Abstract
ADP-ribosyltransferases (ARTs) are important enzymes that regulate the genotoxic stress response and the maintenance of genome integrity. ARTD1 (PARP1) and ARTD2 (PARP2) are homologous proteins that modify themselves and target proteins by the addition of mono- and poly-ADP-ribose (PAR) moieties. Both enzymes have been described to be involved in the genotoxic stress response. Here, we characterize cellular PAR formation on hydrogen peroxide (H2O2) or N-methyl-N′-methyl-nitro-N-nitrosoguanidine (MNNG) stress, in combination with application of the RNA polymerase I inhibitor Actinomycin D (ActD), known to cause accumulation of short RNA polymerase I-dependent rRNA transcripts. Intriguingly, co-treatment with ActD substantially increased H2O2- or MNNG-induced PAR formation. In cells, this enhancement was predominantly mediated by ARTD2 and not ARTD1. In vitro experiments confirmed that ARTD2 is strongly activated by RNA and that the N-terminal SAP domain is important for the binding to RNA. Thus, our findings identify a new activator of ARTD2-dependent ADP-ribosylation, which has important implications for the future analysis of the biological role of ARTD2 in the nucleus.
INTRODUCTION
Cells have evolved a complex and diverse arsenal of mechanisms to overcome genotoxic stress and to guarantee genome integrity (1). Depending on the type of stress, different response mechanisms are activated to repair damaged DNA or to prevent its inheritance (2,3). In addition to factors that directly bind and replace incorrect bases and repair DNA strand breaks, a variety of proteins are indirectly involved in the genotoxic stress response by regulating the levels and activities of other proteins or by modulating chromatin structure. ADP-ribosyltransferases (ARTs) are prominent members of this group of enzymes.
ARTs use nicotinamide adenine dinucleotide (NAD+) as a substrate for the synthesis of mono- and poly-ADP-ribose modifications on target proteins (4). The ART protein family is divided into diphtheria toxin-like ARTs (ARTDs) and cholera toxin-like ARTs (ARTCs) (5). In humans, the ARTD (PARP) family currently comprises 18 nuclear and cytoplasmic mono- and poly-ARTs, while ARTCs are mainly extracellular enzymes that only transfer a single ADP-ribose unit to their target proteins (5).
Proteins of the ARTD family have been implicated in a plethora of cellular functions (6,7). Research during the past years has documented numerous functions of ARTD1 (PARP1) and of ADP-ribosylation in general that are not directly linked to DNA repair or the DNA damage response (8,9). The function of ARTD1 in DNA repair is substantiated by the strong activation of ARTD1 activity by DNA in vitro, as well as by the strong induction of poly-ADP-ribosylation on treatment of cells with DNA-damaging agents. Nevertheless, a direct involvement of DNA damage in the activation of ARTD1 in vivo is still largely based on correlations. PAR formation is dependent on the severity of the genotoxic stress and can even lead to cell death due to depletion of NAD+ and ATP (10). The functional involvement of ADP-ribosylation in the DNA damage response has provided the incentive to generate PARP inhibitors as antitumor drugs, which are being developed and tested as novel therapies (11–14). The closest homolog of ARTD1 is ARTD2 (PARP2), which too is able to mono- and poly-ADP-ribosylate itself in addition to target proteins. Although ARTD1 has already been discovered several decades ago, ARTD2 was coincidentally discovered much later in the 1990s because of the detection of residual PAR-forming activity in mouse embryonic fibroblasts (MEFs) from ARTD1 knockout mice (15). Like ARTD1, ARTD2 has also been implicated in various cellular functions, which include genome and chromosome stability, heterochromatin integrity, cell death, differentiation and inflammation (16). Mammalian ARTD2 is a 66.2-kDA protein with a C-terminal catalytic domain that is 69% similar to the homologous domain in ARTD1 (15,17). Despite this common domain, ARTD2 is catalytically much less active than ARTD1, suggesting that ARTD2 may become activated by different and as yet unknown stimuli. While the DNA binding domain of ARTD1 contains two Zn fingers and a Zn-binding domain, the DNA binding element of ARTD2 is represented by the SAF-A/B, Acinus and PIAS DNA-binding (SAP) domain. Furthermore, ARTD2 seems to modify different proteins, suggesting that both enzymes might regulate distinct biological functions (18,19). Thus, the identification of such new ARTD2 activators will likely also reveal novel biological phenomena that are regulated specifically by ARTD2.
