Abstract
The pathogenic yeast Candida glabrata exhibits innate resistance to fluconazole, the most commonly used antifungal agent. By screening a library of 9,216 random insertion mutants, we identified a set of 27 genes which upon mutation, confer altered fluconazole susceptibility in C. glabrata. Homologues of three of these genes have been implicated in azole and/or drug resistance in Saccharomyces cerevisiae: two of these belong to the family of ABC transporters (PDR5 and PDR16), and one is involved in retrograde signaling from mitochondria to nucleus (RTG2). The remaining 24 genes are involved in diverse cellular functions, including ribosomal biogenesis and mitochondrial function, activation of RNA polymerase II transcription, nuclear ubiquitin ligase function, cell wall biosynthesis, and calcium homeostasis. We characterized two sets of mutants in more detail. Strains defective in a putative plasma membrane calcium channel (Cch1-Mid1) were modestly more susceptible to fluconazole but showed a significant loss of viability upon prolonged fluconazole exposure, suggesting that calcium signaling is required for survival of azole stress in C. glabrata. These mutants were defective in calcium uptake in response to fluconazole exposure. The combined results suggest that, in the absence of Ca2+ signaling, fluconazole has a fungicidal rather than a fungistatic effect on C. glabrata. The second set of mutants characterized in detail were defective in mitochondrial assembly and organization, and these exhibited very high levels of fluconazole resistance. Further analysis of these mutants indicated that in C. glabrata a mechanism exists for reversible loss of mitochondrial function that does not involve loss of mitochondrial genome and that C. glabrata can switch between states of mitochondrial competence and incompetence in response to fluconazole exposure.
The incidence of opportunistic fungal infections in the immunocompromised patient population has increased significantly over the last two decades (62, 65). Candida species account for the majority of fungal infections, and these are primarily due to Candida albicans (47, 65). Coinciding with the introduction and widespread prophylactic use of the triazole antifungal fluconazole, there has been a notable increase in the relative proportion of infections due to non-C. albicans Candida species: from 1989 to 1999, the incidence of nosocomial C. albicans bloodstream infections dropped fourfold, whereas the incidence of non-C. albicans bloodstream infections in the same population remained strikingly constant (43, 65). Whereas the rate of infections due to all non-C. albicans species remained constant, there was a statistically significant increase in the rate of infections due to Candida glabrata, which is now the second most common cause of bloodstream and mucosal infections in the United States (6, 48, 65, 69). Perhaps related to its increasing prevalence, C. glabrata is innately resistant to fluconazole (49) and indeed is often isolated as a replacement species in patients receiving fluconazole treatment (47, 48, 69).
The triazole class of compounds are key antifungal agents, with fluconazole being the most commonly used antifungal agent to treat Candida infections due to its high efficiency, low toxicity, and oral availability (18). The triazoles target cytochrome P-450-dependent C14 lanosterol demethylase (encoded by ERG11), an essential enzyme in the ergosterol biosynthesis in fungi (24, 31, 72). The nitrogen atoms in the azole ring bind to the heme group in the active site of the enzyme, thereby blocking the binding of the normal substrate (72). The efficacy of triazole compounds has been proposed to be due to both the depletion of ergosterol in the plasma membrane and the intracellular accumulation of toxic 14α-methylated sterol intermediates, ultimately leading to growth arrest but not cell death (26, 29, 31, 58).
The molecular mechanism of azole resistance has been extensively studied both in the clinical isolates of C. albicans, as well as in the model yeast organism, Saccharomyces cerevisiae. The principal biochemical mechanisms associated with azole resistance include drug target (ERG11) amplification, structural modifications in the Erg11p enzyme, mutations in other ergosterol biosynthesis genes that result in the accumulation of alternative nontoxic sterols, and reduced drug uptake and enhanced drug efflux due to the overexpression of membrane-associated multidrug resistance transporters (25, 27, 30, 40, 51, 61, 66, 70, 71). Recently, studies of global gene expression analysis in the azole-resistant strains of C. albicans or in S. cerevisiae strains exposed transiently to azoles demonstrated upregulation of various genes previously implicated in azole resistance (3, 10, 54, 55). In addition, these studies uncovered new genes involved in oxidative stress response that seem to be coordinately regulated with the multidrug resistance transporter genes, CDR1, CDR2, and CaMDR1 (55).
In addition to the moderate resistance to fluconazole displayed by clinical isolates, C. glabrata, like C. albicans, can become even more resistant after selection in the presence of drug. The major described mechanisms of acquired azole resistance in C. glabrata clinical isolates include transcriptional upregulation of the CgERG11 gene and increased drug efflux due to the upregulation of CDR1 and CDR2 (40, 57, 67).
In addition, a high frequency of acquired azole resistance in vitro in C. glabrata has been linked to a loss of mitochondrial function (11, 56). Although the precise molecular mechanism underlying fluconazole resistance and dysfunctional mitochondria in C. glabrata is not well understood, a likely mechanism involves the over expression in petites of the multidrug resistance pumps, Cdr1p and Cdr2p (56). This is consistent with reports for S. cerevisiae which demonstrate that dysfunctional mitochondria send a signal to the nucleus via a retrograde signaling pathway that leads to the over expression of many genes, including the pleiotropic drug resistance transcriptional factors, PDR1 and PDR3 (21, 73). The activation of PDR3 further leads to transcriptional induction of genes encoding multidrug resistance pumps, including PDR5 (an S. cerevisiae homolog of Candida CDR1) (2, 21, 51, 73). It has also been suggested that functional mitochondria increase azole susceptibility due to the conversion, under conditions of Erg11p inhibition, of nontoxic sterol intermediates to toxic sterol compounds (25, 29). Despite a clear association in vitro between mitochondrial dysfunction and fluconazole resistance, the importance of mitochondrial dysfunction in clinical isolates is not clear.
As mentioned above, the azoles are fungistatic rather than fungicidal, and this lack of fungicidal activity could limit the efficacy of this class of compounds in long-term treatment regimes, even providing a fertile selective environment for the emergence of drug resistant strains. Recently, the immunosuppressive drugs FK506 and cyclosporine have been shown to act synergistically with azole compounds, converting azoles from fungistatic to fungicidal drugs (9, 14, 37) without significantly altering the susceptibility (50% MIC [MIC50]) to the drug per se. Both FK506 and cyclosporine act via the inhibition of a highly conserved Ca2+/calmodulin-dependent serine/threonine protein phosphatase, calcineurin (34), a key component of calcium signaling in yeast (22). Calcineurin is required to survive membrane stress in yeast (7, 9) and is required for tolerance of C. albicans to antifungal compounds (5, 59). Calcium signaling in yeast controls diverse cellular processes, including pheromone arrest, morphogenesis, and cell cycle regulation. It is believed that membrane perturbation by azole treatment leads to an influx of extracellular calcium (Ca2+) through the plasma membrane or its release from intracellular stores into the cytosol (14). Intracellular Ca2+ is compartmentalized in different organelles, including the endoplasmic reticulum (ER), mitochondria, and vacuole.
Cells have evolved many regulatory mechanisms to maintain the cytosolic concentration of Ca2+ required for intracellular signaling by coordinating the efflux and influx of Ca2+ from the extracellular environment (50). One of the mechanisms of Ca2+ influx through the plasma membrane is referred to as capacitative Ca2+ entry (46) and is mediated by a high-affinity Ca2+ channel composed of Cch1p and Mid1p (15, 23, 44). Cch1p is homologous to the catalytic subunit of voltage-gated Ca2+ channels in mammalian cells, and Mid1p is an N-glycosylated, integral plasma membrane protein that functions as the regulatory subunit of this channel (20). Both Cch1p and Mid1p proteins have been shown to be necessary for Ca2+ influx and survival of ER stress in S. cerevisiae (7, 15, 23, 36, 44).
We have utilized here a functional genomic approach to identify the major factors responsible for the limited fluconazole susceptibility of C. glabrata by identifying mutants that display an altered fluconazole susceptibility. One important class of mutants that led to high-level fluconazole resistance resulted from the disruption of genes important in mitochondrial function. We report here that high-frequency (mitochondrion-related) acquired fluconazole resistance is always associated with the loss of the mitochondrial function but, in many isolates, not with the loss of the mitochondrial genome. In such isolates, the mitochondrion-incompetent fluconazole resistance phenotype is in fact a revertible phenotype. In addition, the rate at which mitochondrial incompetence can be reacquired in these isolates is much higher than the expected mutation rate, suggesting that C. glabrata can switch between states of mitochondrial competence and incompetence in response to azole exposure.
We also found, as has been previously reported for C. glabrata (40, 57), that multidrug transporters have a significant role in reducing fluconazole susceptibility. In addition, we identified two mutants implicating calcium signaling with fluconazole susceptibility in C. glabrata. Our study demonstrates for the first time that C. glabrata mutants defective in the Cch1/Mid1p Ca2+ channel lose viability upon prolonged fluconazole exposure, suggesting that calcium signaling is required to survive fluconazole stress in C. glabrata.
MATERIALS AND METHODS
Media and chemicals.
Yeast media were prepared as described previously (63) and 2% agar was added for plates. Yeast extract-peptone-dextrose (YPD) medium contained 10 g of yeast extract/liter and 20 g of peptone/liter supplemented with 2% glucose. Yeast extract-peptone-glycerol (YPG) medium contained 10 g of yeast extract/liter, 20 g of peptone/liter, and 3% glycerol. Selective medium contains yeast nitrogen base supplemented with 0.6% Casamino Acids. Bacterial media was prepared as described previously (1) and 1.5% agar was used for plates. Luria-Bertani (LB) medium contained yeast extract 5 of g/liter, Bacto Peptone (10 g/liter), and NaCl 10 of g/liter supplemented with 30 μg of kanamycin/ml (Km30) where needed. Phosphate-buffered saline (PBS) is composed of 8 of g of NaCl/liter, 0.2 g of KCl/liter, 1.65 g of Na2HPO4·7H2O/liter, and 0.2 g of KH2PO4/liter. YPD plates containing various concentrations of fluconazole were prepared by adding appropriate volume of fluconazole (Diflucan-Pfizer) from a stock solution of 2 mg/ml.
