Abstract
The basic helix-loop-helix transcription factor NeuroD1 is expressed in embryonic and adult mouse olfactory epithelium (OE), as well as during epithelial regeneration, suggesting that it plays an important role in olfactory neurogenesis. We characterized NEUROD1-expressing progenitors, determined their progeny in the adult OE, and identified a subtle phenotype in ΔNeuroD1-knockout mice. All olfactory sensory neurons (OSNs) derive from a NeuroD1-expressing progenitor as shown by recombination-mediated lineage tracing, as do other sensory receptors of the nose, including vomeronasal, nasal septal, and Grunenberg ganglion neurons. NEUROD1-expressing cells are found among the globose basal cell population: they are actively proliferating and frequently coexpress Neurog1, but not the transit amplifying cell marker MASH1, nor the neuronal marker NCAM. As a consequence, NEUROD1-expressing globose basal cells are best classified as immediate neuronal precursors. In adolescent ΔNeuroD1-LacZ knock-in null mice the OE displays subtle abnormalities, as compared to wildtype and heterozygous littermates. In some areas of the OE, mature neurons are absent, or sparse, although those same areas retain immature OSNs and LacZ-expressing progenitors, albeit both of these populations are smaller than expected. Our results support the conclusion that most, if not all, nasal chemosensory neurons derive from NeuroD1-expressing globose basal cells of the immediate neuronal precursor variety. Moreover, elimination of NeuroD1 by gene knockout, while it does not disrupt initial OSN differentiation, does compromise the integrity of parts of the olfactory epithelium by altering proliferation, neuronal differentiation, or neuronal survival there.
INDEXING TERMS: transcription factors, differentiation, lineage tracing, knockout
The olfactory epithelium (OE), by virtue of its life-long capacity to maintain and reconstitute the population of olfactory sensory neurons (OSNs), is an attractive model system for analyzing the regulation of neurogenesis. The cell types of the OE include a heterogeneous group of basal cells that can be subdivided into HBCs, which tightly attach to the basal lamina, and GBCs, which are comparatively simple in shape, multipotent progenitors, and found within both populations (Mackay-Sim and Kittel, 1991; Goldstein et al., 1998; Carter et al., 2004; Chen et al., 2004; Leung et al., 2007). The GBCs can be further subdivided with respect to the expression of transcription factors, including Sox2, Pax6, and several markers of the basic-helix-loop-helix (bHLH) family (Cau et al., 1997, 2002).
Genetic loss-of-function studies have determined the epistatic relationship for a subset of bHLH transcription factors that play important roles in the generation of OSNs. Within a transcription factor cascade, Mash1 expression lies upstream of Neurogenin1 (Neurog1), which, in turn, slightly anticipates but overlaps with the expression of NeuroD1. All of these pro-neural genes precede the onset of neuronal differentiation per se (Cau et al., 1997, 2002). Mash1 appears to mark transit-amplifying cells that are committed neuronal precursors with the capacity for substantial expansion, whereas Neurog1 is thought to identify the immediate neuronal precursor, a more committed and mitotically limited cell than transit amplifying Mash1(+) GBCs (Mumm et al., 1996; Cau et al., 1997, 2002; Murray et al., 2003; Manglapus et al., 2004). In contrast to other bHLH factors, the characteristics of NeuroD1-expressing cells remain poorly defined.
To highlight the importance of Mash1 and Neurog1 within the neurogenic pathway, deletion of either gene produces striking disruptions of the embryonic OE, as well as elsewhere in the nervous system (Cau et al., 1997, 2002; Murray et al., 2003; Wildner et al., 2006). Although loss of NeuroD1 has dramatic effects in other systems, no abnormalities have been reported for the OE of NeuroD1-null mice. Against the apparent absence of an effect in the OE, NeuroD1 promotes the survival of photoreceptor cells of the retina, as well as granule cells of the hippocampus (Pennesi et al., 2003; Gao et al., 2009; Kuwabara et al., 2009). NeuroD1 can induce neural cell differentiation in the embryo, and recently NeuroD1 was identified as a critical factor for the transition from undifferentiated, neural precursors to differentiated neurons by hippocampal, periglomerular, and cochlear progenitor cells (Lee et al., 1995; Kuwabara et al., 2009; Puligilla et al., 2010; Boutin et al., 2010).
By in situ hybridization (ISH) analysis, NeuroD1 expression is roughly contemporaneous with Neurog1 in adult rat and embryonic OE, and anticipates terminal mitosis and neuronal marker expression (Schwob et al., 1995; Cau et al., 1997, 2002; Manglapus et al., 2004). Nonetheless, the NeuroD1-expressing basal cell progenitor is poorly defined otherwise; the questions of whether these progenitors are still actively cycling and whether they are necessary for neuronal development and survival have not been directly addressed until now.
MATERIALS AND METHODS
Animals
Three ΔNeuroD1-Cre BAC transgenic mouse lines (Wang et al., 2007) were generated by BAC recombineering that introduced the Cre recombinase construct within the coding sequence of the NeuroD1 locus. Sequences encoding Cre recombinase that included a nuclear localization signal and simian virus 40 polyadenylation sequences were introduced into a murine BAC clone, RPCI-23188B11 (Invitrogen, La Jolla, CA) containing the NeuroD1 locus, by homologous recombination in E. coli as described previously (Datsenko and Wanner, 2000; Cotta-De-Almeida et al., 2003; Schonhoff et al., 2004). The Cre encoding sequences were inserted into the translation initiation ATG for the NeuroD1 gene. The polymerase chain reaction (PCR) primers used were: sense-TGCTTGCCTCTCTCCCTGTTCAATACAGGAAGTGGAAACATGCCCAAGAAGA AGAGGAA and antisense-GGCTCGCCCATCAGCCCGCTCTCGCTGTATGATTTGGTCATCCTCCTTAGTTCCTATTCCGA. Sequence analysis of the selected clones confirmed correct transgene construction.
Three separate lines of transgenic mice were generated by pronuclear injection of the purified circular NeuroD1-Cre BAC DNA into the pronuclei of fertilized oocytes of B6 × B6D2F1 mice. Founders were identified by genotyping the tail DNA with primers specific for the Cre transgene. No abnormalities were observed in any of the NeuroD1-Cre BAC transgenic lines as a consequence of the small region of genomic duplication. ΔNeuroD1-LacZ knockin mice were generated by Naya et al. (1997) and backcrossed onto a 129S1/SvImJ (stock no. 002448) background to extend the viability of null offspring past birth (Liu et al., 2000). All ΔNeuroD1-LacZ knock-in null mice exhibited severe neurological deficiencies, including reduced body size and ataxia, consistent with previous reports (Miyata et al., 1999; Liu et al., 2000). Although the 129SvJ backcross extended survival of the NeuroD1 null progeny, their lifespan is still abbreviated compared to wildtype mice, leading us to use 4–6-week-old adolescent mice in this study. For purposes of lineage analysis, NeuroD1-Cre mice were crossed to homozygous ΔRosa26-flox-stop-flox-LacZ (B6.129S4-Gt(Rosa)26-Sortm1Sor/J, stock no. 003474) or ΔRosa26-flox-stop-flox-eYFP (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J, stock no. 006418) indicator mice (purchased from Jackson Laboratories, Bar Harbor, ME) to generate a bigenic offspring (designated NeuroD1-Cre/LacZ or NeuroD1-Cre/eYFP) (Soriano, 1999). Wildtype adult C57BL/6J mice were purchased from JAX. All mice were maintained on ad libitum rodent chow and water. The animals were housed in a heat- and humidity-controlled, AALAC-accredited vivarium operating under a 12:12-hour light-dark cycle. All animal-use protocols were approved by the Committee for the Humane Use of Animals at Tufts University School of Medicine, where the animals were housed and experiments were conducted.
Tissue processing
Mice ranging in age from P21 to adult were deeply anesthetized by intraperitoneal (i.p.) injection of a cocktail of ketamine (37.5 mg/kg), xylazine (7.5 mg/kg), and acepromazine (1.25 mg/kg), transcardially flushed with phosphate-buffered saline (PBS) (pH 7.2), and perfused with one of a number of fixatives. The various fixatives include 4% paraformaldehyde (Fisher Scientific, Suwanee, GA) in 0.05 M sodium phosphate buffer, pH 7.2, Zamboni’s Fixative, 1% paraformaldehyde / 0.1% glutaraldehyde in phosphate buffer, and Carnoy’s Fixative; for each the tissue was additionally fixed by immersion under vacuum for 2–4 hours postperfusion. After removal of the soft tissue, the calvaria, and the other heavy bones of the head, the nose and attached olfactory bulbs were rinsed with PBS, cryoprotected with 30% sucrose in PBS, and then frozen in OCT compound (Miles, Elkhart, IN). The nasal skeleton with the olfactory mucosa was sectioned on a Leica cryostat in the coronal plane; 8-μm sections were collected on to ‘Plus’ slides (Fisher Scientific) and stored at −20°C for future applications.
Genotyping
Five-mm diameter ear punches were harvested from all mice in a litter. The punched tissue was digested in 300 μL of 50 mM NaOH for 1 hour at 95°C, vortexed, and supplemented with 16.7 μl of 1M Tris-HCl pH 8. All tissue samples were then vortexed to dissociate the tissue and spun down at 13,000g for 6 minutes. Three μL of supernatant, containing DNA, from the prepared tissue sample were used for each PCR. Sequence-specific primers were used to identify the appropriate amplicons for transgene-carrying pups and are available upon request.
Antibody characterization
Please see Table 1 for a list of all antibodies used. Information on the antibodies is derived from the manufacturers’ description and our own data.
TABLE 1.
