did not deviate much from neutral to slightly hydrophilic, which is similar to what was observed for wild-type hRAG1 (Fig 4, K).33 In contrast, a significant shift toward a charged/hydrophilic profile of CDR-H3 sequences was observed for the more severe R314W and K992E mutants (Fig 4, K), mainly because of abundance of arginine, asparagine, and aspartic acid residues (see Fig E7 in this article’s Online Repository at www.jacionline.org). The higher frequency of these amino acids resulted mainly from N1 additions in the case of the R314W mutant and from N2 additions for the K992E mutant (see Fig E8 in this article’s Online Repository at www.jacionline.org). Moreover, the nature of the RF, which is largely determined by DH genes, also contributed to the skewed use of these amino acids. In particular, the R314W mutant induced preferential use of the neutrally charged RF1 and reduced use of the hydrophobic RF2, whereas the K992E mutant induced more abundant use of the hydrophilic RF3 (Fig 4, L).
Finally, to analyze the diversity of amino acid composition at each residue of the CDR-H3, we used the Shannon entropy index (H), a measure of variability in which the higher the score, the more variable the amino acid use at each position. The CDR-H3 diversity was significantly reduced for the M435V, R314W, and K922E mutants but was only modestly decreased for R699Q and G516A mutants (Fig 4, M). Homozygosity for the K992E mutation in human subjects is associated with OS, whereas the R699Q and G516A mutations have been identified only in patients with CID-G/A (Table I). Altogether, these data indicate that restricted diversity in the immune repertoire correlates with severity of impairment of RAG1 function.
DISCUSSION
An important property of the Rag1−/− Abl pro–B-cell system described here is that it allows in-depth analysis of VDJ recombination activity at the endogenous Ighc locus before selection processes. As expected for pro-B cells, we observed that DH1 and DH4 (commonly known as DFL16 and DQ52, respectively) genes were preferentially targeted for rearrangement.36 Furthermore, significant skewing toward DH4 gene use was observed with the more severe K992E mutant, which reflects impairment of secondary D-replacement rearrangements.
Similarly, the most DH-proximal VH2 and VH5 genes were preferentially targeted in VH-DJH rearrangements, as also seen early during B-cell ontogeny.18,32 Even for these genes, a skewed use by various hRAG1 mutants was observed. Moreover, increased targeting was observed for the other DH-proximal VH11 and VH14 genes. Although the association of individual hRAG1 mutations with preferential targeting of specificVH genes has not been previously reported, significantly reduced DH-JH rearrangement in mice homozygous for a hypomorphic Rag1 mutation had been demonstrated.37 In addition, introduction of an OS-associated mutation into themRag1 gene induced selective impairment in the ability to target certain coding flanks at VH, DH and JH gene segments.38 Along with our findings, these data suggest that reduced RAG1 activity might directly alter the quality of endogenous VDJ rearrangements. Overall, hRAG1 mutants with lower recombination activity preferentially targeted the most proximal DH elements, whereas hRAG1 mutants with relatively higher recombination activity showed the ability to target DH distal VH gene segments, thereby allowing generation of a more diverse repertoire. This correlated with a milder clinical and immunologic phenotype in vivo, with preservation of B-cell development and detectable immunoglobulin levels.
Immune dysregulation is being increasingly recognized as an important clinical phenotype associated with hypomorphic RAG1 mutations (Table I).13–15 The R699Q and G516A mutants induced rearrangements with longer CDR-H3 sequences, a feature that has been previously identified in self-reactive B cells.34,35,39 These data suggest that some hRAG1 mutations might favor generation of a self-reactive B-cell repertoire by skewing the quality of VDJ rearrangements. On the other hand, generation of markedly shorter CDR-H3 sequences caused by increased use of the stop codon–rich RF3 has been observed for the K992E mutant identified in 2 patients with OS.
One caveat of the Rag1−/− Abl pro–B-cell system described here is that it allows in-depth analysis of the recombination properties of one mutant at a time. In contrast, many patients carry compound heterozygous mutations. Use of polycistronic vectors might allow analysis of the combined effect of 2 distinct mutations.