Strains and growth conditions.
Escherichia coli strain BW23473 (Δlac-169 robA1 creC510 hsdR514 ΔuidA::pir endA recA1) (39) was used to rescue the plasmids from C. glabrata insertion mutants. C. glabrata strains BG2 and BG14 (ura3Δ::Tn903Neor) were used for all of the experiments described. Yeast cells were routinely grown at 30°C on standard S. cerevisiae media (63), and E. coli strain was grown at 37°C on yeast extract-tryptone media.
Tn7 mutagenesis.
In vitro mutagenesis of plasmids with a modified Tn7 transposon was carried out as described previously (8). Briefly, we mutagenized fosmid DNAs carrying C. glabrata genomic DNA by using Tn7 in vitro. Pools of insertions in a given fosmid were recovered in Escherichia coli, and mutagenized fosmid DNA was prepared. Genomic DNA was released from the fosmid backbone by restriction endonuclease digestion and introduced into the genome of C. glabrata. The majority of insertions are double crossovers that replace the wild-type allele with a disrupted allele. A total of 10% of insertions are nonhomologous ectopic integrants. These are easily identified (see below) by PCR and were excluded from further analysis.
Yeast transformation.
C. glabrata cells were transformed as described previously (16). Briefly, cells were grown to early log phase in YPD, harvested, and washed with an equal volume of sterile water. The cells were then resuspended in a 1/100 volume of 100 mM lithium acetate, and 50-μl aliquots were used for each transformation. To each tube containing 50 μl of cell suspension, 240 μl of 50% PEG (3500) was added, followed by 36 μl of 1 M lithium acetate, 50 μg of heat-denatured salmon sperm DNA, and 50 μl of transforming DNA. Tubes containing transforming mix were incubated at 30°C for 45 min, and 43 μl of dimethyl sulfoxide (DMSO) was added before heat shock treatment at 42°C for 15 min. The cells were centrifuged, resuspended in H2O, and plated on selective media.
Screening of mutant library.
Mutants were grown in 96-well plates at 30°C for overnight. A 360× dilution from overnight cultures was made in PBS and 5 μl of this cell suspension (about 5,000 cells) was spotted onto agar plates containing different concentrations of fluconazole with a 96-pin replicator. Growth phenotype of mutants was scored after incubation at 30°C for 2 days.
Fluconazole susceptibility.
Susceptibility of C. glabrata mutants to fluconazole was tested by using the broth microdilution assay according to NCCLS protocol M27-A (42) with morpholinepropanesulfonic acid-buffered (pH 6.2) RPMI 1640 medium. Essentially, 104 cells were inoculated in RPMI 1640 medium supplemented with 0.5% ammonium sulfate. Endpoint readings were measured with a microplate reader after 24 h of growth with shaking at 35°C. A fluconazole concentration yielding 80% growth inhibition compared to the growth in drug-free medium was defined as the MIC at which 80% of the isolates tested are inhibited (MIC80). These assays were carried out in duplicate. Susceptibility of the C. glabrata mutants was also tested qualitatively by spotting serial dilutions of yeast cells onto YPD agar plates containing different concentrations of fluconazole. The yeast strains were grown overnight at 30°C in YPD liquid medium, cells were diluted to 2 × 108 cells/ml in PBS, and 5 μl of the cell suspension and 10-fold serial dilutions of the yeast cells were spotted onto different plates, followed by incubation for 1 to 2 days at 30°C. The fluconazole breakpoint on YPD plates containing fluconazole was considered to be the drug concentration at which no mitochondrion-competent individual colonies were observed.
Mapping of Tn7 insertion.
To map Tn7 insertion in C. glabrata mutants, Tn7 transposon from the disrupted locus was rescued along with the flanking genomic fragment. Briefly, genomic DNA was isolated from each mutant, digested with either MfeI or SpeI enzyme, recircularized, and transformed into BW23473 cells by electroporation as described previously (8). The transformants were selected on LB plates containing kanamycin (30 μg/ml), and DNA was prepared by using a Qiagen Miniprep kit. The resulting circular plasmid contains the Tn7 transposon flanked on either side by the gene it disrupts in the genome of C. glabrata mutant. Sequence analysis of the rescued plasmids with outward primers from Tn7 right and left ends revealed the identity of the disrupted genes. The sequence obtained from each plasmid (∼1.2 kb) was blasted against the S. cerevisiae genome database (http://seq.yeastgenome.org/cgi-bin/SGD/nph-blast2sgd) and the C. glabrata Genolevures database (http://cbi.labri.fr/Genolevures/advanced_blast.php3), and the mutant and open reading frame names were designated accordingly.
Regeneration of mutants.
Genomic DNA was isolated from mutant strain and digested with either MfeI or SpeI enzyme, recircularized, and transformed into BW23473 cells by electroporation as described previously (8). The transformants were selected on LB plates containing kanamycin (30 μg/ml), and DNA was prepared by Qiagen Miniprep kit. The rescued plasmid was sequenced with the outward primers from the Tn7 right and left ends. Using the sequence information from each end of the transposon, primers were designed corresponding to the sequence of the rescued genomic loci, and PCR was carried out on genomic DNA from mutants. The presence of a large band corresponding to the locus with a Tn7 insertion indicated disruption of the locus. The presence of both a small wild-type band and a large band carrying Tn7 in these mutants indicated that the transposon had integrated into the genome by nonhomologous integration. Such nonhomologous integration mutants were not further analyzed (for a detailed description of the mutagenesis and analysis of insertion mutants, see reference 8). The original Tn7 insertion mutation was regenerated by transforming MfeI- or SpeI-digested rescued plasmid into C. glabrata BG14 strain. Transformants were colony purified on synthetic minimal medium plates lacking uracil; for all insertions, disruption of the wild-type locus was confirmed by PCR as described above, and fluconazole susceptibility was scored by spotting cells on plates containing fluconazole.
Consideration of multiple mutants derived from a single fosmid.
The expectation is that if a fosmid contains a gene that can mutate to fluconazole susceptibility, then in a pool of 100 transformants derived from that particular fosmid there will likely be multiple fluconazole-susceptible mutants. These multiple mutants most likely represent independent insertions in the same gene. Since our mutant library represents only 20 to 25% of the genome, most fosmids analyzed here will not physically overlap. Therefore, the number of fosmids from which mutants with a phenotype were derived, rather than the number of mutants themselves, gives a good indication of the number of loci identified. This hypothesis was tested experimentally by rescuing multiple fluconazole-susceptible or -resistant mutants from 10 different fosmids; the Tn7 insertions were always found in the same gene in all of the fluconazole-susceptible or -resistant mutants derived from the same fosmid. Thus, for the analysis of fluconazole-susceptible mutants, one representative mutant per fosmid was selected for further characterization.
DAPI staining.
To stain cells with DAPI (4′,6′-diamidino-2-phenylindole), 1 ml of overnight culture was taken and then washed once with PBS, and the cells were suspended in 1 ml of 4% p-formaldehyde. After incubation at room temperature for 2 h, the cells were washed thrice with PBS and resuspended in PBS. Then, 100 μl of cell suspension was incubated with 1 μl of DAPI (0.2 mg/ml) for 30 to 40 min, and the cells were visualized by fluorescence microscopy.
Trypan blue exclusion assay.
Cells were grown in YPD medium at 30°C overnight and inoculated to an optical density at 600 nm (OD600) of 0.01 in minimal medium containing different concentrations of fluconazole. After 24 h of incubation, 1 ml of cells was harvested by centrifugation, washed with PBS, resuspended in PBS, and stained with 0.4% trypan blue for 5 to 10 min. A minimum of 300 cells (viable [unstained] and dead [stained] cells) was counted microscopically for each data set.
Viable count assay.
Cells were grown in YPD medium at 30°C overnight and inoculated to an OD600 of 0.01 in minimal medium containing different concentrations of fluconazole with or without FK506. After 24 h of incubation, 100 μl of cell suspension was taken and diluted in PBS, and the total number of cells was counted with hemocytometer. Appropriate dilutions were plated on YPD and YPG plates to assess the number of viable cells. CFU on YPG plates were counted after 2 days of incubation at 30°C as the final readout, although essentially the same number of CFU was obtained on both YPG and YPD plates if small colonies (petites, as tested by their inability to grow on glycerol) from YPD plates were excluded from the final viable cell counts.
45Ca2+ accumulation assay.
The total intracellular 45Ca2+ levels were measured as described previously (7). Briefly, cells were grown in YPD to log phase, harvested, washed, and suspended in YPD containing 2 μCi of 45CaCl2/ml (ICN Biomedicals). Fluconazole and FK506 were added to a final concentration of 64 and 2 μg/ml, respectively. After the cells were grown at 30°C for 4 h, cells were collected on GF/F filters (Whatman). The filters were washed thrice with buffer (10 mM CaCl2 and 5 mM HEPES-NaOH [pH 6.5]) and then dried at 85°C for 1 to 2 h. The cellular accumulation of 45Ca2+ was quantitated by liquid scintillation counting.
RESULTS
Construction and screening of a C. glabrata mutant library.
A library of 9,216 random insertion mutants covering 25% of the genome of C. glabrata was generated through homologous recombination of Tn7-mutagenized, linearized C. glabrata genomic fragments (8). This mutant library was screened individually in a 96-well format for strains that displayed altered susceptibility to fluconazole. The C. glabrata wild-type strain BG2 showed robust growth on agar plates containing 8 μg of fluconazole/ml, partial inhibition of growth on agar plates containing 16 μg of fluconazole/ml, and essentially no growth on plates containing fluconazole at a concentration of 32 μg/ml (Fig. 1). The mutant library was screened for two classes of mutants: hypersusceptible mutants that showed growth defects on plates with 8 μg of fluconazole/ml and resistant mutants that displayed robust growth in the presence of 32 μg of fluconazole/ml.