Antibodies Used
| Primary antibody | Immunogen | Source, species, clonality and catalog number | Dilution |
|---|---|---|---|
| β-Gal | Bacterial β-galactosidase, 119kDa protein. | Biogenesis, goat polyclonal, cat. no. 4600-1409 | 1:300 (CI) or 1:1,500 (DAB) |
| Cre | Bacteriophage P1 Cre Recombinase, 35kDa protein, native and denatured. | Novagen, rabbit polyclonal, cat. no. 69050-3 | 1:7,000 (DAB) |
| Gustducin | Peptide corresponding to aa 93-112 of rat gustducin | Santa Cruz, rabbit, cat. no. sc-395 | 1:400 (CI) |
| Ki67 | Recombinant Human Ki-67 peptide (clone B56) APKEKAQPLEDLASFQELSQ | BD-Biosciences, mouse monoclonal, cat. no. 556003 | 1:150 (CI) |
| Mash1 | Recombinant-full length rat MASH1 (clone 24B72D11.1) | BD-Biosciences, mouse monoclonal, cat. no. 556604 | 1:2,000 (TSA) |
| NCAM | Raised against the 180, 140 and 120 kDa forms of NCAM derived from adult rat brain. | Dr. J. Covault, rabbit polyclonal | 1:1,200 (CI) |
| NeuroD1 | C-terminal synthetic peptide of mouse NeuroD1 (G-20): GSIFSSGAAAPRCEIPIDNI | Santa Cruz, goat polyclonal, cat. no. sc-1086 | 1:40 (CI) or 1:1,600 (TSA) |
| Neurog1 | N-terminal synthetic peptide of mouse Neurog1 (A-20): ARLQPLASTSGLSVPARRSAK | Santa Cruz, goat polyclonal, cat. no. sc-19231 | 1:1,600 (TSA) |
| OMP | Purified rodent olfactory marker protein | Wako chemicals, goat polyclonal, cat. no. 544-10001 | 1:3,500 (DAB) |
| Pax6 | C-terminal peptide sequence of mouse/rat/ human Pax6: QVPGSEPDMSQYWPRLQ | Chemicon, rabbit polyclonal, cat. no. AB5409 | 1:1,200 (CI) |
| PCNA | C-terminal peptide corresponding to a.a. 243-261 of Human PCNA: DMGHLKYYLAPKIEDEEGS | abcam, rabbit polyclonal, cat. no. ab2426 | 1:200 (CI) |
| PGP9.5 | Purified human PGP9.5 protein from brain | Ultraclone, rabbit polyclonal, cat. no. 31A3 | 1:1,200 (CI) or 1:3,500 (DAB) |
| Sox2 | C-terminal peptide (a.a. 277-293) of human Sox2 (Y-17): YLPGAEVPEPAAPSRLH | Santa Cruz, goat polyclonal, cat. no. sc-17320 | 1:300 (CI) |
| TGFα-mAb213 | Full-length recombinant human TGFα, clone 213-4.4 | Oncogene Science, mouse monoclonal, mAb213 | 1:20 (CI) |
| Tuj1 | Raised against microtubules derived from rat brain. | Covance, mouse monoclonal, cat. no. MMS-435P | 1:200 (CI) |
Visualization by conventional immunofluorescence (CI), by 3,3′-diaminobenzidine (DAB), or by Tyramide Signal Amplification (TSA).
The β-galactosidase antiserum recognizes the LacZ gene product among neurons of the olfactory epithelium with the same cellular distribution as described by Schwarting et al. (2007). The β-gal labeling is restricted to the neural progenitors in NeuroD1-LacZ mice, in a similar pattern to NeuroD1 mRNA and protein expression (Manglapus et al., 2004, Guo et al., 2010).
The Cre antiserum recognizes a single 35-kDa band corresponding to the bacterial enzyme in western blots. Cre immunolabeling is specifically detected and costains, along with GFP, among cells that express a Cre-IRES-eGFP transgene from a genetically engineered mammalian promoter in the retina (Taranova et al., 2006).
The gustducin antiserum (Gulbransen et al., 2008) is the canonical antibody marker for solitary chemoreceptor cells in the nasal epithelium by researchers in the field (Ogura et al., 2010). Western blots show a 45-kDa band that is specific for gustducin (Miura et al., 2007)
The Ki67 antiserum reacts with a 345/395 kDa protein doublet on western blots. In addition, binding to cells is blocked by the canonical anti-Ki67 monoclonal antibody, MIB 1.
The Mash1 monoclonal antibody recognizes a single 34-kDa band on western blots of fetal brain. Mash1 antibody labeling is also coexpressed along with green fluorescent protein (GFP) in the spinal cord of transgenic mice that have eGFP knocked into the Mash1 locus (Wildner et al., 2006).
The NCAM antiserum recognizes the three modified forms of the protein, derived from rat brain, corresponding to their 120, 140, and 180-kDa molecular weights by western blots. NCAM antiserum stained a pattern of cellular morphology and distribution in the rodent olfactory epithelium that is identical with previous reports (Jang et al., 2007).
The NeuroD1 antiserum recognizes a single 50-kDa band on western blots, and the post-MeBr lesion timing and pattern of labeled cells corresponds well with ISH studies (Manglapus et al., 2004). NeuroD1 protein is also detected in neural progenitor cells of the rat olfactory epithelium in a pattern that reflects its mRNA expression pattern (Guo et al., 2010).
The Neurog1 antiserum recognizes a single 27-kDa band on western blots; the timing, number, and pattern of antiserum-labeled cells during recovery from MeBr lesions all fit with ISH studies in the olfactory epithelium (Manglapus et al., 2004).
The OMP antiserum selectively labels mature olfactory sensory neurons in mouse nasal mucosa along with all of the β-gal-positive neurons derived from OMP-LacZ-expressing mice (Walters et al., 1996). After epithelial injury in OMP-LacZ mice, immunodetectable OMP disappears concomitant with the elimination of β-gal labeled neurons.
The Pax6 antiserum recognizes a single 50-kDa band on western blots of fetal mouse brain. It exclusively labels the transcription factor in nuclei of pancreatic endocrine cells; however, mutants defective in Pax6 nuclear localization are detected in the cytoplasm due to sensitive immunolabeling (Dames et al., 2010). Pax6 is also detected in rodent olfactory epithelium in an identical pattern using another antibody raised in a different host species.
The proliferating cell nuclear antigen (PCNA) antiserum recognizes a 29-kDa band on western blots of human epithelial carcinoma cell lines within the nuclear fraction.
The PGP9.5 antiserum recognizes a single 27-kDa band on western blots of mouse brain. The subcellular labeling for PGP9.5 is cytoplasmic and restricted to neural populations in mice, including within the vomeronasal organ and olfactory epithelium (Ishida et al., 2008; Guo et al., 2010).
The Sox2 antiserum recognizes a single 34-kDa band in western blots of mouse and human embryonic stem cell lysates. This antibody generates an identical staining pattern in rat olfactory epithelium as another Sox2 antibody raised in different host species. Conditional deletion of Sox2 extinguishes immunolabeling in mouse cortex by the antibody (Favaro et al., 2009).
The mAb213 (TGF alpha) monoclonal antibody labels a distinct subset of neurons within the olfactory epithelium in both rats (Ring et al., 1997; Guo et al., 2010) and mice (current publication). As demonstrated in the original article from this laboratory (Ring et al., 1997), the antibody is not binding to TGF-alpha in the necklace neurons, but some other, as yet uncharacterized epitope, that is limited to the necklace neurons, making this antibody a useful marker for them.
The Tuj1 monoclonal antibody is well characterized and highly reactive to neuron-specific Class III β-tubulin (βIII). Tuj1 does not identify β-tubulin found in glial cells.
The following secondary antibodies were used, Alexa-Fluor488-donkey antigoat IgG, AlexaFluor488-donkey antimouse IgG, AlexaFluor488-donkey antirabbit IgG, AlexaFluor594-donkey antigoat IgG, AlexaFluor594-donkey antimouse IgG (all from Invitrogen/Molecular Probes, Eugene, OR) each used at a dilution of 1:250. Additionally, AlexaFluor488-conjugated streptavidin and Alexa-Fluor594-conjugated streptavidin (Invitrogen/Molecular Probes) were used at dilutions of 1:250 for conventional immunofluorescence, but at a dilution of 1:1,250 for tyramide signal amplification (TSA). The remaining secondary antibodies were used at a dilution of 1:50 and were purchased from Jackson Immunoresearch (West Grove, PA): biotin-conjugated donkey antigoat IgG, biotin-conjugated donkey antimouse IgG, biotin-conjugated donkey antirabbit IgG, FITC-conjugated donkey antigoat IgG, FITC-conjugated donkey antirabbit IgG, AMCA-conjugated donkey antimouse IgG, Texas Red-conjugated donkey antigoat IgG.