Nucleoli are sites of ribosomal RNA (rRNA) gene transcription, rRNA maturation and ribosome production, and assemble around the nucleolar organizer regions (20,21). A human diploid cell contains ∼400 rRNA genes, which are all organized in head-to-tail tandem repeats on five different chromosomes (21). However, as in highly metabolically active cells, only a subset of these genes is actively transcribed (22,23). The remaining rRNA genes are silenced in a tissue- and cell type-specific manner (24). Active rRNA genes are transcribed by RNA polymerase I (Pol I) to synthesize a 45S pre-rRNA. For the initiation of rRNA transcription, Pol I has to be part of a multi-protein complex that includes factors such as UBF, SL1 and TIF-IA (25). The production of ribosome subunits is heavily influenced by diverse stress stimuli and metabolic changes (26). The cell reacts to nutrient starvation, oxidative stress or inhibition of protein synthesis with a decrease of rRNA transcription, whereas growth factors and proliferation-stimulating agents increase rRNA transcription. Actinomycin D (ActD) inhibits rRNA synthesis already at low concentrations (50 ng/ml), whereas RNA polymerases II (Pol II) and III (Pol III) are inhibited only at higher concentrations (Pol II > 1 µg/ml, Pol III > 5 µg/ml) (27,28). ActD intercalates into CG-rich regions of the DNA, and thus stabilizes covalent topoisomerase I-DNA complexes, preventing progression of RNA polymerase, thereby inhibiting RNA synthesis (29). The intercalation mainly takes place downstream of rDNA transcription start sites, thus inhibiting transcription during elongation, which leads to an immense accumulation of short RNA transcripts over time (30,31). In Drosophila, ARTD1 has been reported to regulate ribosomal biogenesis at the posttranscriptional pre-rRNA processing level (32). Furthermore, ARTD1 and ARTD2 have both been shown to co-localize with B23/nucleophosmin and nucleolin, nucleolar proteins involved in several processes including rRNA transcription and elongation, ribosome assembly and rRNA processing (33,34). However, no direct effect of ARTD1 and ARTD2 on Pol I transcription was described in these studies. More recently, ARTD1 has been linked to heterochromatin formation, specifically to silencing of rRNA genes in the nucleolus (35,36).
Here, we characterize the nucleolar function of ARTD2 on different stresses. Hydrogen peroxide (H2O2) or N-methyl-N’-methyl-nitro-N-nitrosoguanidine (MNNG) treatment of different cell lines induced PAR formation in the nucleolus. Co-treatment with low doses of ActD enhanced PAR formation. Surprisingly, this co-treatment mainly activated ARTD2. In vitro experiments confirmed that ARTD2 is strongly activated by short rRNA and other single-stranded RNAs, but not by double-stranded DNA, through its SAP domain. Thus, our findings reveal a new activator of ARTD2, which opens new possible implications for the future analysis of the biological role of ARTD2.
MATERIALS AND METHODS
Cell culture
T24 bladder tumor cells were cultivated in McCoy’s 5 A medium (Gibco, Invitrogen, CA, USA) at 37°C. NIH/3T3, HeLa cells and MEFs were cultivated in Dulbecco’s modified Eagle’s medium (PAA, Pasching, Austria). MD-MBA-231 cells were cultivated in Roswell Park Memorial Institute medium (Gibco, Invitrogen, CA, USA). All media were supplemented with 1% (v/v) Penicillin/Streptomycin and 10% (v/v) fetal calf serum (Gibco, Invitrogen, CA, USA).
siRNA transfection
Negative control allstars (siMock), human siPARP1 (SI02662996), human siPARP2 (SI02664725), human siPARG (SI00677782), mouse siPARP1 (SI02731428) and mouse siPARP2 (SI02735670) were all obtained from Qiagen (Hilden, Germany). Cells were seeded 1 day before transfection (5 × 105 cells per 6 cm plate) and transfected with 40 nmol siRNA per plate and RNAi MAX lipofectamine (Invitrogen, Carlsbad, CA, USA).
Antibodies
The following antibodies were used: from Santa Cruz Biotechnology, Inc (Dallas, TX, USA): PARP1/ARTD1 (H-250, sc-7150, rabbit), Pol I (RPA 194, H-300, sc-28714, rabbit). From Active motif (Carlsbad, CA, USA): PARP2 (39744, rabbit). From Sigma Aldrich (St. Louis, MO, USA): tubulin (T6199, mouse). From Cell Signaling (Danvers, MA, USA): fibrillarin (C13C3). From Merck-Millipore (Billerica, MA, USA): phosphor-Histone H2A.X (Ser139). From Jackson ImmunoResearch Laboratories (Suffolk, UK): secondary FITC-conjugated AffiniPure goat anti-mouse, secondary Cy3TM -conjugated AffiniPure goat anti-mouse. From Invitrogen (Gibco, Invitrogen, CA, USA): Alexa-Fluor 488 anti-rabbit IgG (A11008). The antibody 10H anti-PAR was used to identify PAR by immunofluorescence (IF).
RNA analysis
RNA was purified using TRIzol® RNA Isolation Reagent (500 µl, Life Technologies, CA, USA) and reverse transcribed according to the supplier’s protocol (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, Foster City, CA, USA). Quantitative real-time polymerase chain reactions (qPCR) were performed with SYBR® green SensiMix SYBR Hi-ROX Kit (Bioline Reagents Ltd., London, UK) and a Rotor-Gene Q 2plex HRM System (Qiagen, Hilden, Germany).