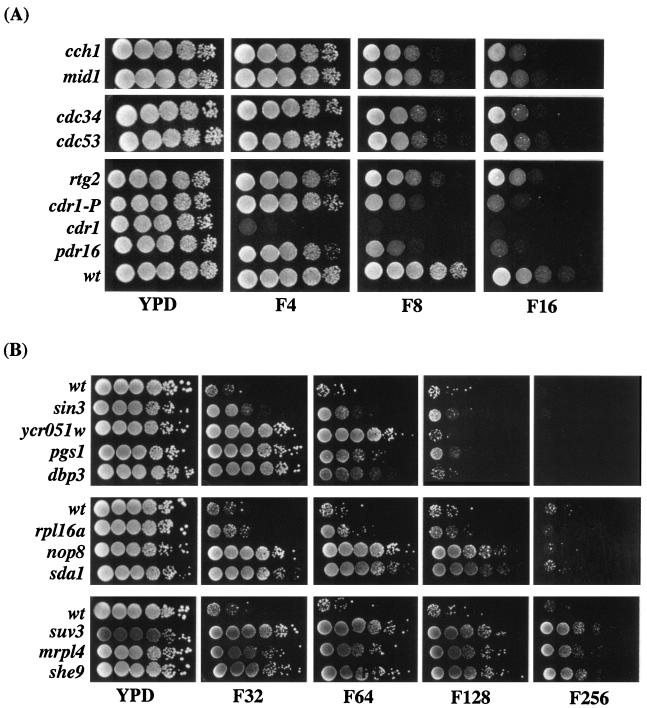
FIG. 1.
Fluconazole susceptibility profiles of C. glabrata mutants. Wild-type and mutant strains were grown in YPD for overnight at 30°C, the OD600 was normalized, and 5 μl of 10-fold serial dilutions were spotted onto YPD plates with or without various concentrations of fluconazole (4 to 256 μg/ml) as indicated. Plates were photographed after 2 days of growth at 30°C. (A) Growth profiles of fluconazole-susceptible mutants on different concentrations of fluconazole. cdr1-P mutant has a Tn7 insertion in the promoter region of CDR1 and displays susceptibility to 8 μg of fluconazole/ml compared to the cdr1 mutant (Tn7 insertion in the open reading frame), which is hypersusceptible even to 4 μg of fluconazole/ml. (B) Growth profiles of fluconazole-resistant mutants in the presence of various concentrations of fluconazole.
The screen yielded 73 hypersusceptible mutants, corresponding to 19 genomic loci, that showed an increased susceptibility to fluconazole (8 μg/ml). Of these 73 mutants, 14 were also unable to grow at 4 μg of fluconazole/ml, whereas the rest showed growth phenotypes only at a concentration of 8 μg of fluconazole/ml (Fig. 1A and data not shown). Two were found to be due to nonhomologous integration events (see Materials and Methods) and were not further analyzed; the remaining 17 were analyzed as described in next section.
The screen for fluconazole-resistant mutants yielded 68 mutants (corresponding to 26 loci) that displayed robust growth in the presence of 32 μg of fluconazole/ml (data not shown). A total of 39 mutants (representing 13 loci) showed resistance to higher levels of fluconazole (64 μg/ml) and were selected for further analysis (Fig. 1B and data not shown). Of these, two were found to be due to nonhomologous integration events (see Materials and Methods) and were not further analyzed. Within the class of mutants resistant to 64 μg of fluconazole/ml, 22 of the mutants (all derived from one mutagenized fosmid) were found to be in the same locus, a 2-kb noncoding region. The sequence of this intergenic fragment showed no homology with or similarity to known sequences in basic local alignment search tool (BLAST) searches (data not shown), and these mutants have not been further analyzed here. The remaining 15 mutants were tested for resistance to fluconazole concentrations ranging from 128 to 512 μg/ml. Of the 15 mutants (representing 10 loci) in total, 6 mutants (representing four loci) were resistant to 64 μg of fluconazole/ml, 5 mutants (representing three loci) were resistant to 128 μg of fluconazole/ml, and 4 mutants (representing three loci) were resistant to 256 μg of fluconazole/ml (Table 2). One mutant for each locus (10 total) was chosen and analyzed as described in the following sections.
TABLE 2.
Mutants identified in a screen for fluconazole resistance in C. glabrataa
| Wild type or mutant | Fluconazole MIC80 (μg/ml) | Breakpoint on fluconazole plates (μg/ml) | CAGL-CDS (C. glabrata CDS) | Position (aa) of Tn7 insertion | Protein length (aa) |
|---|---|---|---|---|---|
| Wild type | 16 | 16 | |||
| Mutants | |||||
| Ribosomal biogenesis | |||||
| Cgdbp3 | 32 | 64 | CDS1837.1 | 101 | 540 |
| Cgrpl16a | 128 | 128 | CDS4527.1 | 190 | 190 |
| Cgnop8 | 128 | 128 | CDS1989.1 | 169 | 514 |
| Mitochondrial biogenesis | |||||
| Cgsuv3 | 256 | 256 | CDS1015.1 | 53 | 724 |
| Cgmrpl4 | 128 | 256 | CDS3457.1 | 278 | 315 |
| Cgshe9 | 128 | 256 | CDS2737.1 | 126 | 405 |
| Others | |||||
| Cgsin3 | 64 | 64 | CDS0107.1 | 845 | 1,493 |
| Cgsda1 | 16 | 128 | CDS0953.1 | Promoter | 745 |
| Cgpgs1 | 128 | 64 | CDS2039.1 | 45 | 506 |
| Cgycr051w | 64 | 64 | CDS4399.1 | 84 | 206 |
aa, amino acids.
We verified that the mutants we identified showed changes in fluconazole susceptibility as measured by the standard MIC assay (42). Overall, the breakpoint as measured in liquid correlated extremely well with that seen on plates (Tables 1 and 2). The one significant exception was a mutant in the SDA1 gene that showed resistance on plates containing 128 μg of fluconazole/ml, whereas the breakpoint measured in liquid was no different than wild type (Table 2).
TABLE 1.
Mutants identified in a screen for fluconazole sensitivity in C. glabrata
| Wild type or mutant | Fluconazole MIC80 (μg/ml) | Breakpoint on fluconazole plates (μg/ml) | CAGL-CDS (C. glabrata CDS) | Position (aa)a of Tn7 insertion | Protein length (aa) |
|---|---|---|---|---|---|
| Wild type | 16 | 16 | |||
| Mutants | |||||
| ABC transporters | |||||
| Cgcdr1 | 4 | 4 | CDS0106.1 | 141 | 1,499 |
| Cgpdr16 | 8 | 8 | CDS3202.1 | Promoter | 344 |
| Calcium transporters | |||||
| Cgcch1 | 8 | 8 | CDS0026.1 | 1,137 | 2,124 |
| Cgmid1 | 8 | 8 | CDS1553.1 | 316 | 591 |
| RNA polymerase II coactivators | |||||
| Cgspt20 | 8 | 8 | CDS1138.1 | 22 | 685 |
| Cgsrb8 | 8 | 8 | CDS0156.1 | 745 | 1,358 |
| Cgrgr1 | 8 | 8 | CDS0391.1 | 67 | 1,037 |
| Cgnut1 | 8 | 8 | CDS0322.1 | 502 | 1,099 |
| Cgrtg2 | 16 | 8 | CDS1501.1 | 280 | 600 |
| Nuclear ubiquitin complex | |||||
| Cgcdc34 | 8 | 8 | CDS3640.1 | 281 | 295 |
| Cgcdc53 | 8 | 8 | CDS0757.1 | 360 | 822 |
| ATPases | |||||
| Cgstv1 | 8 | 8 | CDS0587.1 | 213 | 910 |
| Cgvma13 | 4 | 8 | CDS 2045.1 | 89 | 505 |
| Miscellaneous | |||||
| Cgynr047w | 8 | 8 | CDS0607.1 | 107 | 895 |
| Cgecm7 | 8 | 8 | CDS2498.1 | 183 | 437 |
| Cgpom152 | 8 | 8 | CDS0180.1 | 114 | 1,317 |
| Cginp53 | 8 | 8 | CDS0327.1 | 182 | 1,096 |
aa, amino acids.
Genes disrupted in mutants with altered fluconazole susceptibility.
In order to identify the genes disrupted in the various mutants, we rescued the Tn7 transposon and flanking DNA for at least one mutant at each disrupted locus (see Materials and Methods). For each mutant, genomic DNA was isolated, digested, recircularized, and transformed into E. coli (8). The resulting circular plasmid contains the Tn7 transposon flanked on either side by the gene it disrupts in the C. glabrata genome. Sequence analysis of the rescued plasmids with primers complementary to the ends of Tn7 revealed the identity of the disrupted genes. For the 17 susceptible and 10 resistant mutants, we regenerated the Tn7 insertion (see Materials and Methods) in the wild-type parental background. In all cases, the fluconazole-susceptible or -resistant phenotype was regenerated, demonstrating that the Tn7 disruption was in fact responsible for the altered fluconazole susceptibility phenotype (data not shown). The disrupted strains had no general growth defects. We measured the generation time for all 27 mutants in YPD media and found that, for 23 mutants, the range was 1.0 to 1.1 h, which was close to the wild-type generation time of 1.0 h. We found that nop8 (1.3 h), sin3 (1.2 h), sda1 (1.2 h), and vma13 (1.2 h) mutants had slightly longer doubling times (data not shown).
The disrupted genes giving rise to sensitive phenotypes included genes encoding multidrug transporters, a calcium channel, components of RNA polymerase II mediator, and nuclear ubiquitin ligase complex (Table 1). The disrupted genes giving rise to resistant phenotypes included genes involved in phospholipid biosynthesis, transcription, and actin skeleton dynamics; three genes were involved in ribosomal biogenesis, and three were involved in mitochondrial assembly and function (Table 2).