Histochemistry and immunohistochemistry
For histochemical detection of β-gal, tissue was reacted with 5-bromo-4-chloro-3-indolyl-d-D-galactopyra-noside solution (X-gal, Sigma, St. Louis, MO) according to previously published procedures (Huard et al., 1998). Standard immunohistochemical protocols were used to detect the expression pattern of individual proteins in normal OE and mutant OE (Table 1). Adequate labeling with a number of the antibodies required a series of pre-treatments to the sections prior to incubation with the reagents. Briefly, sections were rinsed in PBS for 5 minutes to remove OCT, puddled with 0.01M citric acid buffer (pH 6.0), then heated by placing in a commercial food steamer (Oster, model no. 5712) for 10 minutes (“steaming”). After cooling, sections were rinsed with PBS briefly before incubating with blocking solution (10% serum + 5% nonfat dry milk + 4% bovine serum albumin [BSA] + 0.01% Triton X-100) for 30 minutes at room temperature. The analyses conducted here depended on a number of double- and triple-immunohistochemical staining approaches. In all cases the sections were incubated with primary antibodies overnight at 4°C. In Table 1 immunolabeling methods are classified as one of three types: 1) conventional immunofluorescence detected with fluorescent probes either directly conjugated to secondary antibodies or to fluorescently conjugated streptavidin in conjunction with biotin conjugated secondary antibodies; 2) use of 3,3′-diaminobenzidine (DAB) as chromogen; or 3) use of TSA to enhance a weak signal or permit staining with two antibodies from the same species. In all cases the specificity of the secondary antibody was confirmed by staining in the absence of primary antibody as a control. For conventional immunofluorescence, following primary antibody incubation, sections were washed thoroughly and incubated with secondary antibodies for 1 hour at room temperature. After washing, sections were either coverslip-mounted in p-phenylenedi-amine (PPD) or incubated with fluorescently conjugated streptavidin molecules (after which they were mounted). For staining with the chromogen DAB, after incubation with primary antibody sections were washed and incubated in the corresponding biotin-conjugated secondary antibody, incubated for 1 hour at room temperature. After washing, sections were incubated in avidin-bHRP conjugate (Elite ABC Kit, Vector Laboratory, Burlingame, CA) and DAB as chromogen. For staining using TSA, sections were washed after incubation with primary antibody and incubated in the corresponding biotin-conjugated secondary antibody for 1 hour at room temperature. After washing, sections were treated with reagents provided by the TSA kit according to kit instructions (Renaissance TSA Biotin System, Perkin-Elmer, Boston, MA, cat. no. NEL700A).
Photography/image processing
Sections were imaged with a Spot RT2 color digital camera attached to Nikon 800 E microscope. Image preparation, assembly, and analysis were performed in Adobe Photoshop CS2 (San Jose, CA). In the vast majority of photos, only balance, contrast, and evenness of the illumination were altered. On rare occasions the photos were cropped to eliminate off-the-section debris.
RESULTS
NeuroD1 expression marks the olfactory mucosa throughout adulthood
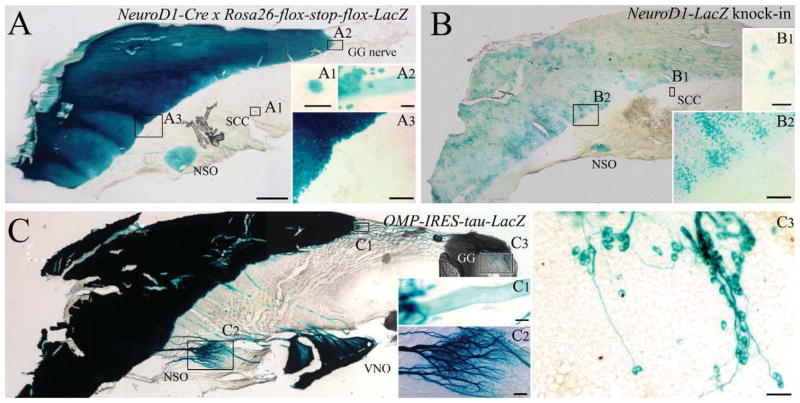
The progeny of NeuroD1-expressing precursor cells were identified in the adult OE by expression of β-gal in ΔNeuroD1-Cre × Rosa26-flox-stop-flox-LacZ (NeuroD1-Cre/LacZ) bigenic mice and by the detection of eYFP in ΔNeuroD1-Cre × Rosa26-flox-stop-flox-eYFP (NeuroD1-Cre/eYFP). In the X-gal-stained whole mounts of the nasal septal mucosa from NeuroD1-Cre/LacZ mice β-gal(+)/ NeuroD1-derived cells fill the olfactory mucosa at a high density, up to a sharp border with respiratory epithelium (RE) (Fig. 1A). The neuronal labeling is as extensive and dense as that observed in ΔOMP-IRES-tau-LacZ mice, where β-gal is selectively expressed by mature OSNs (compare Fig. 1A, C). Situated anteroventral to the olfactory/respiratory boundary, the nasal septal organ (NSO) is also densely labeled (Fig. 1A). Deep to the respiratory epithelium and rostral to the NSO, fascicles of the vomer-onasal nerve are X-gal-stained, in agreement with Suzuki et al. (2003), and are easily identified by location and appearance with reference to the ΔOMP-IRES-tau-LacZ nasal septum (compare Fig. 1A, C). In addition, X-gal labels nerve fascicles that extend from the dorsal anterior nasal septum toward the olfactory/respiratory boundary in the NeuroD1-Cre/LacZ mice (Fig. 1A). The fascicles are composed of axons projecting from the Grunenberg ganglion at the far rostral edge of the nasal septum, as shown by comparison with OMP-driven LacZ expression (Fig. 1C). The labeling of axons of the vomeronasal nerve and of the projection from the Grunenberg ganglion suggest that the neurons themselves also derive from a NeuroD1-expressing progenitor. That interpretation was confirmed subsequently by examination of cryosections from NeuroD1-Cre/eYFP mice. Finally, rare X-gal-labeled cells are scattered throughout the respiratory epithelium of the bigenic NeuroD1-Cre/LacZ mice (Fig. 1A). The rare NeuroD1-derived cells in respiratory epithelium do not appear to be solitary chemoreceptor cells (SCCs) (Finger et al., 2003). They are fewer in number than the solitary chemoreceptors and, in cryosections of NeuroD1-Cre/ eYFP or NeuroD1-Cre/LacZ bigenic mice, gustducin(+) solitary chemoreceptors do not express the marker protein at detectable levels (in contrast to the foregoing neuronal populations) (Fig. 2); well over 100 gustducin(+) SCCs were examined in multiple locations of the NeuroD1-Cre/eYFP mice including the opening of the duct of the vomeronasal organ (Ogura et al., 2010), the nonsensory epithelium of the vomeronasal organ, and the respiratory epithelium within the main nasal cavity.
Figure 1.
Expression of NeuroD1 by GBCs and the progeny of NeuroD1-expressing GBCs are generally restricted to the sharply bounded domain of the olfactory epithelium and persist throughout adulthood. A–C: Adult mouse septal whole mounts from ΔNeuroD1-Cre × Rosa26-flox-stop-flox-LacZ (A), ΔNeuroD1-LacZ (B) and ΔOMP-IRES-tau-LacZ (C) mice stained with X-gal. X-gal-labeled neurons fill the entirety of the OE (A3, B2 inset) and NSO (A–C), while axons from the VNO (A, C) and GG (A2, C1) are apparent as well. Scattered cells in the respiratory epithelium (A1, B1) are likely SCCs, previously described by Finger et al. (2003). X-gal labeled cells are clustered and largely limited to OE (B2); however, the NSO and putative SCCs (B1) also react with X-gal. Mature, OMP-expressing neurons are provided for anatomical orientation, depicting the NSO (C2), VNO, and GG (C1, C3) by X-gal stain. Scale bars = 500 μm in A (applies to B, C); = 20 μm in A1, A2, B1; 100 μm in A3, B2, C2; 50 μm in C1; 25 μm in C3.
Figure 2.
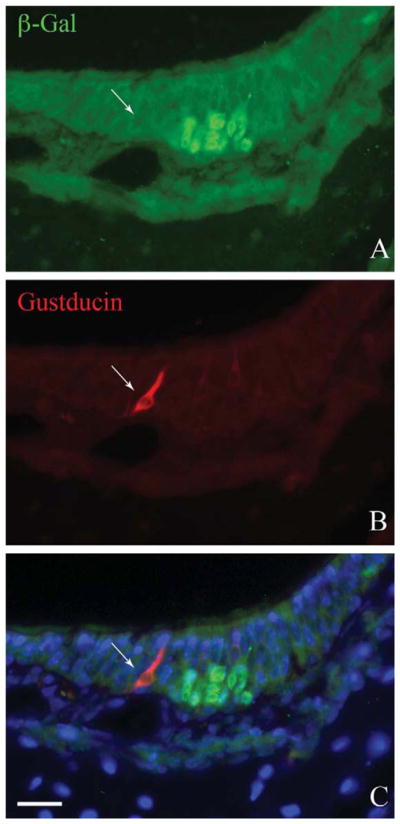
The gustducin(+) solitary chemoreceptor cells apparently do not derive from a NeuroD1-expressing progenitor. Section through a cul-de-sac showing a small patch of olfactory epithelium. A: Anti-β-gal staining marks the cells of the olfactory epithelium that derive from a NeuroD1-expressing progenitor. B: Anti-gustducin marks a solitary chemoreceptor cell in the respiratory epithelium adjacent respiratory epithelium, which shows no evidence of β-gal labeling. C: Merged image including Hoechst nuclear counterstain (blue). Scale bar = 20 μm.
The foregoing pattern of expression is identical in two of three independently generated NeuroD1-Cre/LacZ BAC transgenic driver lines. In the third line the density of labeling in the OE was both substantially less and variable across the OE, although the labeled neurons extended throughout the OE and respected similar boundaries as the other two BAC lines. The X-gal pattern in the third line suggests that some aspects of BAC transgene expression are under the influence of the locus of transgene integration. As a consequence, no further analysis was done on that line.
We compared the distribution of NeuroD1-derived progeny across the epithelial sheet with the expression of the NeuroD1 gene at the time of tissue harvest, using ΔNeuroD1-LacZ reporter animals. In these mice the LacZ gene was knocked into the NeuroD1 locus, rendering it null and putting LacZ expression under the control of the endogenous NeuroD1 promoter. Thus, the detection of β-gal labeling serves as a reporter for NeuroD1 expression during the period leading up to perfusion, recognizing that there is likely to be some perdurance of LacZ by comparison with the NEUROD1 protein. Clusters of X-gal labeled cells are distributed throughout the olfactory domain, up to its boundary with the respiratory epithelium (as depicted above, Fig. 1B). Likewise, X-gal-labeled cells are also seen in the nasal septal organ of the ΔNeuroD1-LacZ reporter mice (Fig. 1B). Finally, scattered cells are also detectable throughout the respiratory epithelium with a similar distribution as the NeuroD1-Cre/LacZ mice. The clustering of X-gal-positive cells within the plane of the OE is consistent with the description of NeuroD1-expressing progenitor cells in the adult OE shown by ISH (Manglapus et al., 2004).