Cell lysis, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blot analysis
Whole cell lysis was performed either with trypsinized cells or directly on plates by using RIPA lysis buffer (50 mM Tris, pH 8, 400 mM NaCl, 0.5% NP40, 1% DOC, 0.1% SDS, 1 µg/ml pepstatin, 1 µg/ml bestatin, 1 µg/ml leupeptin, 2 mM PMSF, 10 mM β-Glycerophosphate, 1 mM NaF and 1 mM dithiothreitol; 10 min, 4°C). Proteins were quantified using the Lowry assay and, if not otherwise indicated, 30 µg of protein extract was loaded and separated on a 10 or 12% SDS-polyacrylamide gel (120 V, 2 h). The gel was blotted on a polyvinylidene difluoride membrane and analysed by using protein specific antibodies.
Immunofluorescence microscopy
Cells were seeded on coverslips (105 cells per well in a 24-well-plate) and grown overnight. After H2O2 (1 mM in FCS-free medium, 10 min), MNNG (500 µM in FCS-free medium, 30 min) or medium-only treatment, cells were fixed (methanol: acetic acid 3:1, 5 min on ice), washed twice with phosphate buffered saline (PBS) and incubated with antibody (1:350) in PBS (containing 5% milk and 0.05% Tween, 1 h at room temperature or overnight at 4°C). When indicated, 4% PFA was used as fixation method (15 min, room temperature). Cells were then incubated with antibody for 1 h at room temperature. After washing with PBS, coverslips were mounted with Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). Conventional microscopy was carried out using a Leica DMI 6000B light microscope (Leica microsystems GmbH, Wetzlar, Germany). Confocal laser scanning microscopy was carried out with a Leica SP 5 resonant APD system (Leica microsystems GmbH, Wetzlar, Germany).
The mean PAR signal intensity per cell was quantified with ImageJ (37) in at least 100 cells per replicate. The color threshold was set for Max entropy and B&W. According to the intensity of the picture, the threshold was set for each experiment the same way; thus, different replicates were comparable with each other.
Co-localization was quantified with Imaris (Bitplane, Belfast, UK) using the co-loc application. The thresholds were set for all three channels (DAPI, blue; PAR, red; and fibrillarin, green) and kept for the whole analysis. At least 50 cells were analyzed for H2O2 treated samples (from three biological replicates) and at least 25 cells were analyzed for H2O2 ActD treated samples (from two biological replicates).
In vitro RNA transcription
Linearized vectors containing rRNA sequences (mouse rRNA from −16 to +130 and −232 to −1) and control sequences (36) were used to in vitro transcribe RNA using T7 polymerase. After DNase I treatment, transcripts were double purified using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
In vitro ARDT1 or ARTD2 activity assay
Ten picomoles of baculovirus-purified MYC-hARTD1(wt)-HIS or purified human ARTD2(fl)-HIS was incubated with NAD+ in the reaction buffer (250 mM Tris–HCl, pH 8, 20 mM MgCl2, 1.25 mM dithiothreitol, 5 µg/ml P/B/L–proteinase inhibitors, 30°C, 10 min). Five picomoles of EcoRI linker DNA or different concentrations as indicated of short RNA transcripts were added to the reaction. For ADP-ribosylation reactions, radioactive NAD+ (32P, final concentration 100 nM) and nonradioactive NAD+ (final concentration 1.6 or 16 µM) were added. Reactions were terminated by adding Laemmli buffer and subsequent boiling of the samples (5 min, 95°C). SDS-PAGE was performed; gels were stained with coomassie, destained and subjected to film exposure. The autoradiography was quantified with the software GelEval (http://www.frogdance.dundee.ac.uk/).
In vitro poly-ADP-ribose-glycohydrolase (PARG) assay
ARTD1 auto-ADP-ribosylation was carried out as described before and the reaction mix was purified over illustra MicroSpin G-50 Columns (GE Healthcare GmbH Europe, Freiburg, Germany). Equal amounts of reaction mix were added to prechilled tubes containing baculo-purified hPARG-(fl) (2 pmol). Reactions were carried out in the presence or absence of the indicated molecules.
Nothern blot and Northwestern analysis
Purified RNA from NIH/3T3 or T24 cells pretreated with ActD (50 ng/ml for 0.5 h, 4 h or 20 h) and rRNA transcripts were monitored by hybridization to a 32P-labeled riboprobe complementary to +1/+130 mouse or human rRNA sequences. Northwestern analysis was performed as described previously (36).