Disruption of CDR1 leads to a highly fluconazole-susceptible phenotype.
Fourteen mutants were susceptible to fluconazole even at concentrations as low as 4 μg/ml. One of these mutants was derived from a nonhomologous recombination event (as described in previous section) and was not analyzed further. The other 13 mutants were independent insertions derived from mutagenesis of a single fosmid F160; two of these were rescued. Both were Tn7 insertions in CDR1 (for Candida drug resistance 1) (57), a homologue of the S. cerevisiae multidrug transporter, PDR5 (2). The mutagenesis of fosmid F160 (carrying the CDR1 gene) had also resulted in one mutant showing a growth defect at 8 μg of fluconazole/ml (Fig. 1A) and wild-type growth at 4 μg of fluconazole/ml. Sequence analysis revealed that this mutant contained a Tn7 insertion in the promoter region of the CDR1 gene, suggesting that the mild fluconazole-sensitive phenotype of this mutant is due to a reduction in CDR1 transcription and thereby in the levels of Cdr1p.
Mutants in the putative calcium channel (Cch1-Mid1p) show increased killing by fluconazole.
Two of our mutants contained insertions in the CCH1 and MID1 genes. These mutants were of particular interest to us since the S. cerevisiae homologues of CCH1 and MID1 are thought to encode subunits of a plasma membrane gated channel involved in Ca2+ uptake (15, 23, 44). Cch1p is an N-glycosylated integral membrane protein which likely constitutes the catalytic subunit of the channel, whereas Mid1p is thought to be a regulatory subunit required for Cch1p function (36, 44). In C. albicans, functional calcineurin has been shown to be important for survival upon treatment with agents causing membrane stress, including azole agents (9, 60), and in S. cerevisiae, cch1 and mid1 mutants lose viability upon treatment with tunicamycin (which causes ER stress) (7). In light of these results, we examined whether the C. glabrata cch1 and mid1 mutants are unable to survive fluconazole stress.
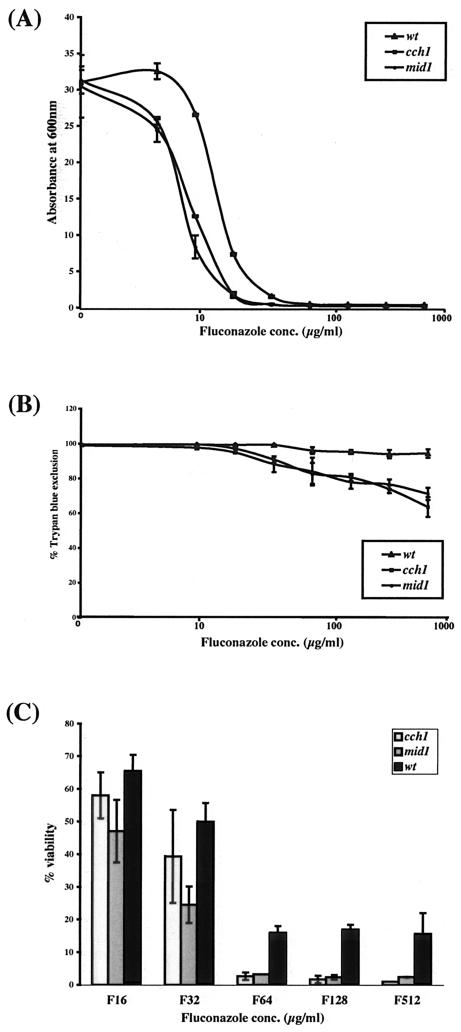
We first compared the growth of the cch1 and mid1 mutant and wild-type strains in liquid after 24 h in the presence of different concentrations of fluconazole. At all concentrations of fluconazole, all strains—the wild type and the cch1 and mid1 mutants—showed some growth (four to five doublings) before fluconazole inhibited the cell growth (data not shown) as measured by OD600. However, the final cell density of the cch1 and mid1 mutant cells at all inhibitory concentrations of fluconazole was consistently lower than that of wild-type cells, suggesting that cch1and mid1 cells either fail to replicate after a few generations or undergo cell death in the presence of fluconazole. Consistent with their slightly increased susceptibility to fluconazole, growth of the cch1 and mid1 mutants was significantly inhibited at 8 μg of fluconazole/ml, whereas the growth of the wild-type strain remained unaffected at this concentration of fluconazole (Fig. 2A).
FIG. 2.
C. glabrata cch1 and mid1 mutants lose viability upon prolonged fluconazole stress. (A) Growth of the C. glabrata cch1 and mid1 mutants is inhibited by fluconazole. Wild-type cch1and mid1 mutant cells were grown at 30°C in minimal medium supplemented with or without various amounts of fluconazole (4 to 512 μg/ml), and absorbance at 600 nm was monitored after 24 h of fluconazole exposure. (B) Trypan blue exclusion assay for the viability assessment of the wild type and the cch1 and mid1 mutants in fluconazole. Cells were grown at 30°C in minimal medium supplemented with or without various amounts of fluconazole as indicated. After 24 h of fluconazole treatment, cells were stained with trypan blue for 5 to 10 min, and the number of stained (dead) and unstained (viable) cells was assessed microscopically. A minimum of 300 cells total (viable [unstained] and dead [stained] cells) were counted for each datum point. Cell survival data were plotted as the percentage of trypan blue exclusion and represent the mean of three experiments (± the standard deviation [SD]). (C) Viable cell count assay to assess the viability of wild typeand cch1 and mid1 mutants in fluconazole. Cells were grown at 30°C in minimal medium supplemented with or without various amounts of fluconazole as indicated. After 24 h of fluconazole treatment, the total number of cells was counted by using a hemocytometer, and the number of viable cells was determined by plating appropriate dilutions on YPD and YPG plates. CFU from YPG plates were counted as the final readout, although essentially the same number of CFU was obtained on both YPG and YPD plates if small colonies (petites, as determined by their inability to grow on glycerol) on YPD plates were excluded from the final viable cell counts. Cell survival data were plotted as the percentage of viability and represent the mean of three experiments (± the SD).
Next, we assessed the viability of wild-type, cch1, and mid1 cells after 24 h of growth in the presence of fluconazole by a trypan blue exclusion assay. Cells were stained with trypan blue after fluconazole exposure, and the percentage of staining cells was assessed microscopically. As shown in Fig. 2B, by this measure the cch1 and mid1 mutants displayed a significantly lower viability than the wild-type strain after exposure to concentrations of fluconazole ranging from 64 to 512 μg/ml, suggesting that these mutants are killed at a higher rate than the wild type upon prolonged fluconazole treatment. However, this difference was not seen at lower inhibitory concentrations of fluconazole (8 or 16 μg/ml) (Fig. 2B) nor when cells were exposed to fluconazole (all concentrations) for short (8-h) time periods (data not shown). This observation suggests that in C. glabrata a functional Cch1-Mid1p channel is required for survival under long-term fluconazole exposure.
We made an independent more quantitative assessment of cell viability after fluconazole treatment by quantitating the CFU of the cch1, mid1, and wild-type strains recovered in the presence of different concentrations of fluconazole. C. glabrata wild-type, cch1, and mid1 cells were grown in minimal medium containing different concentrations of fluconazole. After growth, total cell number was assessed by counting them with a hemocytometer, and the CFU were then measured by plating the cells on YPD or YPG plates (CFU were counted from YPG plates to exclude the petite [mitochondrion-incompetent] cells, since C. glabrata petites are fluconazole resistant). No notable differences were seen between recoverable CFU of the cch1, mid1, or the wild-type strain when cells were grown at lower concentrations of fluconazole (<32 μg/ml) (Fig. 2C and data not shown). However, consistently 10- to 20-fold lower CFU were observed for the cch1 and mid1 mutants compared to the wild-type strain when cells were treated with fluconazole at concentrations from 64 to 512 μg/ml (Fig. 2C), reconfirming the earlier result (Fig. 2B) that both cch1 and mid1 cells are unable to survive the prolonged fluconazole exposure.
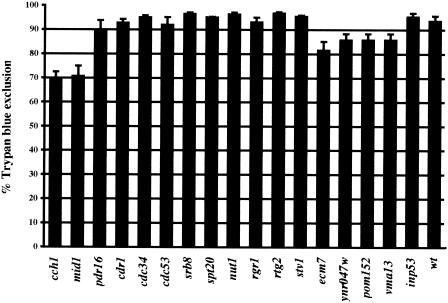
To classify our other fluconazole-susceptible mutants, particularly in light of the relatively modest change in their fluconazole susceptibility, we sought to determine whether any of the other fluconazole-susceptible mutants also display a loss of viability upon fluconazole treatment. We initially carried out a trypan blue exclusion assay on all mutants. C. glabrata cch1and mid1 mutants were used as positive controls in this assay. As shown in Fig. 3, most of the mutants (11 of 17 tested) displayed a wild-type-like survival rate of 90 to 95%. The cch1 and mid1 mutants exhibited 70 to 75% survival as determined by trypan blue exclusion assay, a finding consistent with earlier results, whereas four more mutants—ecm7, ynr047w, pom152, and vma13—were borderline cases, with 80 to 85% survival (Fig. 3). The results of the trypan blue assay were also confirmed by carrying out the viable count assay on two of the mutants that showed trypan blue exclusion of 80 to 85% (ecm7 and ynr047w) and two of the mutants that showed 90 to 95% trypan blue exclusion (cdc34 and pdr16). Viable counts from the cdc34, pdr16, ecm7, and ynr047w mutants ranged from 16 to 22%, which is essentially the same as for the wild-type strain (20% survival) (Fig. 2C and data not shown). Thus, a significant loss of viability was seen only with the calcium channel mutant strains. The loss of viability phenotype for the cch1 and mid1 mutants was not due to any enhanced fluconazole susceptibility since all of the fluconazole-susceptible mutants (including those with no loss of viability phenotype) showed similar susceptibility profiles on plates and in liquid culture (Fig. 1, Table 1, and data not shown). Therefore, the loss of viability phenotype is a specific phenotype associated with a small subset of fluconazole-susceptible mutants.