In conclusion, this study represents the most comprehensive study of the expression and function of hRAG1 mutants identified in patients with a spectrum of clinical and immunologic presentations. Using this platform, we have demonstrated genotype-phenotype correlation and have provided novel molecular insights that might help explain the phenotypic diversity of this disease. The results presented in this study could serve as a reference for recombination activity of known RAG1 variants. Moreover, the fluorescence-activated cell sorting–based assay described here could represent a helpful cellular platform to assess the pathogenicity of newly identified RAG1 variants. This would be especially important in the diagnostic approach to asymptomatic or presymptomatic subjects with RAG1 mutations, including those identified at birth through newborn screening. Note added in proofs: Yu et al40 have recently demonstrated restricted TCRβ diversity in patients with OS, and preserved diversity, but skewed usage of V, D and J elements in patients with CID-G/A.
Supplementary Material
Key messages.
We describe a cellular platform that permits rapid analysis of expression and recombination activity of RAG1 mutant variants.
RAG1 recombination activity correlates with the severity of the clinical and immunologic phenotype observed in vivo.
Mutant RAG1 proteins differ in the efficiency and quality of V(D)J recombination activity, indicating that skewing of the immune repertoire might occur independently from in vivo selection.
Acknowledgments
Supported in part by National Institutes of Health grants U54AI082973 (to L.D.N.) and 1P01AI076210-01A1 (to L.D.N., F.W.A., and R.S.G.). F.W.A is an investigator of the Howard Hughes Medical Institute. This work received additionally contributions from the March of Dimes (grant 1-FY-13-500 to L.D.N.), the Jeffrey Modell Foundation (to L.D.N.), the Translational Research Program grant TRP 2009 042809 (to L.D.N.), the Dubai Harvard Foundation for Medical Research (to L.D.N. and R.S.G.), and the Manton Foundation (to Y.N.L. and L.D.N.).
Y. N. Lee has received research support from the Manton Foundation and the March of Dimes and is employed by Boston Children’s Hospital. F. Frugoni, J. Puck, and F. Alt have received research support from the National Institutes of Health (NIH). J. E. Walter is employed by Massachusetts General Hospital and receives research support from the National Institute of Allergy and Infectious Disease. L. A. Henderson has received travel support from the American College of Rheumatology and the American Academy of Pediatrics. S.-Y. Pai is employed by Boston Children’s Hospital and receives research support from the Translational Investigator Service Award from Boston Children’s Hospital. P. Soler-Palacin has consultant arrangements with CSL Behring; has provided expert testimony on behalf of CSL Behring; has received research support from Baxter, CSL Behring, and Octapharma; has received payment for lectures from CSL Behring; and has received travel support from the European Society of Immunodeficiencies. R. S. Geha has received research support from the NIH and the Dubai Harvard Foundation for Medical Research. L. D. Notarangelo has received research support from the NIH, the Jeffrey Modell Foundation, the March of Dimes, the Dubai Harvard Foundation for Medical Research, and the Manton Foundation; has board memberships with the Program in Molecular and Cellular Medicine and Pediatric University Hospital “Meyer” in Florence, Italy; is employed by Boston Children’s Hospital; and has received royalties from UpToDate.
We thank Y. I. Avnir, PhD, for critical input in the design and data analysis for Ighc rearrangements determined by using deep sequencing. We thank H. C. Su, MD, PhD, for critical evaluation and care of patients. We also thank all the patients and their families who agreed to take part in this study.