RESULTS
H2O2 treatment predominantly induces nucleolar PAR formation
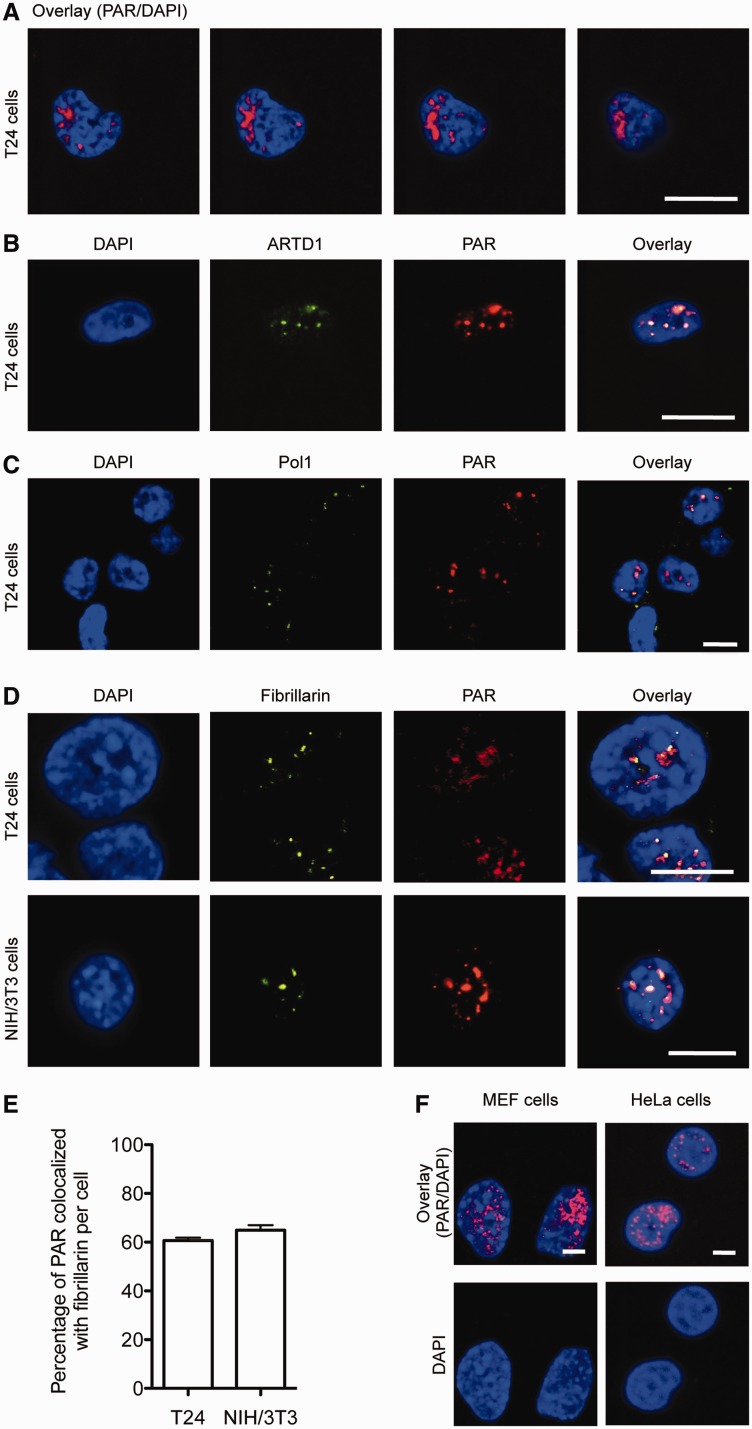
To investigate the localization and molecular mechanism of PAR formation, T24 cells were treated with H2O2 (1 mM for 10 min). PAR formation was analysed by IF using a 10H anti-PAR antibody and both conventional and confocal fluorescence microscopy. The PAR signal was localized mainly to regions of the nucleus that were stained weakly with DAPI, indicating that nucleoli are sites of PAR formation (Figure 1A). The PAR signal in T24 cells treated with H2O2 co-localized with ARTD1, which suggests that ARTD1 is at least partly responsible for PAR synthesis in response to oxidative stress in T24 cells (Figure 1B). Co-staining with anti-Pol I and anti-fibrillarin antibodies suggested partial co-localization with the PAR signal (Figure 1C and D). Computerized analysis of the acquired images revealed that 60–65% of the fibrillarin co-localized with a strong PAR signal (Figure 1E). Similar levels of co-localization of fibrillarin with PAR signals were observed in H2O2-treated NIH/3T3 (Figure 1D and E). H2O2-treated HeLa cells and MEFs displayed a similar location of the PAR signal in nucleoli (Figure 1F). Taken together, these results indicate that the nucleoli are the main location of cellular PAR formation during oxidative stress.
Figure 1.
H2O2 treatment induces nucleolar poly-ADP-ribose formation. Confocal immunofluorescence microscopy of PAR (red) was performed after H2O2 treatment (1 mM, 10 min). (A) Z-Stack-resolution of T24 cells, bar = 10 μM. (B) Double-staining of T24 cells: PAR (red), ARTD1 (green), bar = 10 µM. (C) Double-staining of T24 cells: PAR (red), RNA Pol 1 (green), bar = 10 µM. (D) Double staining of T24 (upper row) and 3T3 (lower row) cells after H2O2 treatment (1 mM, 10 min): PAR (red) and fibrillarin (green), bar = 10 µM. (E) Quantification of the co-localization of the PAR signal and fibrillarin staining in T24 and NIH/3T3 cells. (F) Confocal IF microscopy of PAR (red) after H2O2 treatment (1 mM, 10 min) was analyzed in MEF and HeLa cells. Bar = 10 µM.