FIG. 3.
Trypan blue exclusion assay to assess the viability of all fluconazole-susceptible mutants in fluconazole. Cells were grown in minimal medium containing 512 μg of fluconazole/ml for 24 h and stained with trypan blue. A minimum of 300 cells total (viable [unstained] and dead [stained] cells) were counted microscopically for each mutant. Cell survival data were plotted as the percentage of trypan blue exclusion and represent the mean of two experiments (± the SD).
Mutants in the putative calcium channel (Cch1-Mid1) are defective in Ca2+ uptake upon fluconazole exposure.
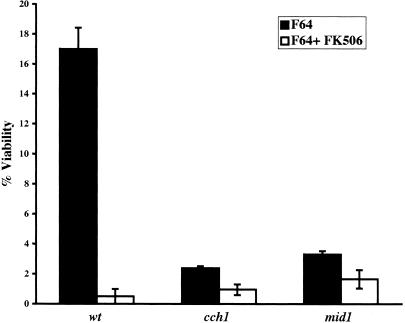
Fluconazole and FK506 (a calcineurin inhibitor) in combination have been reported to act synergistically in a variety of yeast cells (9, 14, 37). In C. glabrata, a mild synergistic effect of the two drugs has been reported based on disk diffusion assays (9). Because of the role in S. cerevisiae of Cch1p and Mid1p in Ca2+ uptake, we considered whether the loss of viability phenotype for the C. glabrata cch1 and mid1 mutants in our assays was also related to calcium signaling. We first examined whether fluconazole and FK506 act synergistically in C. glabrata. C. glabrata wild-type cells were grown in the presence of fluconazole alone, FK506 alone, or fluconazole and FK506 together, and the CFU were counted after 24 h of drug treatment. Consistent with the reports for other yeasts, a combination of fluconazole and FK506 was found to be fungicidal since 10- to 15-fold-fewer CFU were obtained when the cells were treated with both drugs versus when they were treated with fluconazole alone (Fig. 4). No drop in viability was seen when cells were treated with FK506 alone (data not shown). This 10- to 15-fold drop in the viability of wild-type cells upon fluconazole and FK506 treatment is of the same magnitude as the drop (relative to wild type) seen with cch1 and mid1 mutants treated with fluconazole (Fig. 2C), a finding consistent with the mutants affecting the same pathway. In addition, the cch1 and mid1 mutants were no more susceptible than the wild-type strain to treatment with a combination of fluconazole and FK506 (Fig. 4), a result consistent with the cch1 and mid1 mutants being compromised in the calcineurin signaling pathway, targeted by FK506.
FIG. 4.
Fluconazole and FK506 act synergistically and lead to cell death in wild-type C. glabrata cells. Cells were grown in minimal medium supplemented with either fluconazole (64 μg/ml) alone or fluconazole (64 μg/ml) plus FK506 (2 μg/ml) as indicated. After 24 h of drug treatment, the total number of cells was counted by using a hemocytometer, and CFU counts were determined by plating cells onto YPG plates. Cell survival data were plotted as the percentage of viable cells in each culture and represent the mean of two experiments (± the SD).
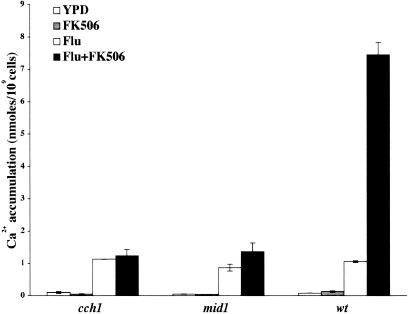
To directly test whether the cch1 and mid1 mutants had defects in the transport of Ca2+, we measured the accumulation of Ca2+ in wild-type and mutant strains. ER stress has been shown to lead to increased Ca2+ uptake in S. cerevisiae (7). First, we tested whether fluconazole exposure would lead to the uptake of extracellular Ca2+ in C. glabrata. C. glabrata wild-type cells were grown in the presence of fluconazole, fluconazole plus FK506, FK506 alone, or solvent (DMSO) alone. FK506 was used (to block calcineurin) because, in S. cerevisiae, calcineurin signals a feedback inhibition of the Cch1-Mid1p channel (36, 41). Total intracellular levels of Ca2+ were measured after 1 to 4 h of drug treatment. Although no Ca2+ accumulation was seen when cells were treated with FK506, a 10- to 12-fold increase in Ca2+ uptake was seen for all strains when cells were grown in the presence of fluconazole (Fig. 5). This Ca2+ influx was significantly elevated (70- to 75-fold) when wild-type cells were treated with both fluconazole and FK506. However, no such increase in Ca2+ levels was seen for the cch1 and mid1 mutants under similar conditions (Fig. 5), suggesting that these mutants are profoundly compromised for Ca2+ uptake in response to azole stress.
FIG. 5.
Fluconazole exposure in C. glabrata induces Ca2+ influx via Cch1-Mid1 channel. 45Ca2+ accumulation in the wild type and the cch1 and mid1 mutants was measured after 4 h of treatment with either fluconazole (64 μg/ml) alone, FK506 (2 μg/ml) alone, fluconazole (64 μg/ml) plus FK506 (2 μg/ml), or DMSO (solvent) in YPD medium containing tracer amounts of 45CaCl2. The bars represent the mean of two experiments (± the SD); each experiment was performed in triplicate; thus, the bars represent the mean of six assays.
We also analyzed two other fluconazole-susceptible mutants, rtg2 and spt20 (which do not exhibit loss of viability upon fluconazole treatment), as negative controls. Both mutants exhibited the wild-type 60- to 90-fold increase in Ca2+ uptake when treated with fluconazole and FK506 together (data not shown).
Fluconazole-resistant mutants affecting mitochondrial function.
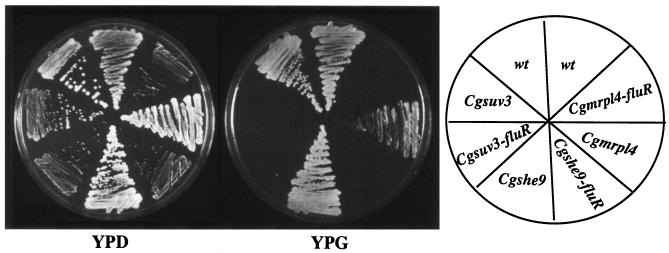
The C. glabrata mutants exhibiting the highest level of fluconazole resistance (256 μg/ml) had insertions in genes (SUV3, MRPL4, and SHE9) whose orthologues in S. cerevisiae are involved in mitochondrial function (Table 1) (12, 17, 38, 64). The corresponding S. cerevisiae mutants are all petites (17, 38, 64). We tested whether the C. glabrata counterparts (suv3, mrpl4, and she9, respectively) were also in fact petites. Surprisingly, only one of the C. glabrata mutants, suv3, was unable to grow on YPG plates, whereas mrpl4 and she9 were able to grow on YPG plates (Fig. 6).
FIG. 6.
Fluconazole resistance of C. glabrata suv3, mrpl4, and she9 mutants is due to the loss of mitochondrial function. Various mutant strains and their fluconazole-resistant derivatives (fluR) were streaked on YPD and YPG plates, and growth was recorded after 2 days of incubation at 30°C. Fluconazole-resistant derivatives of each mutant refer to the mutant colonies selected on YPD plates supplemented with fluconazole.
In conjunction with YPG growth assays, the function of mitochondria for the various mutants was assessed by staining cells with a mitochondria specific dye, Mito-Tracker Green FM-250. This dye is specifically sequestered inside active mitochondria. Consistent with the growth data on YPG plates, bright staining of mitochondria was seen for C. glabrata wild-type cells and for the mrpl4 and she9 mutants, whereas the suv3 mutant showed only faint background staining, indicating the lack of functional mitochondria (data not shown).
Although the C. glabrata mrpl4, she9, and suv3 mutants all showed the same apparent high level of fluconazole resistance, only the suv3 mutant appeared phenotypically to be petite (Fig. 6). However, fluconazole-resistant colonies arising from the C. glabrata mrpl4 and she9 did fail to grow on YPG plates (Fig. 6) showing that fluconazole-resistant derivatives of these mutants were functionally petite. Thus, despite initial appearances, the fluconazole resistance phenotype of the mrpl4 and she9 mutants is due to a loss of mitochondrial function. In contrast, the other seven fluconazole-resistant mutants (both the parental mutant strains and the single colonies taken from fluconazole-containing plates for these same mutants) exhibited normal growth on YPG plates, indicating that they all had functional mitochondria (data not shown). Thus, the fluconazole resistance phenotype for these other mutants is not due to the loss of mitochondrial function.
From the data above, the apparent fluconazole-resistant phenotype of the mrpl4 and she9 mutants was in fact due to a high frequency loss of mitochondrial function in these mutants. To quantify this, we carried out plating assays onto plates containing inhibitory concentrations of fluconazole (64 to 512 μg of fluconazole/ml). As expected, the plating efficiency of wild-type C. glabrata was low (ranging between 0.25 and 0.45%) at all inhibitory concentrations of fluconazole. These low-frequency fluconazole-resistant colonies were functionally petite and unable to grow on YPG plates. This result is consistent with the findings of Sanglard et al. (56), who originally reported that acquired fluconazole resistance in C. glabrata is due to the loss of functional mitochondria (56). For the mrpl4 and she9 mutants, the plating efficiency onto plates containing fluconazole was approximately 10% (data not shown). For both mutant strains, the fluconazole-resistant colonies were, as expected, functionally petite and unable to grow on YPG plates. Thus, our data suggested that the mrpl4 and she9 mutants, which display apparent very high levels of resistance to fluconazole, are not in fact innately resistant to the drug but become resistant after losing mitochondrial function at a high frequency.