Abbreviations used
- A-MuLV
Abelson murine leukemia virus
- CDR
Complementarity-determining region
- CID-G/A
Combined immune deficiency with granuloma and/or autoimmunity
- γδ-T
SCID with expansion of γδ T lymphocytes
- GFP
Green fluorescent protein
- HBR
Heptamer-binding region
- hRAG1
Human RAG1
- ICL
Idiopathic CD4+ T-cell lymphopenia
- mRag1
Mouse Rag1
- NBR
Nonamer-binding region
- OS
Omenn syndrome
- PON-P
Pathogenic Or Not Pipeline
- RAG
Recombination-activating gene
- RF
Reading frame
- RSS
Recombination signal sequence
- SCID
Severe combined immune deficiency
- TCR
T-cell receptor
Footnotes
Disclosure of potential conflict of interest:
The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 2.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz K, Gauss GH, Ludwig L, Pannicke U, Li Z, Lindner D, et al. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 4.Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, et al. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 5.Corneo B, Moshous D, Gungor T, Wulffraat N, Philippet P, Le Deist FL, et al. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T−B− severe combined immune deficiency or Omenn syndrome. Blood. 2001;97:2772–2776. doi: 10.1182/blood.v97.9.2772. [DOI] [PubMed] [Google Scholar]
- 6.Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. RAG-dependent primary immunodeficiencies. Hum Mutat. 2006;27:1174–1184. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- 7.Pasic S, Djuricic S, Ristic G, Slavkovic B. Recombinase-activating gene 1 immunodeficiency: different immunological phenotypes in three siblings. Acta Paediatr. 2009;98:1062–1064. doi: 10.1111/j.1651-2227.2009.01250.x. [DOI] [PubMed] [Google Scholar]
- 8.Dalal I, Tasher D, Somech R, Etzioni A, Garti BZ, Lev D, et al. Novel mutations in RAG1/2 and ADA genes in Israeli patients presenting with T-B-SCID or Omenn syndrome. Clin Immunol. 2011;140:284–290. doi: 10.1016/j.clim.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Cassani B, Poliani PL, Moratto D, Sobacchi C, Marrella V, Imperatori L, et al. Defect of regulatory T cells in patients with Omenn syndrome. J Allergy Clin Immunol. 2010;125:209–216. doi: 10.1016/j.jaci.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 11.de Villartay JP, Lim A, Al-Mousa H, Dupont S, Dechanet-Merville J, Coumau-Gatbois E, et al. A novel immunodeficiency associated with hypomorphic RAG1 mutations and CMV infection. J Clin Invest. 2005;115:3291–3299. doi: 10.1172/JCI25178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehl S, Schwarz K, Enders A, Duffner U, Pannicke U, Kuhr J, et al. A variant of SCID with specific immune responses and predominance of gamma delta T cells. J Clin Invest. 2005;115:3140–3148. doi: 10.1172/JCI25221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuetz C, Huck K, Gudowius S, Megahed M, Feyen O, Hubner B, et al. An immunodeficiency disease with RAG mutations and granulomas. N Engl J Med. 2008;358:2030–2038. doi: 10.1056/NEJMoa073966. [DOI] [PubMed] [Google Scholar]
- 14.De Ravin SS, Cowen EW, Zarember KA, Whiting-Theobald NL, Kuhns DB, Sandler NG, et al. Hypomorphic Rag mutations can cause destructive midline granulomatous disease. Blood. 2010;116:1263–1271. doi: 10.1182/blood-2010-02-267583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson LA, Frugoni F, Hopkins G, de Boer H, Pai SY, Lee YN, et al. Expanding the spectrum of recombination-activating gene 1 deficiency: a family with early-onset autoimmunity. J Allergy Clin Immunol. 2013;132:969–971.e2. doi: 10.1016/j.jaci.2013.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuijpers TW, Ijspeert H, van Leeuwen EM, Jansen MH, Hazenberg MD, Weijer KC, et al. Idiopathic CD4+ T lymphopenia without autoimmunity or granulomatous disease in the slipstream of RAG mutations. Blood. 2011;117:5892–5896. doi: 10.1182/blood-2011-01-329052. [DOI] [PubMed] [Google Scholar]
- 17.Lieber MR, Hesse JE, Lewis S, Bosma GC, Rosenberg N, Mizuuchi K, et al. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 18.Couedel C, Roman C, Jones A, Vezzoni P, Villa A, Cortes P. Analysis of mutations from SCID and Omenn syndrome patients reveals the central role of the Rag2 PHD domain in regulating V(D)J recombination. J Clin Invest. 2010;120:1337–1344. doi: 10.1172/JCI41305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Sanders CM, Yang Q, Schroeder HW, Jr, Wang E, Babrzadeh F, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc Natl Acad Sci U S A. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alamyar E, Giudicelli V, Li S, Duroux P, Lefranc MP. IMGT/HighV-QUEST: the IMGT(R) web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 2012;8:26. [Google Scholar]