β-Gal expression marks the vast majority of OSNs
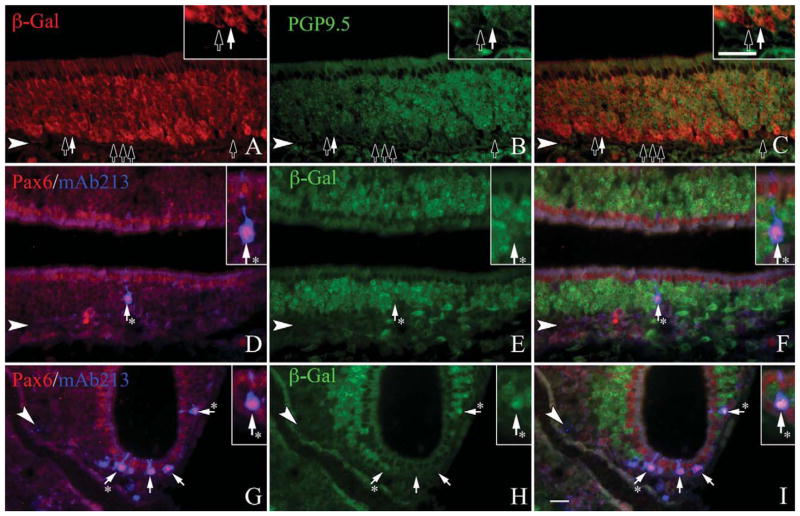
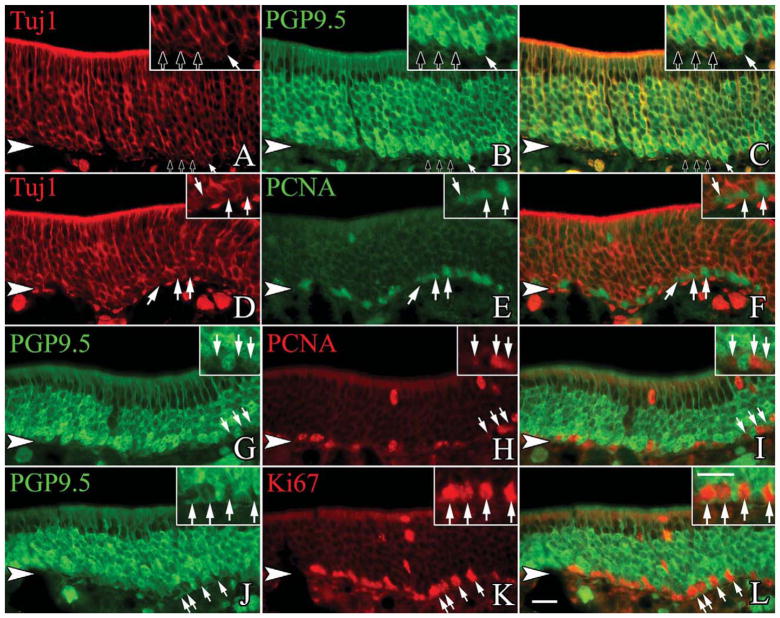
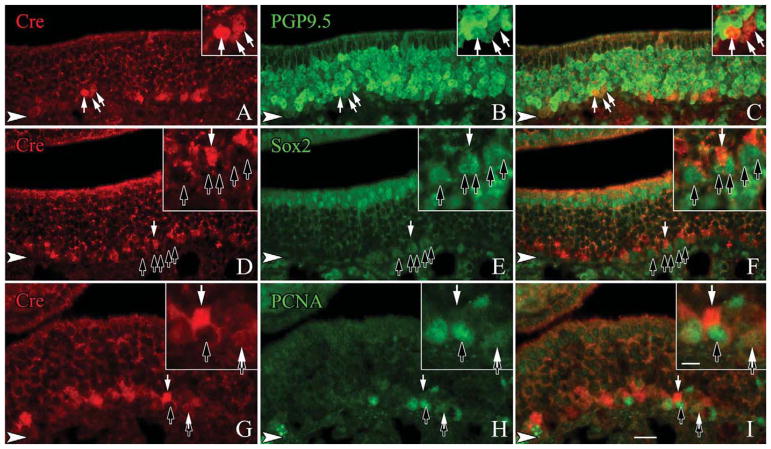
The density of β-gal labeling in the whole mounts of the NeuroD1-Cre/LacZ bigenic mice suggests that most, if not all, OSNs are labeled via this lineage tracing approach. We assessed the extent to which β-gal was coexpressed with neuronal markers in sections of OE from the NeuroD1-Cre/LacZ mice. Tissue sections were stained with anti-β-gal and anti-PGP9.5, a marker of the neuronal lineage in the OE. As expected, anti-PGP9.5 labels the full height of the stratum occupied by OSNs, leaving sustentacular cells unstained above them and basal cells below (Fig. 3A–C). Anti-β-gal also labels all, or nearly all, of the cells that are heavily labeled with anti-PGP9.5, but staining for β-gal is not detectable in weakly PGP9.5(+) cells, which are situated at the deep edge of the neuronal strata. Many of the weakly PGP9.5(+)-cells are mitotically cycling, judging by coexpression of Ki-67 and PCNA (Fig. 4G–L). The weakly PGP9.5(+) cells also stain with the antineuron-specific tubulin (NST) monoclonal antibody TuJ1 but not with anti-NCAM, suggesting that expression of PGP9.5 and NST anticipates terminal mitosis and the onset of postmitotic neuronal differentiation in the mouse. In light of data described below indicating that NEUROD1 is expressed by proliferating GBCs, the absence of detectable levels of β-gal in the lightly stained PGP cells most likely reflects the need for β-gal to accumulate to a level sufficient for detection and/or the latency between Cre expression and Cre-mediated recombination. We were able to demonstrate Cre labeling among the basal cells and immature neurons as well (Fig. 5A–I). Taken together, we interpret these data as indicating that all typical OSNs pass through a NeuroD1-expressing stage during differentiation.
Figure 3.
NeuroD1 is expressed in the vast majority of OSNs at some point during their differentiation. A–C: Sections from the olfactory epithelium of adult, ΔNeuroD1-Cre × Rosa26-flox-stop-flox-LacZ mice are stained with β-gal and PGP9.5, a marker of all typical OSNs. DI: Sections stained with Pax6/mAb213, a marker of atypical OSNs. Most PGP9.5-labeled cells colocalize with β-gal (A–C, white arrows); however, some do not (A–C, black arrows). In the septum (D–F) and the cul-de-sacs of the OE (G–I), the majority of β-gal-expressing cells colabel with anti-Pax6 and mAb213 (white arrows with asterisks); rarely, staining for β-gal is undetectable (black arrows). The arrowheads mark the basal lamina in all panels. Scale bar = 20 μm.
Figure 4.
A small subset of the PGP9.5 and Tuj1-labeled population express markers of the cell cycle. Sections from adult, wildtype mice are stained with anti-PGP9.5, Tuj1, and cell cycle markers. A–C: Tuj1-labeled cells at the base of the neuronal layer also show heavy expression of PGP9.5 on occasion (white arrow), although the vast majority of basally situated, Tuj1-labeled cells are only very lightly labeled for PGP9.5 (black arrows). D–F: Of the basal-most Tujl-labeled cells, a large percentage of them express PCNA, a marker of S-phase (white arrows). G–I: The basally located, lightly labeled, PGP9.5-positive cells also coexpress PCNA frequently (white arrows). J–L: Additionally, a large number of the light PGP9.5-labeled cells coexpress Ki67, a broad marker for cells in the mitotic cycle (white arrows). The arrowheads mark the basal lamina in all figures. A magenta-green version of this figure is available online as Supporting Information Figure 1. Scale bar = 20 μm.
Figure 5.
The vast majority of the Cre-expressing cells are likely to be immediate neuronal precursors as they are only lightly labeled with PGP9.5. Sections from adult, wildtype mice are stained with anti-Cre and markers of undifferentiated neuronal precursors. A–C: A large number of basally situated, lightly labeled, PGP9.5-positive cells coexpress Cre (white arrows). D–F: In contrast, none of the SOX2-labeled GBCs label with Cre (black arrows) although Cre-only labeled cells are seen (white arrow). G–I: Cre is generally not found at a detectable level in PCNA-labeled, cycling cells (black arrows), except rarely when a PCNA-positive cells is also lightly labeled with Cre (white arrow). Occasionally, light Cre labeling is detected in cells that also have light labeling for PCNA (tandem black and white arrows). The arrowheads mark the basal lamina in all figures. A magenta-green version of this figure is available online as Supporting Information Figure 2. Scale bar = 20 μm.
In addition to the typical OSNs, the density of labeling suggests that NeuroD1-expressing progenitors can give rise to atypical OSNs as well. One such set of atypical OSNs is the group of neurons that target the necklace glomeruli (Ring et al., 1997; Cockerham et al., 2009; Zufall and Munger, 2010). The necklace glomeruli encircle the olfactory peduncle at the caudal edge of the glomerular sheet; these neurons use a distinct signal transduction pathway and play a role in the detection of social cues (Ring et al., 1997; Cockerham et al., 2009; Zufall and Munger, 2010). Typical markers for necklace neurons (or a subset thereof), include monoclonal antibody 213-4.4 (mAb213) and Pax6 (Ring et al., 1997; Guo et al., 2010). Both markers were applied to the NeuroD1-Cre/LacZ-derived tissue used for lineage tracing. mAb213(+)/Pax6(+) necklace neurons are concentrated in the OE lining the caudal septum and the cul-de-sacs of the nasal cavity, and our analysis focused there. In the NeuroD1-Cre/LacZ mice the vast majority of necklace neurons in the septal epithelium display detectable expression of β-gal (Fig. 3D–F). Pax6(+) and MAb213(+) necklace neurons in the cul-de-sacs show variable β-gal expression ranging from none to very dense staining, but most of them are visibly labeled with anti-β-gal (Fig. 3G–I). Those necklace neurons that do not label with anti-β-gal may reflect the delay between onset of LacZ expression and accumulation of sufficient β-gal as seen earlier with the typical neurons of the OE (Fig. 3A–C). Taken together, these data suggest that the vast majority of typical and atypical OSNs descend from a NeuroD1-expressing progenitor at some point during their differentiation.