Mechanisms of loss of mitochondrial function in wild-type and mutant C. glabrata.
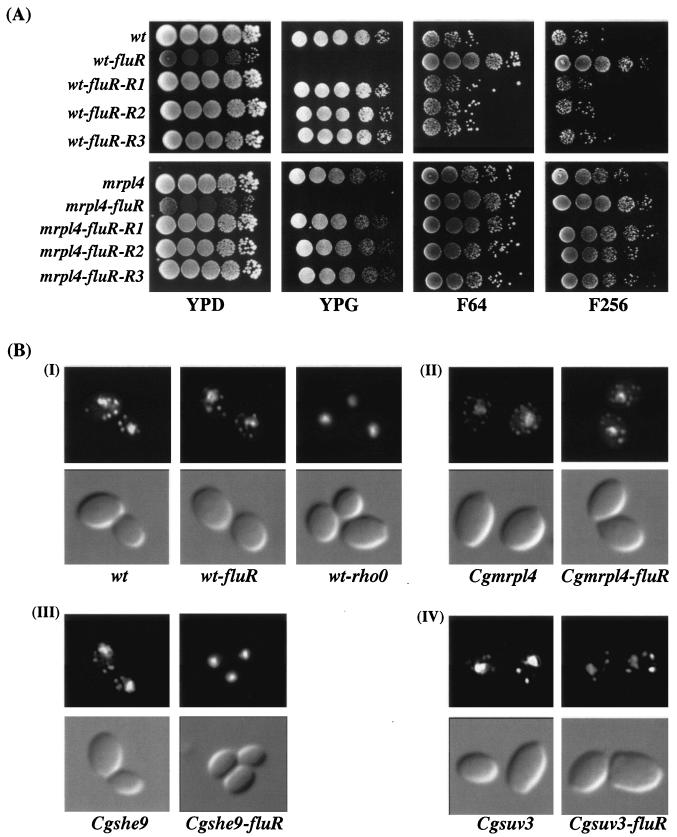
To gain an insight into the molecular mechanisms underlying the loss of mitochondrial competence in wild-type and mutant C. glabrata strains, we examined whether the petite phenotype of these derivative strains was reversible. We grew fluconazole-resistant derivatives of the wild-type parent and of the mrpl4, she9, and suv3 mutant strains in YPD, plated the cells on YPG plates, and scored the number of colonies that are able to grow on YPG. We used a rho0 derivative of the wild-type strain as a control. This rho0 derivative was generated by treatment of the wild-type strain with ethidium bromide. Petite mutants generated in this fashion lack mitochondrial DNA. We found that the wt-rho0 strain, the suv3 mutant and its fluconazole-resistant derivatives, and the fluconazole-resistant derivatives of the she9 mutant were nonrevertible petites (data not shown). Seven of thirteen fluconazole-resistant derivatives of the wild-type strain and three of six fluconazole-resistant derivatives of the mrpl4 mutant were also nonrevertible petites. However, six of thirteen wild-type derivatives and three of six mrpl4 derivatives were able to revert to growth on YPG plates at a frequencies of 1.5 × 10−2 and 2.5 × 10−4, respectively (data not shown). For both strains, the mitochondrion-competent revertants were indistinguishable from the original nonpetite parental strains on the basis of their fluconazole susceptibility profiles (Fig. 7A).
FIG. 7.
Mitochondrion-associated acquired fluconazole resistance in C. glabrata is reversible and is not associated with loss of the mitochondrial genome. (A) Revertants of fluconazole-resistant derivatives of the C. glabrata wild type and mrpl4 mutant exhibit fluconazole susceptibility like that of the original parental strain. Cells were grown in YPD medium overnight at 30°C, and 5 μl of 10-fold serial dilutions were spotted onto YPG and YPD plates with or without fluconazole at a given concentration (64 and 256 μg/ml) as indicated. Three independent revertant colonies (R1 to R3) were tested for each strain. Plates were photographed after 2 days of incubation at 30°C. (B) The revertible fluconazole-resistant derivatives of C. glabrata wild type and mrpl4 mutant possess mitochondrial genome. Nuclear and mitochondrial genomes were stained with DAPI as described in Materials and Methods, and cells were visualized by fluorescence microscopy. Subpanel groups: I, reversible (wt-fluR) and nonreversible (wt-rho0) fluconazole-resistant derivatives of wild type; II, mrpl4 mutant and reversible (mrpl4-fluR) fluconazole-resistant derivative of mrpl4; III, she9 mutant and its nonreversible fluconazole-resistant derivative she9-fluR; IV, suv9 mutant, along with its nonreversible fluconazole-resistant derivative (suv3-fluR).
Revertible fluconazole resistance is not due to mitochondrial genome loss.
We examined the various wild-type and mutant fluconazole-resistant strains for the presence of the mitochondrial genome by DAPI staining. DAPI staining of the wild-type strain exhibited the wild-type pattern of a large nuclear genome and small dotted mitochondrial nucleoids (Fig. 7B). The ethidium bromide-derived wt-rho0 strain, which is a nonreverting petite, showed no staining of punctuate cytoplasmic bodies, indicating a loss of the mitochondrial genome (Fig. 7B). The suv3 petite strains are nonrevertible because the Suv3p protein product, disrupted by Tn7, is required for mitochondrial function and, as expected, these mutants still showed staining of mitochondrial nucleoids. Although the she9 mutant displayed distinct mitochondrial nucleoids upon DAPI staining, the derivative nonreverting fluconazole-resistant colonies (she9-fluR) lacked these staining nucleoids (Fig. 7C), indicating loss of the mitochondrial genome. That was also true for the nonreverting fluconazole resistant derivatives of the wild-type and mrpl4 strain (data not shown). In contrast, the revertible fluconazole-resistant derivatives of the wild-type strain (wt-fluR) and of the mrpl4 mutant (mrpl4-fluR) showed the normal DAPI staining pattern, with a large nuclear genome and small dotted mitochondrial nucleoids (Fig. 7B), demonstrating that the petite phenotype in these isolates is not due to the loss of mitochondrial genome loss.
In conclusion, we showed that fluconazole-associated loss of mitochondrial function, leading to acquired fluconazole resistance, in wild-type C. glabrata is often reversible and, in these instances, not associated with the loss of the mitochondrial genome. This finding was reinforced by the behavior of the mrpl4 mutant, in which the fraction of cells able to become petite without losing the mitochondrial genome is much increased.
DISCUSSION
The innate fluconazole resistance and the acquisition of elevated resistance in C. glabrata complicates the use of azole therapy for this organism. We have undertaken a functional genomic analysis addressing fluconazole susceptibility in C. glabrata. By screening a library of 9,216 random insertion mutants (representing approximately 25% of C. glabarata genome) individually for their susceptibility to fluconazole, we identified 27 genes that when mutated by Tn7 insertion alter susceptibility to fluconazole. Mutations in three of these genes (CDR1, PDR16, and RTG2) have been previously demonstrated to result in changes in drug or azole susceptibility in various yeast species. The remaining 24 genes are novel and are implicated for the first time in the azole susceptibility in any yeast. We tested the fluconazole susceptibility of S. cerevisiae strains with deletions of the orthologues of the genes shown in Table 2 and found that of seven mutants tested (NOP8, PGS1, and SDA1 are essential genes in S. cerevisiae), only mutants in SUV3 and MRPL4 showed a slight (<2-fold) increase in drug resistance (data not shown). This finding emphasizes the importance of carrying out analysis of drug resistance mechanisms in clinically relevant species.
Azole-susceptible mutants.
Of the 17 mutants that increase susceptibility to fluconazole, two disrupt the multidrug efflux pumps, Cdr1p and Pdr16p. Cdr1p belongs to the family of ABC transporters and is a close homologue of S. cerevisiae Pdr5p and C. albicans Cdr1p (2, 4, 51). Cdr1p in C. glabrata has been implicated in fluconazole resistance in vitro, and its expression has also been found to be significantly elevated in many of clinical fluconazole-resistant strains (57). PDR16 is another member of the ABC transporter family and is under the regulation by the pleiotropic drug resistance transcriptional factor, PDR1 (13, 68). In S. cerevisiae, PDR16 has been shown to regulate sterol metabolism and is specifically required for resistance to the azole compounds miconazole and ketoconazole (68).
Mutants of Cch1p and Mid1p, subunits of the putative plasma membrane channel, constitute the second interesting class of fluconazole-susceptible mutants. Although mutants in these two genes were mildly susceptible to fluconazole, they were notably defective in survival of long-term fluconazole exposure, as measured by trypan blue exclusion and viable counts. Both the trypan blue exclusion assay and the viable count assay make clear that there is a significant difference in survival of the wild-type and mutant strains; however, for the cch1 mutant, mid1 mutant, and wild-type strains, noteworthy differences lie in the numbers of calculated viable cells (recovered CFU) relative to cells that exclude trypan blue. For example, whereas the trypan blue assay shows that 90 to 95% of wild-type cells exclude dye after 24 h of fluconazole treatment, only 15 to 20% of the cells are viable by the CFU assay (Fig. 2). This discrepancy likely means that the trypan blue assay underestimates the number of dead cells; alternatively, there may be significant numbers of cells that are metabolically viable after fluconazole treatment but which are unable to divide and form colonies when placed in nonselective media.