ActD treatment enhances nucleolar PAR formation on H2O2 and MNNG stimulation
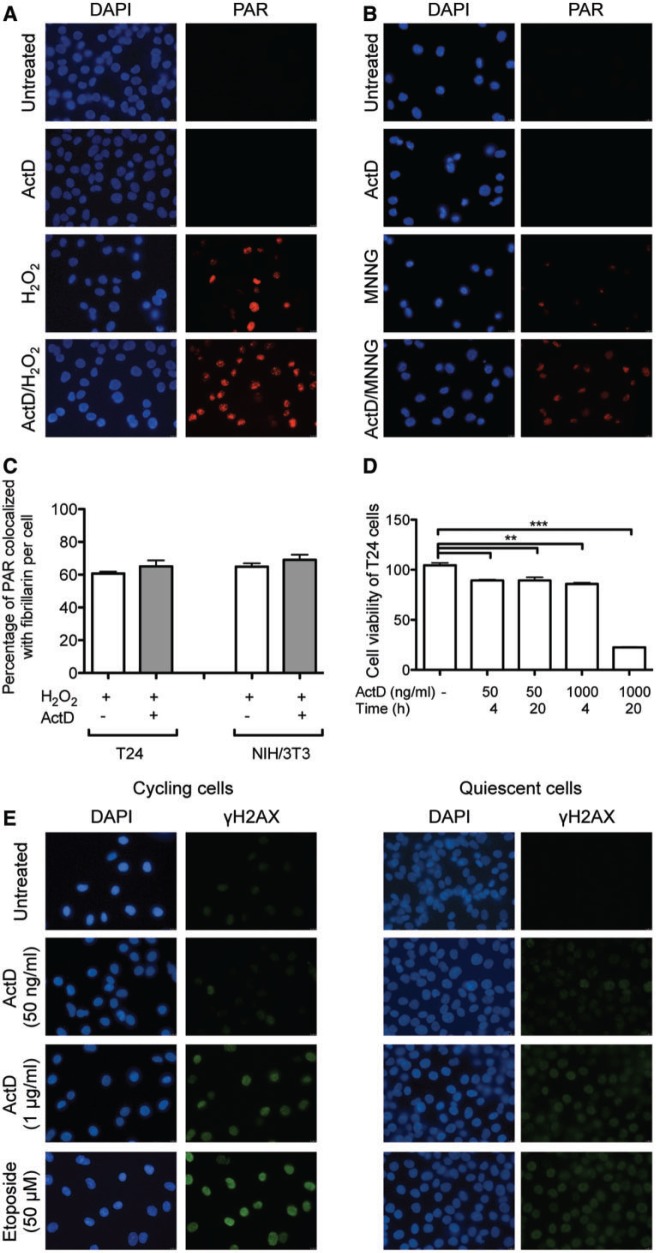
Although H2O2- and MNNG-induced PAR formation has been attributed to DNA damage, a link between stress-induced PAR synthesis and Pol I-dependent transcription has not been documented previously. To investigate this possibility, we pretreated cells for 20 h with a low dose of ActD (50 ng/ml), which inhibits Pol I-dependent transcription (38), before the exposure to H2O2 or MNNG. Intriguingly, pretreatment with ActD followed by exposure to H2O2 or MNNG increased both the number of PAR-positive T24 cells and the intensity of the PAR signal in comparison with cells not pretreated with ActD (Figure 2A and B). A similar effect of ActD on the PAR signal was also observed in cells fixed with PFA, thus excluding the possibility that the results obtained by the methanol/acetic acid fixation protocol were an artifact (Supplementary Figure S1A, upper part).
Figure 2.
ActD treatment increases nuclear poly-ADP-ribose formation on H2O2 and MNNG stimulation. (A) Immunofluorescence microscopy of T24 cells was examined after H2O2 (1 mM, 10 min) and/or ActD treatment (50 ng/ml, 20 h) and stained for PAR formation (red). (B) Immunofluorescence microscopy of T24 cells after MNNG (500 µM, 30 min) and/or ActD treatment (50 ng/ml, 20 h) was examined. (C) Quantification of the co-localization of the PAR signal and fibrillarin staining in T24 and NIH/3T3 cells pretreated with ActD. (D) Viability assay (Alamar blue assay) of T24 cells treated with ActD (50 and 1000 ng/ml) for 4 h or 20 h [n = 3, one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test]. (E) Immunofluorescence microscopy of T24 cells stained for γ-H2AX after ActD treatment (50 and 1000 ng/ml for 20 h) and etoposide (50 µM for 16 h), either in cycling or in quiescent T24 cells.