NeuroD1 expression is largely confined to cells that lie upstream of OSNs
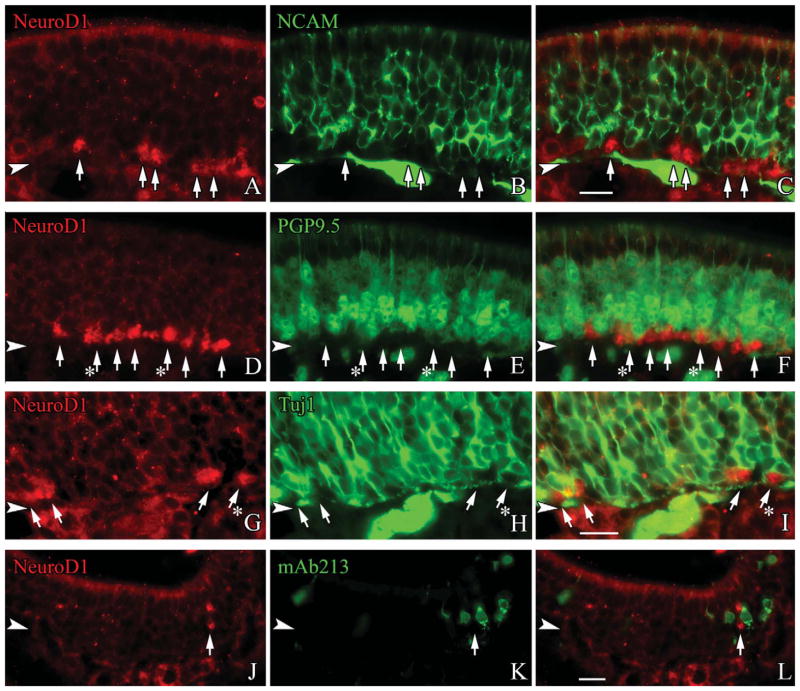
The foregoing results do not tell us which cells near the base of the OE express NeuroD1, only that they are upstream of differentiating and fully differentiated OSNs. Accordingly, we used double immunofluorescence labeling with anti-NEUROD1 and a variety of other markers to characterize the NEUROD1-expressing cells of the OE. NEUROD1-labeled cells are present in and limited to the basal part of the epithelium, and are found in clusters and in singletons, through all regions of the OE. Where the NEUROD1(+) cells sit, i.e., immediately superficial to the HBC monolayer, suggests that they must be either GBCs or very immature neurons. In comparison with the neuronal markers NCAM, Tuj1, and PGP9.5, the NEUROD1(+) basal cells show no evidence of labeling with anti-NCAM, which stains the membrane of bona fide, postmitotic neurons (Fig. 6A–C). However, the NEUROD1(+) cells often exhibit faint but detectable labeling for PGP9.5 and weak, but stronger staining for Tuj1 (Fig. 6D–I). As noted above, both PGP9.5 and Tuj1 anticipate terminal mitosis since we can identify PGP9.5 and Tuj1 marked cells that label with PCNA, Ki67, and other markers of proliferating cells (Fig. 5A–L).
Figure 6.
NEUROD1 is predominantly expressed prior to the onset of neuronal differentiation. Sections from adult, wildtype mice are stained with anti-NeuroD1 and a range of neuronal markers. A–C: NeuroD1-labeled GBCs always lack the mature neuronal marker NCAM (white arrows). D–F: Lightly PGP9.5-labeled cells at the basal margin of the neuronal layer typically lack NEUROD1 (white arrows), although occasionally there is coexpression of NEUROD1 in these lightly PGP9.5-labeled cells (white arrows with asterisks). G–I: While some NEU-ROD1-labeled cells lack any detectable Tuj1-reactivity (white arrows), many NEUROD1(+) positive cells are weakly labeled with TuJ1 (white arrows with asterisks). J–L: mAb213, a marker for atypical necklace neurons is undetectable in NEUROD1-labeled GBCs (white arrows). The arrowheads mark the basal lamina in all figures. A magenta-green version of this figure is available online as Supporting Information Figure 3. Scale bar = 20 μm.
Since necklace neurons also derive from a NeuroD1-expressing precursor, we assessed whether NeuroD1 anticipated necklace neuron differentiation. We find that labeling for NEUROD1 and mAb213 are mutually exclusive (Fig. 6J–L). The results with typical and atypical neurons, and lack of overlap with NEUROD1 and definitive neuronal markers, suggest that NEUROD1 marks a subset of GBCs that lie upstream of the postmitotic sensory neuronal population.
NeuroD1 is largely confined to proliferating GBCs that are at the immediate neuronal precursor stage
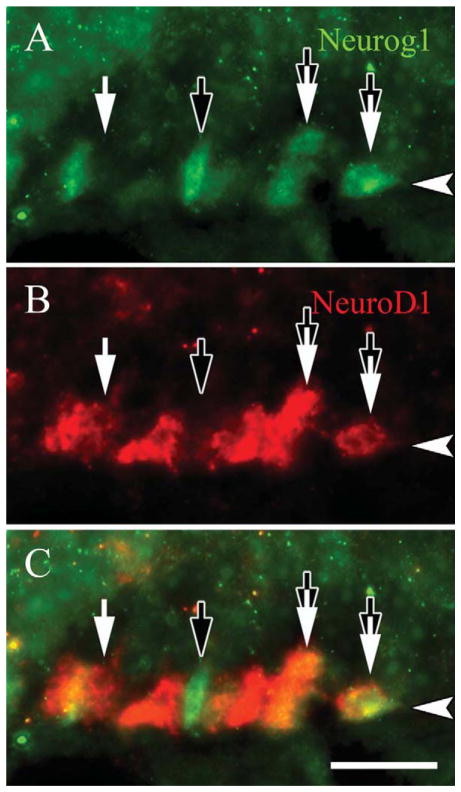
Roughly 90% of more of the mitotically active basal cells are GBCs (Graziadei and Graziadei, 1979; Schwartz Levey et al., 1991; Huard and Schwob, 1995; Goldstein et al., 1998). The foregoing data suggest that some GBCs express NeuroD1 and are mitotically active. We assayed directly whether NEUROD1-expressing basal cells are proliferating by using PCNA, a marker of the replication fork, as a marker of active cycling. The vast majority of NEUROD1(+) basal cells are PCNA-labeled (Fig. 7A–C). The NEUROD1(+) GBCs were also compared to the expression of other transcription factors that are found within the GBC population. For example, SOX2 marks a broad subset of GBCs including likely multipotent progenitors, transit-amplifying cells, and a fraction of immediate neuronal precursors that express NEUROG1 (Guo et al., 2010). We show here that roughly one-third of NEUROD1-labeled cells are SOX2(+) (Fig. 7D–F). Using MASH1 expression to identify the putative transit amplifying GBCs of the epithelium, we find that NEUROD1 and MASH1 label completely separate populations of basal cells (Fig. 7G–I). Likewise, we compared expression of MASH1 with anti-β-gal labeling in the NeuroD1-Cre/LacZ tissue, and saw no overlap of the two markers or of MASH1 and either PGP9.5 or Tuj1-staining (data not shown). Neurog1 expression slightly anticipates that of NeuroD1 in embryological development, but they are roughly contemporaneous during the regeneration of the adult OE that follows epithelial injury (Cau et al., 1997, 2002; Manglapus et al., 2004). We directly compared their expression patterns and find cells that are NEU-ROG1(+)-only and NEUROD1(+)-only, but at least half of the population is colabeled for both (Fig. 8). These data directly demonstrate that NEUROD1 is expressed by proliferating GBCs and suggest that the NEUROD1(+) GBCs lie downstream of MASH1(+) transit amplifying cells (since there is no overlap between MASH1 and β-gal labeling in the bigenic mice and no overlap between MASH1 and TuJ1 or PGP9.5), but upstream of differentiating neurons.
Figure 7.
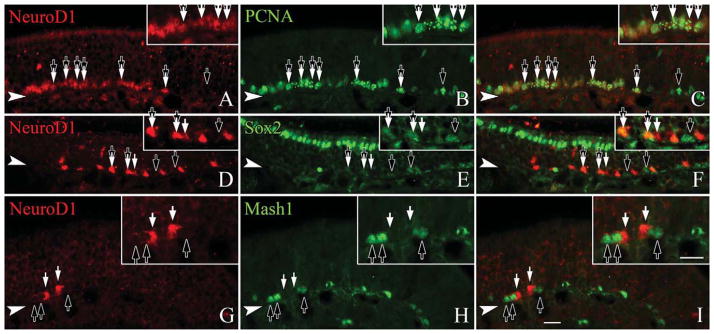
NeuroD1 is expressed in GBCs that retain cell cycle markers and lie downstream of Mash1. Sections from adult, wildtype mice are stained with anti-NeuroD1 and anti-PCNA, a marker of cell cycle progression (A–C), SOX2 (D–F), or MASH1 (G–I), as markers of GBCs. A–C: Nearly all NEUROD1-labeled cells coexpress PCNA (tandem black and white arrows), although not all PCNA-labeled cells are NEU-ROD1-positive (black arrows). D–F: Roughly one-third of the NeuroD1 labeled cells coexpress SOX2 (tandem black and white arrows), a marker for multiple subsets of GBCs, although some NEUROD1-labeled cells lack SOX2 (black arrows). G–I: NEUROD1-labeled cells (white arrows) are not labeled by anti-MASH1 (black arrows), a marker for transit amplifying GBCs. The filled arrowhead marks the basal lamina in all figures. A magenta-green version of this figure is available online as Supporting Information Figure 4. Scale bar = 20 μm.