In S. cerevisiae, Cch1p and Mid1p constitute the catalytic and the regulatory subunits of a high-affinity plasma membrane-based calcium channel (15, 23, 36, 44). Intracellular levels of Ca2+ are important for many signaling cascades in both lower and higher eukaryotes. It has been postulated in S. cerevisiae, that membrane perturbation along with other stresses, leads to the influx of Ca2+ either from the extracellular media or via mobilization of intraorganellar stores of Ca2+ (14). In addition, the Ca2+-dependent protein phosphatase, calcineurin, has been shown to be important for long-term survival under ER stress in S. cerevisiae and for the survival of membrane stress, virulence, and antifungal tolerance in C. albicans (5, 7, 9, 60). Although calcineurin, Cch1p, and Mid1p have been shown to be required for long-term survival of cells treated with agents causing ER stress in S. cerevisiae (7), this is the first study reporting that fluconazole treatment in C. glabrata leads to Cch1-Mid1-dependent Ca2+ uptake and that this uptake is essential for survival upon prolonged fluconazole treatment. Our findings are consistent with those of Bonilla et al. (7), who reported that in S. cerevisiae Cch1-Mid1p was required for survival of ER stress and that FK506 treatment of C. glabrata resulted in sensitivity to tunicamycin.
It is notable that the cch1 and mid1 mutants were not wholly unable to take up Ca2+ in response to fluconazole exposure (Fig. 5). Indeed, when exposed to fluconazole alone, there was a marked increase in Ca2+ uptake in wild-type and both mutant strains. In wild-type cells, there was an additional stimulation of Ca2+ uptake in response to fluconazole in the presence of FK506, which inhibits calcineurin. This additional stimulation depended entirely on the Cch1p-Mid1p channel since the mutant strains showed no such increase. Studies in S. cerevisiae have shown that the Cch1p-Mid1p channel is subject to a rapid downregulation of channel function in a calcineurin-dependent fashion. In S. cerevisiae, as in C. glabrata, Ca2+ influx as a result of membrane stress can only be observed experimentally if feedback regulation of the channel is inhibited by FK506 (36, 41). Experimental considerations aside, our data shows that in C. glabrata, as in S. cerevisiae, there are CCH1/MID1-dependent and -independent mechanisms of Ca2+ influx in response to fluconazole. Based on the loss of viability seen in the cch1 and mid1 mutants, the Cch1-Mid1p-dependent pathway is essential for azole stress survival.
Lastly, five of the fluconazole susceptible mutants encode components of RNA polymerase II coactivator complexes. Srb8, Nut1, and Rgr1 are all components of Mediator (19, 28, 32, 35, 53). This complex of at least 20 proteins is required for activated transcription mediated by RNA polymerase II (28). This suggests that Mediator is required for the regulation of at least some of the genes important in the fluconazole response. Potential targets of mediator include the ERG genes, as well as genes encoding drug transporters (CDR1 and PDH1/CDR2 in particular). Since the RNA polymerase II mediator mutants show only mildly fluconazole-sensitive phenotypes, mediator is unlikely to be absolutely required for transcription of those genes. We also identified mutations in SPT20 and RTG2. Spt20 is a component of the SAGA complex which, among other activities, has histone acetyltransferase activity. Rtg2 is a cytosolic protein which in S. cerevisiae is involved in retrograde signaling from the mitochondria to the nucleus (33, 45). Recently, Rtg2p has been shown to be a constituent of the SLIK complex (SAGA-like complex), which is thought to be involved in chromatin modification and reorganization and is important in transcription by RNA polymerase II of certain loci (52). Intriguingly, rtg2 mutants in S. cerevisiae exhibit a mild sensitivity to cycloheximide and have a defect in the activation of multidrug resistance pumps in response to dysfunctional mitochondria (21). The identification of a rtg2 mutant with a mildly fluconazole-sensitive phenotype suggests that a complex analogous to SLIK may be important in drug-induced activation of multidrug resistance pumps in C. glabrata.
Mitochondrial dysfunction and acquisition of fluconazole resistance.
Three mutants, which exhibited the highest levels of fluconazole resistance, were disrupted for mitochondrial function. Suv3p is a mitochondrial RNA helicase and is required for processing of mitochondrial RNA transcripts (64); suv3 mutant strains therefore inherently lack functional mitochondria. In the case of the mrpl4 and she9 mutants, the mutants had functional mitochondria but lost mitochondrial function efficiently upon exposure to fluconazole. She9p is an integral mitochondrial inner membrane protein and is required for the organization and biogenesis of mitochondria (38). She9p is also required for mitochondrial genome stability since she9 mutants lose their mitochondrial genome at an elevated frequency relative to the wild type. Mrpl4p is a putative structural component of mitochondrial large ribosomal subunit (17). Its disruption caused an increase in the rate of mitochondrial function loss but did not cause a notable destabilization of the mitochondrial genome. Our studies point out that mutants that do not themselves result in a petite phenotype can nonetheless be linked to acquired fluconazole resistance if they result in an increased loss of mitochondrial function.
We also found that loss of mitochondrial function occurs at an appreciable rate in wild-type cells. Dysfunctional mitochondria have been previously linked with drug resistance in both S. cerevisiae and C. glabrata (21, 56). We have extended these earlier studies by demonstrating that acquired fluconazole resistance is not always due to the irreversible loss of mitochondrial genome but rather to a reversible loss of mitochondrial function. Sanglard et al. (56) analyzed upregulation of the multidrug transporter genes CDR1 and CDR2 in cultures of fluconazole-associated petites. These authors reported, upon culturing of these isolates and concomitant with an increase in the growth rate, a downregulation of the transporter genes (56). This might be due to the reversion of their original respiratory-deficient mutants (elevated levels of CDR1 and CDR2) to respiratory-competent cells (reduced CDR1 and CDR2 levels).
Fluconazole-associated petites from the wild-type background can revert to respiratory competence at a rate of 2.5 × 10−2 in our study. This makes it highly unlikely that a fluconazole-associated loss of mitochondrial competence is due to any genetic change and may suggest an epigenetic mechanism for switching between respiratory-competent and -incompetent states. In yeast, there are well established pathways of cross talk between dysfunctional mitochondria and the nucleus (33, 45).
From a clinical perspective, there is a poor correlation in C. glabrata between the level of azole resistance measured in vitro and treatment success or failure. Epigenetic switching between mitochondrially competent (fluconazole-susceptible) and incompetent (fluconazole-resistant) states during infection might help explain how apparently sensitive isolates could be refractory to therapeutic treatment.
Acknowledgments
We thank Kyle Cunningham and members of the Cormack laboratory for helpful discussions during the course of this study. We are grateful to Jun Liu for the gift of FK506.
This study was funded by the National Institutes of Health grant RO1 AI46223 to B.P.C.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Balzi, E., M. Wang, S. Leterme, L. Van Dyck, and A. Goffeau. 1994. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269:2206-2214. [PubMed] [Google Scholar]
- 3.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bissinger, P. H., and K. Kuchler. 1994. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product: a yeast ABC transporter conferring mycotoxin resistance. J. Biol. Chem. 269:4180-4186. [PubMed] [Google Scholar]
- 5.Blankenship, J. R., F. L. Wormley, M. K. Boyce, W. A. Schell, S. G. Filler, J. R. Perfect, and J. Heitman. 2003. Calcineurin is essential for Candida albicans survival in serum and virulence. Eukaryot. Cell 2:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodey, G. P., M. Mardani, H. A. Hanna, M. Boktour, J. Abbas, E. Girgawy, R. Y. Hachem, D. P. Kontoyiannis, and Raad, II. 2002. The epidemiology of Candida glabrata and Candida albicans fungemia in immunocompromised patients with cancer. Am. J. Med. 112:380-385. [DOI] [PubMed] [Google Scholar]
- 7.Bonilla, M., K. K. Nastase, and K. W. Cunningham. 2002. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 21:2343-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castano, I., R. Kaur, S. Pan, R. Cregg, L. Penas Ade, N. Guo, M. C. Biery, N. L. Craig, and B. P. Cormack. 2003. Tn7-based genome-wide random insertional mutagenesis of Candida glabrata. Genome Res. 13:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz, M. C., A. L. Goldstein, J. R. Blankenship, M. Del Poeta, D. Davis, M. E. Cardenas, J. R. Perfect, J. H. McCusker, and J. Heitman. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J. 21:546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Backer, M. D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Defontaine, A., J. P. Bouchara, P. Declerk, C. Planchenault, D. Chabasse, and J. N. Hallet. 1999. In vitro resistance to azoles associated with mitochondrial DNA deficiency in Candida glabrata. J. Med. Microbiol. 48:663-670. [DOI] [PubMed] [Google Scholar]
- 12.Dimmer, K. S., S. Fritz, F. Fuchs, M. Messerschmitt, N. Weinbach, W. Neupert, and B. Westermann. 2002. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol. Biol. Cell 13:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.do Valle Matta, M. A., J. L. Jonniaux, E. Balzi, A. Goffeau, and B. van den Hazel. 2001. Novel target genes of the yeast regulator Pdr1p: a contribution of the TPO1 gene in resistance to quinidine and other drugs. Gene 272:111-119. [DOI] [PubMed] [Google Scholar]
- 14.Edlind, T., L. Smith, K. Henry, S. Katiyar, and J. Nickels. 2002. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signaling. Mol. Microbiol. 46:257-268. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, M., N. Schnell, J. Chattaway, P. Davies, G. Dixon, and D. Sanders. 1997. The Saccharomyces cerevisiae CCH1 gene is involved in calcium influx and mating. FEBS Lett. 419:259-262. [DOI] [PubMed] [Google Scholar]