In PFA-fixed cells, PAR formation assessed by confocal microscopy was also localized to DAPI-poor regions (Supplementary Figure S1A, lower part). However, the apparent extent of PAR and fibrillarin co-localization in PFA-fixed cells was less than that in methanol/acetic acid-fixed cells. This discrepancy could be due to PFA fixation leading to masking of the epitope for antibody recognition, an observation that we also made for other nucleolar proteins, which can only be detected in nucleoli when the denaturating action of methanol fixation is used (R.S., unpublished observation). ActD enhanced H2O2- or MNNG-induced PAR formation also in NIH/3T3, HeLa and MDA-MB-231 cells (Supplementary Figure S1B–D), indicating that the observed ActD effect is biologically conserved. ActD on its own did not induce PAR formation, indicating that ActD caused increased PAR formation by enhancing the signals induced by H2O2 or MMNG. Furthermore, ActD did not alter localization of PAR to nucleoli, as indicated by the unchanged co-staining with fibrillarin (Figure 2C and Supplementary Figure S1D).
To analyze whether ActD-induced PAR formation was due to the induction of cell death, cell viability was monitored. ActD pretreatment with the low concentration of 50 ng/ml did not significantly affect cell viability, while higher ActD concentrations (1 μg/ml) strongly reduced cell viability in T24 cells (Figure 2D). To exclude DNA damage as the stimulus for PAR formation, the levels of histone H2AX phosphorylation (γH2AX), a modification that occurs at sites flanking DNA double-strand breaks (DSBs) (39), were analyzed. Both of the positive controls, the topoisomerase II inhibitor etoposide (50 μM, 16 h), as well as high doses of ActD, induced γH2AX formation in T24 cells (Figure 2E). We thus conclude that the strong PAR signal induced by the pretreatment of T24 cells with the low concentration of ActD (50 ng/ml) was not due to DNA damage or cell death. As PAR formation has previously been reported to occur under replicative stress (40), we also analyzed the PAR signal as well as the γH2AX formation in quiescent (G0 phase) T24 cells, on combined ActD and H2O2, or ActD, etoposide and MNNG treatment (Figure 2E and Supplementary Figure S2A). Comparable with nonarrested cells, quiescent cells treated with the low ActD concentration also displayed PAR formation, but did not show induction of γH2AX, indicating that enhanced PAR formation on ActD treatment does not depend on replicative stress and cell proliferation.
We next tested whether enhanced PAR formation depends on Pol I transcription. The analysis of 45S pre-rRNA levels confirmed that ActD was effective in impairing pre-rRNA synthesis (>90%), while H2O2 or MNNG reduced pre-rRNA levels only by 25–40% under the tested conditions (Supplementary Figure S2B). In contrast to ActD, pretreatment with the RNA Pol II inhibitor α-amanitin did not induce PAR formation (Supplementary Figure S2C), indicating that formation of ActD-mediated PAR formation depends on RNA Pol I transcription. Because PAR formation was inhibited by Olaparib, an inhibitor of ARTDs including ARTD1 and ARTD2 [Supplementary Figure S2D, (41)], ARTD1 and/or ARTD2 could potentially be involved in the synergistic effect between stress signaling and the interference of 45S pre-rRNA synthesis for PAR accumulation in the nucleolus.
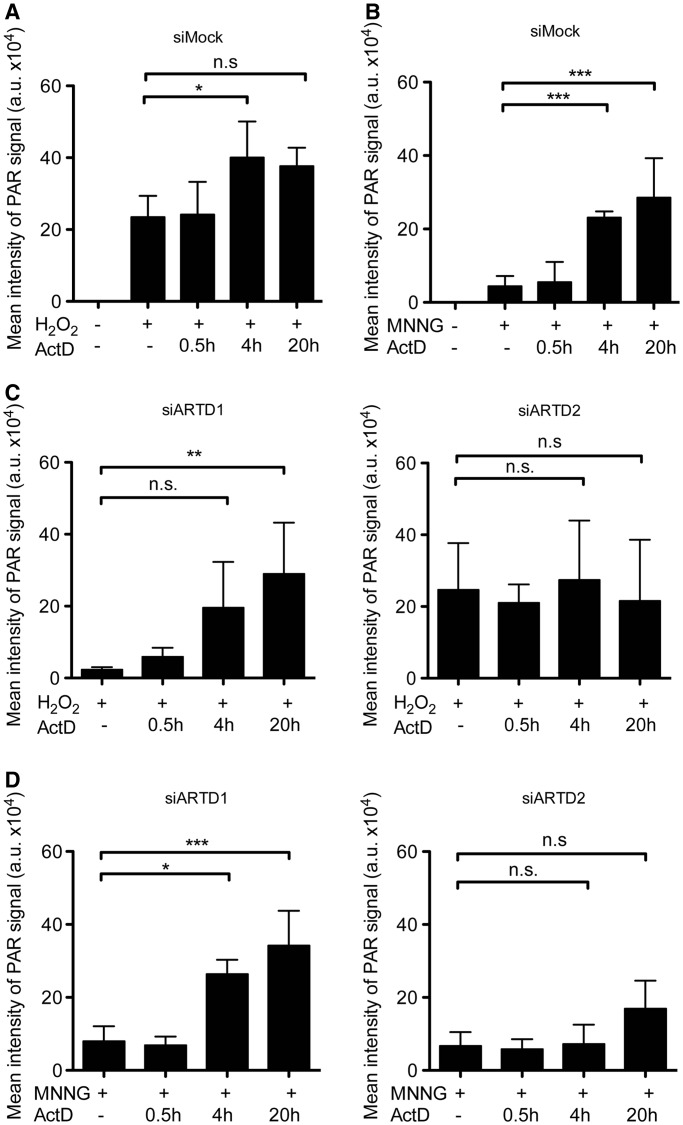
ARTD2, but not ARTD1, is responsible for the ActD-dependent enhancement of PAR formation after H2O2 or MNNG treatment