Figure 8.
NeuroD1 is coexpressed in immediate neuronal precursors that are NEUROG1-positive. Sections from adult, wildtype mice are stained with (A) anti-NeuroD1 and (B) NEUROG1, which are both reputed to be markers of immediate neuronal precursors in the OE. C: Roughly half of the NeuroD1-labeled GBCs colabel for NEUROG1 (tandem black and white arrows), although some of the GBCs stain for only NEUROD1 (white arrows) or for only NEU-ROG1 versa (black arrows). The arrowheads mark the basal lamina in all figures. A magenta-green version of this figure is available online as Supporting Information Figure 5. Scale bar = 20 μm.
Effects of NeuroD1 knockout on adult OE
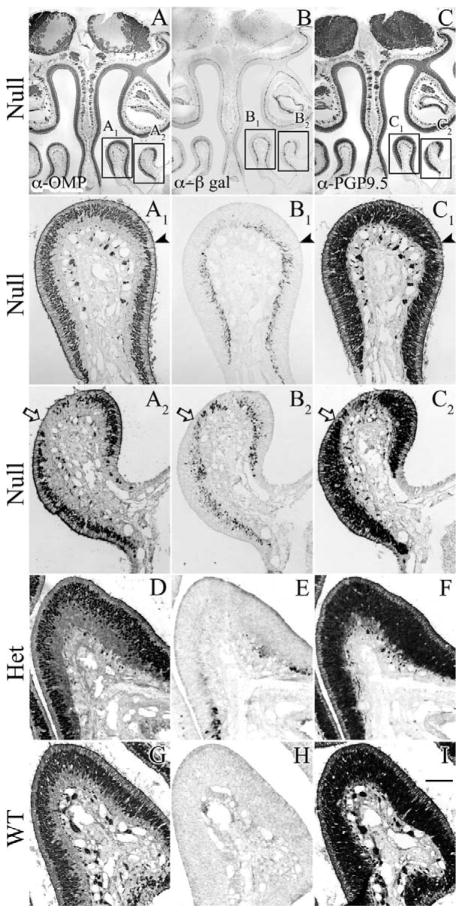
Since NeuroD1 is apparently expressed in immediate neural precursors just prior to the transition to differentiated OSNs, we determined whether mutation of the NeuroD1 gene, rendering it null, has an effect on neuronal differentiation. The ΔNeuroD1-LacZ knockin/knockout mice were bred to homozygosity and analyzed at postnatal day (P)25–P36. We screened a panel of cell type and neuronal stage-specific markers in order to determine whether neuronal progenitor development and renewal were intact despite the loss of NeuroD1 gene function. At this timepoint, most of the OE is indistinguishable from wild-type or heterozygous control mice with respect to dense labeling for PGP9.5 (to mark both immature and mature OSNs) and for OMP (to mark mature OSNs selectively). Thus, the absence of NEUROD1 function does not compromise the establishment of a robust population of OSNs. However, on closer examination some subtle abnormalities are seen in the NeuroD1−/− ice; at its most extreme, areas of the OE have a scant-to-absent population of mature neurons, without a compensatory increase in the population of immature neurons (Fig. 9A2–C2). In other areas, the stratum occupied by OMP(+) mature neurons is substantially thinner by comparison with wildtype or heterozygous mice and even by comparison with the same spot on the contralateral side of the null mouse (Fig. 9A1, C1, D–I). Abnormalities are noted in the null mice that have been examined to date. Although NeuroD1 has been linked to neuronal survival in adult, differentiated cells, there is no indication of accelerated cell death in the affected OE, since staining for cleaved caspase-3 (Pennesi et al., 2003; Gao et al., 2009) shows no evidence of caspase activation here (data not shown). β-Gal-reactive cells are found in the affected areas, indicating that NeuroD1-expressing GBCs have not disappeared from there. However, their numbers are less than would be seen in experimental circumstances where the neuronal population is depleted to a similar extent (Fig. 9A2–C2).
Figure 9.
Null mutation of NeuroD1 results in discrete areas that lack or have thinner layers of mature neurons. Sections from 4-week-old (A–C) ΔNeuroD1-LacZ null (D–F) ΔNeuroD1-LacZ heterozygous, and (G–I) wildtype mice. A, D, G: Anti-OMP staining to mark mature neurons. B, E, H: Anti-β-Gal staining to mark the cells that are, or recently derived from, a NeuroD1-expressing progenitor. C, F, I: Anti-PGP9.5 staining to mark both immature and mature neurons. In its most severe case, loss of NeuroD1 results in an epithelium containing isolated patches that lack mature neurons, while retaining a population of immature neurons and detectible NeuroD1-expressing progenitors, determined by LacZ expression (A2-C2, white arrow). In its least severe case, affected areas of the OE have a thinning in the labeling for mature neurons, while retaining ample expression of β-gal indicative of NeuroD1-expressing progenitors (A1–C1, black arrowhead). In contrast, within the heterozygote, loss of one functional allele has no overt effect similar to wildtype OE (D–I). Scale bar = 20 μm.
DISCUSSION
The current work demonstrates that all typical OSNs and most, if not all, neurons targeting the necklace glomeruli are descendents of a NeuroD1-expressing progenitor cell, as are neurons of the nasal septal organ, the vomeronasal organ, and the Grunenberg ganglion. NEUROD1 protein is found in immediate neuronal precursors that lack MASH1, occasionally colabel with SOX2, and frequently colabel with NEUROG1. NEUROD1(+) precursors are actively in the cell cycle and lack definitive neuronal markers. NeuroD1 is not required for the generation of OSNs since most areas of the epithelium have an apparently normal complement of neurons; however, isolated patches within the OE of mutant mice have an absence, or thinning, of the mature neuronal layer, suggesting that NeuroD1 plays a role in the progression to, or survival of, mature olfactory sensory neurons in some fashion under some circumstances. It is not yet clear whether, with time, all areas of the epithelium would undergo some detectable degree of degeneration. Such a determination would require conditional, i.e., OE-specific deletion of the NeuroD1 gene to allow for the extended survivals required.
Lineage tracing by way of NeuroD1-Cre/LacZ mice reveals near-universal marking of the neuronal population by β-gal expression throughout the whole OE, yet the population of cells that label for NEUROD1 by immunohistochemistry or for LacZ in the ΔNeuroD1-LacZ knockin/ knockout mice is far smaller. The discrepancy in numbers is fully expected from prior results examining the expression of NeuroD1 in the embryonic mice epithelium or adult regenerating OE, in which the NeuroD1(+) cells are few (Cau et al., 1997; Manglapus et al., 2004). The abundance of labeled neurons merely suggests that NeuroD1 is expressed in a small population of progenitor cells before being switched off prior to the onset of postmitotic differentiation and that the expression of the targeted gene is far more transient than that of the recombined and translated LacZ transgene in the ROSA locus of the NeuroD1-Cre/LacZ mice. We cannot fully exclude the possibility that low levels of NeuroD1, below the level of detection, may be expressed in the neuronal population in amounts that manage to catalyze the recombination of the reporter by way of Cre expression. This latter explanation seems unlikely, since more stable proteins expressed from the same promoter are also undetectable in the neuronal layer, i.e., as seen in the ΔNeuroD1-LacZ knockin mice, which do not show detectable levels of β-gal in neurons. As additional support of the notion that NeuroD1 is transiently expressed by some neuronal progenitors, the small population of NEUROD1(+) cells is found among the globose basal cell population, and none of them appear to be immunodetectable neurons.
The lineage tracing also shows a difference in the expression patterns of neuronal markers and β-gal labeling, such that we detect some bona fide neurons at the deep margin of the neuronal layer that express PGP9.5 and Tuj1 densely, but lack detectable β-gal via cytochemical or immunohistological visualization. That NEUROD1 is found in proliferating GBCs, yet β-gal is undetectable in them or at least some of the youngest neurons, may seem something of a conundrum. The extensive overlap of NEUROG1 and NEUROD1 expression both here and during recovery from MeBr lesion suggests that our assignment of NeuroD1 to GBCs is correct (Manglapus et al., 2004). Hence, we favor the interpretation that the absence of a detectable label is caused by the slow accumulation of β-gal from the recombined Rosa26 locus, which, in our experience (unpublished data), is poorly expressed in the neuronal lineage of the OE and the consequent delay in reaching detectability. Also in favor of our explanation is the perfect coexpression of β-gal and neuronal markers in all neurons of the upper four-fifths of the neuronal stratum. In other words, the absence of β-gal label from the most deeply situated neurons is not an indication that they do not arise via a NeuroD1-expressing precursor. The delay between expression of NeuroD1 and detection of β-gal may also reflect in part a lag between the onset of Cre expression and its accumulation in the nucleus, an explanation that is supported by the discrepancy between NeuroD1 expression and anti-Cre labeling in the nucleus. A delay in accumulation of demonstrable levels of β-gal may also be characteristic of mAb213(+) necklace neurons, since a small percent of them also lack detectable β-gal. Alternatively, the population of mAb213(+) necklace neurons may be heterogeneous with respect to progenitor origin; however, this seems unlikely given the result for other OSNs.