- 16.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graack, H. R., L. Grohmann, M. Kitakawa, and S. Goldschmidt-Reisin. 1995. Gene MRP-L4, encoding mitochondrial ribosomal protein YmL4, is indispensable for proper non-respiratory cell functions in yeast. Gene 152:107-112. [DOI] [PubMed] [Google Scholar]
- 18.Grant, S. M., and S. P. Clissold. 1990. Fluconazole: a review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in superficial and systemic mycoses. Drugs 39:877-916. [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson, C. M., L. C. Myers, J. Beve, H. Spahr, M. Lui, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 1998. Identification of new mediator subunits in the RNA polymerase II holoenzyme from Saccharomyces cerevisiae. J. Biol. Chem. 273:30851-30854. [DOI] [PubMed] [Google Scholar]
- 20.Gustin, M. C., B. Martinac, Y. Saimi, M. R. Culbertson, and C. Kung. 1986. Ion channels in yeast. Science 233:1195-1197. [DOI] [PubMed] [Google Scholar]
- 21.Hallstrom, T. C., and W. S. Moye-Rowley. 2000. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:37347-37356. [DOI] [PubMed] [Google Scholar]
- 22.Hemenway, C. S., and J. Heitman. 1999. Calcineurin: structure, function, and inhibition. Cell Biochem. Biophys. 30:115-151. [DOI] [PubMed] [Google Scholar]
- 23.Iida, H., H. Nakamura, T. Ono, M. S. Okumura, and Y. Anraku. 1994. MID1, a novel Saccharomyces cerevisiae gene encoding a plasma membrane protein, is required for Ca2+ influx and mating. Mol. Cell. Biol. 14:8259-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly, S. L., A. Arnoldi, and D. E. Kelly. 1993. Molecular genetic analysis of azole antifungal mode of action. Biochem. Soc. Trans. 21:1034-1038. [DOI] [PubMed] [Google Scholar]
- 25.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, H. Hebart, U. Schumacher, and H. Einsele. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta 5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 26.Kelly, S. L., J. Rowe, and P. F. Watson. 1991. Molecular genetic studies on the mode of action of azole antifungal agents. Biochem. Soc. Trans. 19:796-798. [DOI] [PubMed] [Google Scholar]
- 27.Kenna, S., H. F. Bligh, P. F. Watson, and S. L. Kelly. 1989. Genetic and physiological analysis of azole sensitivity in Saccharomyces cerevisiae. J. Med. Vet. Mycol. 27:397-406. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y. J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 29.Kontoyiannis, D. P. 2000. Modulation of fluconazole sensitivity by the interaction of mitochondria and erg3p in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 46:191-197. [DOI] [PubMed] [Google Scholar]
- 30.Lamb, D. C., D. E. Kelly, W. H. Schunck, A. Z. Shyadehi, M. Akhtar, D. J. Lowe, B. C. Baldwin, and S. L. Kelly. 1997. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J. Biol. Chem. 272:5682-5688. [DOI] [PubMed] [Google Scholar]
- 31.Lees, N. D., M. Bard, and D. R. Kirsch. 1999. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol. 34:33-47. [PubMed] [Google Scholar]
- 32.Li, Y., S. Bjorklund, Y. W. Jiang, Y. J. Kim, W. S. Lane, D. J. Stillman, and R. D. Kornberg. 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92:10864-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao, X., and R. A. Butow. 1993. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72:61-71. [DOI] [PubMed] [Google Scholar]
- 34.Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807-815. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Y., J. A. Ranish, R. Aebersold, and S. Hahn. 2001. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 276:7169-7175. [PubMed] [Google Scholar]
- 36.Locke, E. G., M. Bonilla, L. Liang, Y. Takita, and K. W. Cunningham. 2000. A homolog of voltage-gated Ca2+ channels stimulated by depletion of secretory Ca2+ in yeast. Mol. Cell. Biol. 20:6686-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchetti, O., P. Moreillon, M. P. Glauser, J. Bille, and D. Sanglard. 2000. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob. Agents Chemother. 44:2373-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Messerschmitt, M., S. Jakobs, F. Vogel, S. Fritz, K. S. Dimmer, W. Neupert, and B. Westermann. 2003. The inner membrane protein Mdm33 controls mitochondrial morphology in yeast. J. Cell Biol. 160:553-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 40.Miyazaki, H., Y. Miyazaki, A. Geber, T. Parkinson, C. Hitchcock, D. J. Falconer, D. J. Ward, K. Marsden, and J. E. Bennett. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller, E. M., E. G. Locke, and K. W. Cunningham. 2001. Differential regulation of two Ca2+ influx systems by pheromone signaling in Saccharomyces cerevisiae. Genetics 159:1527-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 43.Nguyen, M. H., J. E. Peacock, Jr., A. J. Morris, D. C. Tanner, M. L. Nguyen, D. R. Snydman, M. M. Wagener, M. G. Rinaldi, and V. L. Yu. 1996. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am. J. Med. 100:617-623. [DOI] [PubMed] [Google Scholar]
- 44.Paidhungat, M., and S. Garrett. 1997. A homolog of mammalian, voltage-gated calcium channels mediates yeast pheromone-stimulated Ca2+ uptake and exacerbates the cdc1(Ts) growth defect. Mol. Cell. Biol. 17:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parikh, V. S., M. M. Morgan, R. Scott, L. S. Clements, and R. A. Butow. 1987. The mitochondrial genotype can influence nuclear gene expression in yeast. Science 235:576-580. [DOI] [PubMed] [Google Scholar]
- 46.Patterson, R. L., D. B. van Rossum, and D. L. Gill. 1999. Store-operated Ca2+ entry: evidence for a secretion-like coupling model. Cell 98:487-499. [DOI] [PubMed] [Google Scholar]
- 47.Pfaller, M. A., D. J. Diekema, R. N. Jones, S. A. Messer, and R. J. Hollis. 2002. Trends in antifungal susceptibility of Candida spp. isolated from pediatric and adult patients with bloodstream infections: SENTRY Antimicrobial Surveillance Program, 1997 to 2000. J. Clin. Microbiol. 40:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, R. J. Hollis, S. A. Messer, et al. 1998. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and antifungal susceptibilities of isolates collected in 1997 in the United States, Canada, and South America for the SENTRY Program. J. Clin. Microbiol. 36:1886-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaller, M. A., S. A. Messer, R. J. Hollis, R. N. Jones, G. V. Doern, M. E. Brandt, and R. A. Hajjeh. 1999. Trends in species distribution and susceptibility to fluconazole among blood stream isolates of Candida species in the United States. Diagn. Microbiol. Infect. Dis. 33:217-222. [DOI] [PubMed] [Google Scholar]
- 50.Pozzan, T., R. Rizzuto, P. Volpe, and J. Meldolesi. 1994. Molecular and cellular physiology of intracellular calcium stores. Physiol. Rev. 74:595-636. [DOI] [PubMed] [Google Scholar]
- 51.Prasad, R., P. De Wergifosse, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 52.Pray-Grant, M. G., D. Schieltz, S. J. McMahon, J. M. Wood, E. L. Kennedy, R. G. Cook, J. L. Workman, J. R. Yates III, and P. A. Grant. 2002. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 22:8774-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers, P. D., and K. S. Barker. 2002. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans by cDNA microarray analysis. Antimicrob. Agents Chemother. 46:3412-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogers, P. D., and K. S. Barker. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob. Agents Chemother. 47:1220-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanglard, D., F. Ischer, and J. Bille. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanglard, D., F. Ischer, O. Marchetti, J. Entenza, and J. Bille. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis, and virulence. Mol. Microbiol. 48:959-976. [DOI] [PubMed] [Google Scholar]
- 60.Sanglard, D., F. Ischer, T. Parkinson, D. Falconer, and J. Bille. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuman, P., J. D. Sobel, S. E. Ohmit, K. H. Mayer, C. C. Carpenter, A. Rompalo, A. Duerr, D. K. Smith, D. Warren, R. S. Klein, et al. 1998. Mucosal candidal colonization and candidiasis in women with or at risk for human immunodeficiency virus infection. Clin. Infect. Dis. 27:1161-1167. [DOI] [PubMed] [Google Scholar]
- 63.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 64.Stepien, P. P., S. P. Margossian, D. Landsman, and R. A. Butow. 1992. The yeast nuclear gene suv3 affecting mitochondrial posttranscriptional processes encodes a putative ATP-dependent RNA helicase. Proc. Natl. Acad. Sci. USA 89:6813-6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trick, W. E., S. K. Fridkin, J. R. Edwards, R. A. Hajjeh, and R. P. Gaynes. 2002. Secular trend of hospital-acquired candidemia among intensive care unit patients in the United States during 1989-1999. Clin. Infect. Dis. 35:627-630. [DOI] [PubMed] [Google Scholar]
- 66.Vanden Bossche, H., P. Marichal, J. Gorrens, D. Bellens, H. Moereels, and P. A. Janssen. 1990. Mutation in cytochrome P-450-dependent 14 alpha-demethylase results in decreased affinity for azole antifungals. Biochem. Soc. Trans. 18:56-59. [DOI] [PubMed] [Google Scholar]
- 67.Vanden Bossche, H., P. Marichal, F. C. Odds, L. Le Jeune, and M. C. Coene. 1992. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 36:2602-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van den Hazel, H. B., H. Pichler, M. A. do Valle Matta, E. Leitner, A. Goffeau, and G. Daum. 1999. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem. 274:1934-1941. [DOI] [PubMed] [Google Scholar]
- 69.Vazquez, J. A., J. D. Sobel, G. Peng, L. Steele-Moore, P. Schuman, W. Holloway, J. D. Neaton, et al. 1999. Evolution of vaginal Candida species recovered from human immunodeficiency virus-infected women receiving fluconazole prophylaxis: the emergence of Candida glabrata? Clin. Infect. Dis. 28:1025-1031. [DOI] [PubMed] [Google Scholar]
- 70.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White, T. C., S. Holleman, F. Dy, L. F. Mirels, and D. A. Stevens. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida, Y., and Y. Aoyama. 1987. Interaction of azole antifungal agents with cytochrome P-45014DM purified from Saccharomyces cerevisiae microsomes. Biochem. Pharmacol. 36:229-235. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, X., and W. S. Moye-Rowley. 2001. Saccharomyces cerevisiae multidrug resistance gene expression inversely correlates with the status of the F(0) component of the mitochondrial ATPase. J. Biol. Chem. 276:47844-47852. [DOI] [PubMed] [Google Scholar]