To investigate whether ActD exerts its synergistic effect through ARTD1 and/or ARTD2, T24 cells were depleted of either ARTD1 or ARTD2 by siRNA (Supplementary Figure S2E), pretreated with ActD as indicated and PAR formation was induced by H2O2 or MNNG. PAR formation was quantified by evaluating the mean signal intensity of immunostained cells. PAR levels were more strongly induced after H2O2 treatment as compared with MNNG treatment (Figure 3A and B). ActD pretreatment for 4 or 20 h enhanced the mean PAR intensity per cell irrespective of whether H2O2 or MNNG was used, while short pretreatment of only 30 min had no effect. This enhancement of PAR formation on ActD treatment was more prominent in MNNG-treated cells as compared with H2O2-treated cells under the tested conditions (Figure 3B). Knockdown of ARTD1 almost completely abolished PAR formation in T24 cells treated with H2O2, indicating that ARTD1 is responsible for most of the H2O2-induced PAR formation (Figure 3C). However, prolonged (4–20 h, 50 ng/ml) pretreatment of ARTD1-silenced cells with ActD strongly enhanced H2O2-induced PAR formation, despite the strong sustained reduction of ARTD1 protein levels (Figure 3C). These results indicate that the synergistic effect between oxidative stress and the inhibition of 45S pre-rRNA synthesis on PAR accumulation in the nucleolus is not mediated by ARTD1. In contrast, compared with siMOCK-treated cells, no additional increase in PAR formation was observed after 4–20 h of ActD pretreatment in ARTD2-depleted T24 cells (Figure 3C), indicating that the stimulatory ActD effect on H2O2-induced PAR formation was mainly regulated by ARTD2. The presence of ARTD2, but not of ARTD1, also seemed to be responsible for the increased PAR formation observed after co-treatment of ActD and MNNG in T24 cells after 4 h (Figure 3D). The slight, but not significant, increase of the PAR signal in cells pretreated with ActD for 20 h and co-treated with MNNG may hint at the delayed and attenuated stimulation of ARTD1, or an additional ARTD family member that catalyzes additional PAR formation under these conditions. H2O2 or MNNG treatment in combination with ActD in NIH/3T3 cells and subsequent quantification of the PAR formation by IF or visualization by western blot further confirmed that ARTD2 is responsible for the stimulatory ActD effect also in mouse cells (Supplementary Figure S3A–H for H2O2 or Supplementary Figure S4A and B for MNNG). Together, these results indicate that ARTD2 is involved in PAR formation in response to H2O2- or MNNG-treatment in combination with ActD in T24 and NIH/3T3 cells.
Figure 3.
ARTD2 is responsible for the ActD-dependent increase in poly-ADP-ribose formation. Quantitative PAR analysis of siMock-, siARTD1- and siARTD2-treated T24 cells was performed after treatment with ActD (50 ng/ml for 0.5, 4 and 20 h) and H2O2 (1 mM, 10 min) or MNNG treatment (500 µM, 30 min). At least 100 cells were analyzed per replicate (for 0.5 h; n = 2 for siARDT1 and siARTD2 samples, for other conditions n = 3–5, one-way ANOVA with Tukey’s post hoc test was performed). (A) siMock T24 cells were treated with H2O2. (B) siMock T24 cells were treated with MNNG. (C) H2O2 treated siARTD1 (left) and siARTD2 (right) T24 cells. (D) MNNG-treated siARTD1 (left) and siARTD2 (right) T24 cells. n.s.: P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
ARTD2 activity is stimulated by rRNA in vitro
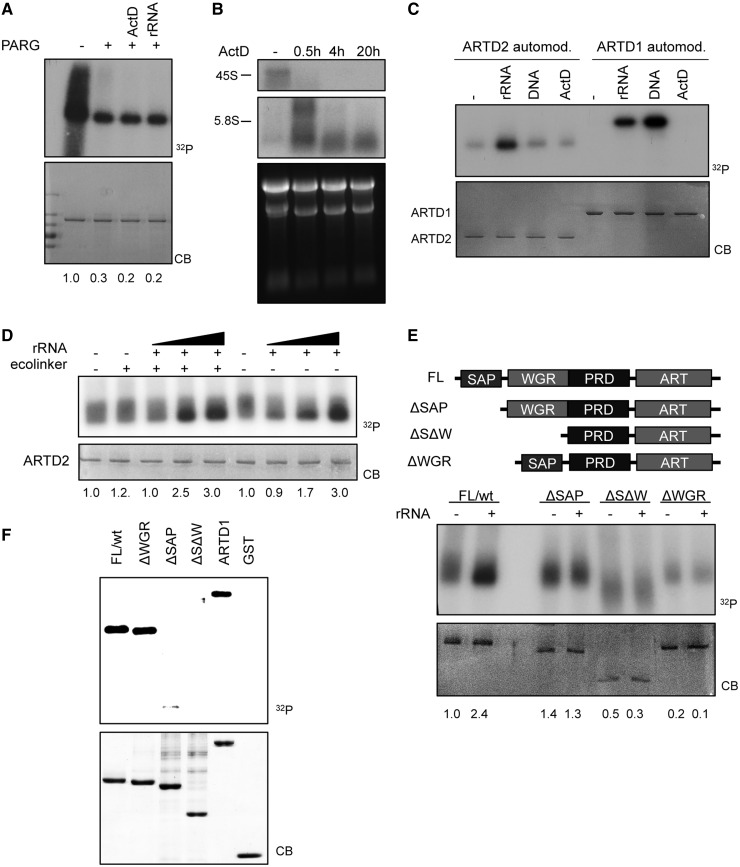