It is remarkable that NeuroD1 is functioning so broadly across the nasal epithelium including the neuronal progenitors of the OE, the NSO, the VNO, and the Grunen-berg ganglion. In addition, the scattered LacZ(+) cells found in the respiratory epithelium suggest that yet another type of cell, almost certainly not SCCs but otherwise undefined, also takes origin from a NeuroD1-expressing progenitor. The common use of a NeuroD1-expressing precursor in all of these tissues may reflect, at least in part, the evolution, development, and differentiation from a common ancestor, i.e. the olfactory placode, upon which more specialized sensory elements are imposed, as is seen in the brain, limbs, and other sensory systems (Sordino et al., 1995; Fritzsch and Beisel, 2001; Lall and Patel, 2001; Reichert and Simeone, 2001; Davidson and Erwin, 2006).
Role of NeuroD1 in the OE
Several features of the expression of NeuroD1 in the OE are different from its role in other neurogenic regions. Elsewhere, exogenous expression of NeuroD1 results in immediate neuronal differentiation; likewise, only a small fraction of NeuroD1-expressing precursors are active in the mitotic cycle, suggesting that expression of NEU-ROD1 leads to an immediate exit from the cycle (Lee et al., 2000; Gao et al., 2009; Kuwabara et al., 2009; Puligilla et al., 2010; Boutin et al., 2010). However, we find that nearly all the NEUROD1-labeled cells in the OE coexpress markers of cycling cells such as PCNA, Ki67, and Edu (not shown) and are GBCs, suggesting the NeuroD1-expressing progenitors of the OE exhibit a greater progenitor cell capacity than previously reported in other systems.
In the dentate gyrus, SOX2 tightly regulates NeuroD1 by repressing it and, in turn, NEUROD1-labeled cells coexpress SOX2 rarely, if ever (Gao et al., 2009; Kuwabara et al., 2009). In this respect, too, NEUROD1(+) OE progenitors are subtly different; we find that anti-SOX2 labels a significant fraction of NEUROD1-labeled GBCs, as we previously reported in rat OE (Guo et al., 2010). Given that a canonical role for Sox2 is repression of neuronal differentiation, its coexpression with NeuroD1, a factor that pushes those same progenitors toward neuronal differentiation, suggests a broader function for Sox2 in the OE (Graham et al., 2003; Ellis et al., 2004; Bani-Yaghoub et al., 2006; Boutin et al., 2010; Puligilla et al., 2010). Any repression of NeuroD1 by SOX2 can only be partial and may reflect a need for greater amplification of olfactory progenitors than in the hippocampus, to accommodate a greater rate of neurogenesis in the OE.
Previous data on NeuroD1 expression in the OE have placed the factor downstream of Mash1, and suggested that it anticipated neuronal differentiation. These data include the epistatic relationships observed in the OE of Mash1-knockout mice (Cau et al., 1997) and the relative timing of the expression of the two factors during the regeneration of the epithelium after methyl bromide injury in rats (Manglapus et al., 2004) and mice (unpubl. data). However, other data have been interpreted as indicating that NEUROD1 is found in recently born neurons; these findings were based solely on the comparison of immunohistochemical labeling with anti-NEUROD1 with location in the OE, i.e., apical vs. basal (Nibu et al., 1999, 2001; Suzuki et al., 2003; Yasui et al., 2004).
In contrast, the current results are most consistent with the interpretation that NEUROD1 is expressed by a subset of GBCs and are based on molecular marker coexpression, including proteins associated with active progression through the mitotic cycle. Moreover, comparisons with other transcription factors are also most consistent with the existence of multiple discrete stages within the population of GBCs. Such comparisons suggest a hierarchical sequencing of MASH1 followed by NEU-ROG1 and NEUROD1 in the normal adult OE, as in the embryo and the regenerating epithelium, even though lineage data are lacking that would prove more directly that Mash1-expressing GBCs give rise to Neurog1 and NeuroD1-expressing progenitors. Included among these data suggesting a hierarchical relationship are the nonoverlapping patterns of MASH1 and NEUROD1 label in the OE, and of MASH1 and LacZ in NeuroD1-Cre/LacZ mice. Furthermore, the complete overlap of MASH1 with SOX2 (but not the reverse) but only partial overlap of NEUROD1 and SOX2 in normal OE (Guo et al., 2010), both suggest that MASH1 is expressed in advance of NEUROD1, since SOX2 expression precedes that of MASH1 during development or regeneration.
Thus, the current data, when taken as a whole, support the reciprocal regulation of NeuroD1 and Mash1 in OE, as seen in the forebrain, where NeuroD1 and Mash1 transcript patterns are mutually exclusive and where NeuroD1 demonstrates a repressive influence on Mash1 in GABAergic neurons (Osorio et al., 2010; Roybon et al., 2010). Taken together, the mutual exclusion of NEUROD1 and MASH1, the finding that nearly all of the NEUROD1 cells express markers of active cycling, and that NEUROD1 and NEUROG1 are expressed in common by a substantial population of GBCs are most consistent with the interpretation that there are multiple stages in the GBC hierarchy from Mash1 onward (Gordon et al., 1995; Cau et al., 1997, 2002; Nibu et al., 1999, 2001; Suzuki et al., 2003; Yasui et al., 2004; Manglapus et al., 2004).
NeuroD1 null phenotype suggests a role in neuronal survival and/or differentiation
In affected areas of the OE in NeuroD1-null mice we find that OMP(+) mature neurons are grossly depleted despite the maintenance of a PGP9.5(+) population of immature neurons and retained β-gal expression from the ΔNeuroD1-LacZ mutant allele. The finding suggests that neurogenesis is ongoing in the affected areas. Thus, olfactory neuronal survival may be compromised, and/or susceptibility to injury enhanced, to a limited extent in the absence of NEUROD1, although a failure of newborn neurons to progress and mature, also limited in extent, is also a possible explanation for this phenotype. We cannot, at present, rule out the possibility that the systemic effects of NeuroD1 elimination (for example, diabetes mellitus due to the blocking of beta cell differentiation) have contributed to the phenotype. A more definitive assessment will require conditional elimination of the gene selectively in the olfactory epithelium and/or close regulation of the metabolic abnormalities caused by gene knockout. Nonetheless, there is evidence in the literature from other settings in the nervous system to support either type of cell autonomous effect.
With regard to an effect of NeuroD1 mutation on neuronal survival, the gene plays a critical role in maintaining neurons in the inner ear, retina, and dentate gyrus (Liu et al., 2000; Pennesi et al., 2003; Gao et al., 2009). We do not find an increase in apoptotic markers within the affected areas of the OE at the timepoints used in these studies; however, additional investigation will be required to fully resolve this point.
Alternatively, aborted differentiation could also explain what is happening in the OE. In the dentate gyrus, granule cell progenitors fail to progress toward neuronal differentiation in the absence of NeuroD1; instead they are held up in an intermediate phase of their development, the late precursor/immature neuron (Prox1(+)/Nestin(+)) stage (Gao et al., 2009).
The patchiness of the OE abnormalities and its concentration at the tips of some turbinates may be more in line with causation by a greater vulnerability to injury and, consequently, accelerated turnover, since not all areas of the OE appear equally at risk for environmental damage (Maruniak et al., 1989; Loo et al., 1996). The NeuroD1-null phenotype is also patchy in the retina, but tends to be more universal in the cerebellum, where greater than 90% of the external granular layer is eliminated, and in the hippocampus, where the dentate gyrus fails to form (Miyata et al., 1999; Liu et al., 2000; Kim et al., 2001; Pennesi et al., 2003; Cho et al., 2007). It is likely to be the case that alternate mechanisms will overcome the loss of NeuroD1, allowing neurogenesis to proceed, as has been seen with the loss of Neurog1 by mutation (Cau et al., 2002). The redundancy in the system may be due to the heavy dependence on intact olfactory function for survival, such that multiple independent pathways may operate simultaneously to ensure and maintain function.
CONCLUSION
NeuroD1-expressing olfactory progenitors are a universal gateway for many types of sensory neurons in the nose, although NeuroD1 itself is not required for this differentiation to occur. Similarities exist between transcription factor expression by the NeuroD1 progenitors in the OE and elsewhere, which imply parallels with respect to gene function as well. Thus, despite the differences among the progenitor cells across neurogenic matrices, the commonalities make the NeuroD1-expressing precursor in the OE an attractive target for analyzing the genetic and cell biological basis of NEUROD1 function, which may be generally applicable to neurogenesis more broadly.
Supplementary Material
Acknowledgments
The authors thank Dr. J. Covault for the generous gift of the NCAM antibody, Kwok Po-Tse for outstanding technical assistance, and members of the Schwob Lab for helpful discussions related to the preparation of the article.
Grant sponsor: National Institutes of Health; Grant number: R01 DC002167 (to J.E.S.).
Footnotes
Additional Supporting Information may be found in the online version of this article.