- 22.Rogosch T, Kerzel S, Hoi KH, Zhang Z, Maier RF, Ippolito GC, et al. Immunoglobulin analysis tool: a novel tool for the analysis of human and mouse heavy and light chain transcripts. Front Immunol. 2012;3:176. doi: 10.3389/fimmu.2012.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:1–9. [Google Scholar]
- 24.Avila EM, Uzel G, Hsu A, Milner JD, Turner ML, Pittaluga S, et al. Highly variable clinical phenotypes of hypomorphic RAG1 mutations. Pediatrics. 2010;126:e1248–e1252. doi: 10.1542/peds.2009-3171. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Quintal L, Atkinson A, Williams B, Grunebaum E, Roifman CM. Novel RAG1 mutation in a case of severe combined immunodeficiency. Pediatrics. 2005;116:e445–e449. doi: 10.1542/peds.2005-0369. [DOI] [PubMed] [Google Scholar]
- 26.Asai E, Wada T, Sakakibara Y, Toga A, Toma T, Shimizu T, et al. Analysis of mutations and recombination activity in RAG-deficient patients. Clin Immunol. 2011;138:172–177. doi: 10.1016/j.clim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Felgentreff K, Perez-Becker R, Speckmann C, Schwarz K, Kalwak K, Markelj G, et al. Clinical and immunological manifestations of patients with atypical severe combined immunodeficiency. Clin Immunol. 2011;141:73–82. doi: 10.1016/j.clim.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Safaei S, Pourpak Z, Moin M, Houshmand M. IL7R and RAG1/2 genes mutations/polymorphisms in patients with SCID. Iran J Allergy Asthma Immunol. 2011;10:129–132. [PubMed] [Google Scholar]
- 29.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4:31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 30.Olatubosun A, Valiaho J, Harkonen J, Thusberg J, Vihinen M. PON-P: integrated predictor for pathogenicity of missense variants. Hum Mutat. 2012;33:1166–1174. doi: 10.1002/humu.22102. [DOI] [PubMed] [Google Scholar]
- 31.Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, et al. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW., Jr Development of the expressed Ig CDR-H3 repertoire is marked by focusing of constraints in length, amino acid use, charge that are first established in early B cell progenitors. J Immunol. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- 34.Radic MZ, Weigert M. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu Rev Immunol. 1994;12:487–520. doi: 10.1146/annurev.iy.12.040194.002415. [DOI] [PubMed] [Google Scholar]
- 35.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 36.Tsukada S, Sugiyama H, Oka Y, Kishimoto S. Estimation of D segment usage in initial D to JH joinings in a murine immature B cell line. Preferential usage of DFL16.1, the most 5’ D segment and DQ52, the most JH-proximal D segment. J Immunol. 1990;144:4053–4059. [PubMed] [Google Scholar]
- 37.Giblin W, Chatterji M, Westfield G, Masud T, Theisen B, Cheng HL, et al. Leaky severe combined immunodeficiency and aberrant DNA rearrangements due to a hypomorphic RAG1 mutation. Blood. 2009;113:2965–2975. doi: 10.1182/blood-2008-07-165167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong SY, Lu CP, Roth DB. A RAG1 mutation found in Omenn syndrome causes coding flank hypersensitivity: a novel mechanism for antigen receptor repertoire restriction. J Immunol. 2008;181:4124–4130. doi: 10.4049/jimmunol.181.6.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zemlin M, Ippolito GC, Zemlin C, Link J, Monestier M, Schroeder HW., Jr Adult lupus-prone MRL/MpJ2+ mice express a primary antibody repertoire that differs in CDR-H3 length distribution and hydrophobicity from that expressed in the C3H parental strain. Mol Immunol. 2005;42:789–798. doi: 10.1016/j.molimm.2004.07.049. [DOI] [PubMed] [Google Scholar]
- 40.Yu X, Almeida J, Darko S, van der Burg M, DeRavin SS, Malech H, et al. Human syndromes of immune deficiency and dysregulation are characterized by distinct defects in T-cell receptor repertoire development. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2013.11.018. [in press]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.