LITERATURE CITED
- Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR, Sikorska M. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Boutin C, Hardt O, de Chevigny A, Core N, Goebbels S, Seidenfaden R, Bosio A, Cremer H. NeuroD1 induces terminal neuronal differentiation in olfactory neurogenesis. Proc Natl Acad Sci U S A. 2010;107:1201–1206. doi: 10.1073/pnas.0909015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter LA, MacDonald JL, Roskams AJ. Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. J Neurosci. 2004;24:5670–5683. doi: 10.1523/JNEUROSCI.0330-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Chen X, Fang H, Schwob JE. Multipotency of purified, transplanted globose basal cells in olfactory epithelium. J Comp Neurol. 2004;469:457–474. doi: 10.1002/cne.11031. [DOI] [PubMed] [Google Scholar]
- Cho JH, Klein WH, Tsai MJ. Compensational regulation of bHLH transcription factors in the postnatal development of BETA2/NeuroD1-null retina. Mech Dev. 2007;124:543–550. doi: 10.1016/j.mod.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham RE, Leinders-Zufall T, Munger SD, Zufall F. Functional analysis of the guanylyl cyclase type D signaling system in the olfactory epithelium. Ann N Y Acad Sci. 2009;1170:173–176. doi: 10.1111/j.1749-6632.2009.04104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Finger TE, Bottger B, Hansen A, Anderson KT, Alimohammadi H, Silver WL. Solitary chemoreceptor cells in the nasal cavity serve as sentinels of respiration. Proc Natl Acad Sci U S A. 2003;100:8981–8986. doi: 10.1073/pnas.1531172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW. Evolution and development of the vertebrate ear. Brain Res Bull. 2001;55:711–721. doi: 10.1016/s0361-9230(01)00558-5. [DOI] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbels S, Eisch AJ, Hsieh J. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci. 2009;12:1090–1092. doi: 10.1038/nn.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein BJ, Fang H, Youngentob SL, Schwob JE. Transplantation of multipotent progenitors from the adult olfactory epithelium. Neuroreport. 1998;9:1611–1617. doi: 10.1097/00001756-199805110-00065. [DOI] [PubMed] [Google Scholar]
- Gordon MK, Mumm JS, Davis RA, Holcomb JD, Calof AL. Dynamics of MASH1 expression in vitro and in vivo suggest a non-stem cell site of MASH1 action in the olfactory receptor neuron lineage. Mol Cell Neurosci. 1995;6:363–379. doi: 10.1006/mcne.1995.1028. [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Graziadei PP, Graziadei GA. Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol. 1979;8:1–18. doi: 10.1007/BF01206454. [DOI] [PubMed] [Google Scholar]
- Gulbransen B, Silver W, Finger TE. Solitary chemoreceptor cell survival is independent of intact trigeminal innervation. J Comp Neurol. 2008;508:62–71. doi: 10.1002/cne.21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Packard A, Krolewski RC, Manglapus GL, Harris M, Schwob JE. Expression of Pax6 and Sox2 in adult olfactory epithelium. J Comp Neurol. 2010;518:4395–4418. doi: 10.1002/cne.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huard JM, Schwob JE. Cell cycle of globose basal cells in rat olfactory epithelium. Dev Dyn. 1995;203:17–26. doi: 10.1002/aja.1002030103. [DOI] [PubMed] [Google Scholar]
- Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400:469–486. [PubMed] [Google Scholar]
- Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, Reichardt LF, Barth DS, Lee JE. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, Moore L, Nakashima K, Asashima M, Gage FH. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12:1097–1105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lall S, Patel NH. Conservation and divergence in molecular mechanisms of axis formation. Annu Rev Genet. 2001;35:407–437. doi: 10.1146/annurev.genet.35.102401.090832. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Lee JK, Cho JH, Hwang WS, Lee YD, Reu DS, Suh-Kim H. Expression of neuroD/BETA2 in mitotic and postmitotic neuronal cells during the development of nervous system. Dev Dyn. 2000;217:361–367. doi: 10.1002/(SICI)1097-0177(200004)217:4<361::AID-DVDY3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- Liu M, Pleasure SJ, Collins AE, Noebels JL, Naya FJ, Tsai MJ, Lowenstein DH. Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc Natl Acad Sci U S A. 2000;97:865–870. doi: 10.1073/pnas.97.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo AT, Youngentob SL, Kent PF, Schwob JE. The aging olfactory epithelium: neurogenesis, response to damage, and odorant-induced activity. Int J Dev Neurosci. 1996;14:881–900. doi: 10.1016/s0736-5748(96)00046-9. [DOI] [PubMed] [Google Scholar]
- Mackay-Sim A, Kittel P. Cell dynamics in the adult mouse olfactory epithelium: a quantitative autoradiographic study. J Neurosci. 1991;11:979–984. doi: 10.1523/JNEUROSCI.11-04-00979.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglapus GL, Youngentob SL, Schwob JE. Expression patterns of basic helix-loop-helix transcription factors define subsets of olfactory progenitor cells. J Comp Neurol. 2004;479:216–233. doi: 10.1002/cne.20316. [DOI] [PubMed] [Google Scholar]
- Maruniak JA, Lin PJ, Henegar JR. Effects of unilateral naris closure on the olfactory epithelia of adult mice. Brain Res. 1989;490:212–218. doi: 10.1016/0006-8993(89)90239-4. [DOI] [PubMed] [Google Scholar]
- Miura H, Nakayama A, Shindo Y, Kusakabe Y, Tomonari H, Harada S. Expression of gustducin overlaps with that of type III IP3 receptor in taste buds of the rat soft palate. Chem Senses. 2007;32:689–696. doi: 10.1093/chemse/bjm036. [DOI] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Shou J, Calof AL. Colony-forming progenitors from mouse olfactory epithelium: evidence for feedback regulation of neuron production. Proc Natl Acad Sci U S A. 1996;93:11167–11172. doi: 10.1073/pnas.93.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RC, Navi D, Fesenko J, Lander AD, Calof AL. Widespread defects in the primary olfactory pathway caused by loss of Mash1 function. J Neurosci. 2003;23:1769–1780. doi: 10.1523/JNEUROSCI.23-05-01769.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu K, Li G, Zhang X, Rawson NE, Restrepo D, Kaga K, Lowry LD, Keane WM, Rothstein JL. Olfactory neuron-specific expression of NeuroD in mouse and human nasal mucosa. Cell Tissue Res. 1999;298:405–414. doi: 10.1007/s004419900098. [DOI] [PubMed] [Google Scholar]
- Nibu K, Kondo K, Ohta Y, Ishibashi T, Rothstein JL, Kaga K. Expression of NeuroD and TrkB in developing and aged mouse olfactory epithelium. Neuroreport. 2001;12:1615–1619. doi: 10.1097/00001756-200106130-00021. [DOI] [PubMed] [Google Scholar]
- Ogura T, Krosnowski K, Zhang L, Bekkerman M, Lin W. Chemoreception regulates chemical access to mouse vomeronasal organ: role of solitary chemosensory cells. PLoS One. 2010;5:e11924. doi: 10.1371/journal.pone.0011924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio J, Mueller T, Retaux S, Vernier P, Wullimann MF. Phylotypic expression of the bHLH genes Neurogenin2, Neurod, and Mash1 in the mouse embryonic forebrain. J Comp Neurol. 2010;518:851–871. doi: 10.1002/cne.22247. [DOI] [PubMed] [Google Scholar]
- Pennesi ME, Cho JH, Yang Z, Wu SH, Zhang J, Wu SM, Tsai MJ. BETA2/NeuroD1 null mice: a new model for transcription factor-dependent photoreceptor degeneration. J Neurosci. 2003;23:453–461. doi: 10.1523/JNEUROSCI.23-02-00453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci. 2010;30:714–722. doi: 10.1523/JNEUROSCI.3852-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert H, Simeone A. Developmental genetic evidence for a monophyletic origin of the bilaterian brain. Philos Trans R Soc Lond B Biol Sci. 2001;356:1533–1544. doi: 10.1098/rstb.2001.0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring G, Mezza RC, Schwob JE. Immunohistochemical identification of discrete subsets of rat olfactory neurons and the glomeruli that they innervate. J Comp Neurol. 1997;388:415–434. [PubMed] [Google Scholar]
- Roybon L, Mastracci TL, Ribeiro D, Sussel L, Brundin P, Li JY. GABAergic differentiation induced by Mash1 is compromised by the bHLH proteins Neurogenin2, NeuroD1, and NeuroD2. Cereb Cortex. 2010;20:1234–1244. doi: 10.1093/cercor/bhp187. [DOI] [PubMed] [Google Scholar]
- Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocriine and non-endocrine cell types. Dev Biol. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Schwartz Levey M, Chikaraishi DM, Kauer JS. Characterization of potential precursor populations in the mouse olfactory epithelium using immunocytochemistry and autoradiography. J Neurosci. 1991;11:3556–3564. doi: 10.1523/JNEUROSCI.11-11-03556.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE, Youngentob SL, Mezza RC. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J Comp Neurol. 1995;359:15–37. doi: 10.1002/cne.903590103. [DOI] [PubMed] [Google Scholar]
- Sordino P, van der Hoeven F, Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995;375:678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Mizoguchi I, Nishiyama H, Takeda M, Obara N. Expression of Hes6 and NeuroD in the olfactory epithelium, vomeronasal organ and non-sensory patches. Chem Senses. 2003;28:197–205. doi: 10.1093/chemse/28.3.197. [DOI] [PubMed] [Google Scholar]
- Wang Y, Giel-Moloney M, Rindi G, Leiter AB. Enteroendocrine precursors differentiate independently of Wnt and form serotonin expressing adenomas in response to active beta-catenin. Proc Natl Acad Sci U S A. 2007;104:11328–11333. doi: 10.1073/pnas.0702665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildner H, Muller T, Cho SH, Brohl D, Cepko CL, Guillemot F, Birchmeier C. dILA neurons in the dorsal spinal cord are the product of terminal and non-terminal asymmetric progenitor cell divisions, and require Mash1 for their development. Development. 2006;133:2105–2113. doi: 10.1242/dev.02345. [DOI] [PubMed] [Google Scholar]
- Yasui R, Hasegawa M, Doi K, Shimizu K, Ohtuski N, Ishida H, Nibu K. Quantitative analysis of expression of NeuroD, GAP43 and receptor tyrosine kinase B in developing mouse olfactory neuroepithelium. Acta Otolaryngol Suppl. 2004;553:90–94. doi: 10.1080/03655230410017742. [DOI] [PubMed] [Google Scholar]
- Zufall F, Munger SD. Receptor guanylyl cyclases in mammalian olfactory function. Mol Cell Biochem. 2010;334:191–197. doi: 10.1007/s11010-009-0325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.