The described synergistic effect between ActD and H2O2/MNNG on PAR formation could either be due to an inhibition of PAR degradation or a stimulation of PAR synthesis. The former was studied by determining whether ActD affects activity of the PARG, the primary enzyme responsible for degrading protein-bound poly-ADP-ribose (Figure 4A). In vitro 32P-labeled PARylated ARTD1 was incubated with PARG in the presence of ActD or rRNA, and PARylated ARTD1 levels were monitored by autoradiography. Treatment with ActD or rRNA did not prevent degradation of PAR moieties of ARTD1, indicating that neither ActD nor rRNA affects PARG activity.
Figure 4.
ARTD2 activity is stimulated by rRNA in vitro and binds to RNA via the SAP domain (A) In vitro radioactive PARG assay carried out with in vitro modified ARTD1. PARG was added to each reaction in combination with pretreatment of ActD (50 ng/ml) or rRNA (146 nt). The PARG reaction was performed for 15 min at 4 °C. Upper panel shows the autoradiography (32P) and the lower panel the corresponding coomassie blue–stained gel (CB) of a representative experiment. Densitometric analysis of the radioactive signals was performed as described in M&M, and values obtained are indicated at the bottom of the panel. The PARG untreated value was arbitrarily set to 1. (B) Northern blot analysis of NIH/3T3 cells treated with ActD (50 ng/ml for 0.5, 4 and 20 h). Hybridization was performed with a riboprobe corresponding to +1/+130 mouse rRNA sequences. The lower panel shows 28S and 18S rRNA stained with ethidium bromide. (C) In vitro radioactive ARTD2 and ARTD1 activity assay was performed with 100 ng NAD+ (32P) in the presence of in vitro transcribed rRNA piece (146 nt), DNA linker or ActD (50 ng/ml). CB = coomassie blot. Densitometric analysis of the radioactive signals was performed as described in M&M and values obtained are included in the result section. (D) In vitro radioactive ARTD2 activity assay was performed with 1.6 µM NAD+ (32P) and in vitro transcribed rRNA fragment (146 nt) of different amounts (0.5, 5 and 10 pmol) and in presence or absence of 0.5 pmol DNA linker. Reaction was carried out at 30 °C, 10 min. Densitometric analysis of the radioactive signals was performed as in panel A and values obtained are indicated at the bottom of the panel. The untreated sample was arbitrarily set to 1. (E) In vitro radioactive ARTD2 activity assay with ARTD2 mutants was performed with 1.6 µM NAD+ (32P) in presence or absence of 5 pmol rRNA fragment (146 nt). FL/wt = full-length human ARTD2, ARDT2 ΔSAP mutant 95–583 aa, ARTD2 ΔSΔW mutant 231–583 aa, ARTD2 ΔWGR mutant without WGR domain (deleted aa 116–193). Densitometric analysis of the radioactive signals was performed as in panel (A) and values indicated at the bottom of the panel. The untreated wild-type sample was arbitrary set 1. (F) ARTD2 binds to RNA via the SAP domain. Northwestern analysis. Membrane-bound recombinant wild type and the indicated ARTD2 mutants as well as ARTD1 (see coomassie-stained gel, CB) were incubated with radiolabeled rRNA (−16 to +130) and bound RNA was visualized by autoradiography (32P).
The results described above indicate that H2O2 induces PAR formation in the nucleoli (Figure 1). ActD impairs 45S pre-rRNA synthesis by preventing Pol I elongation through the rDNA coding region and leads to an accumulation of short RNA transcripts corresponding to sequences immediately downstream of the transcription start site (+1/+130), as detected by northern blotting (Figure 4B and Supplementary Figure S5A) (30,31). We described above that only prolonged treatment with ActD (4 and 20 h) enhanced formation of PAR in cells treated with H2O2 or MNNG (Figure 3). Because PAR formation correlated with the inhibition of 45S pre-rRNA synthesis (Supplementary Figure S5B), we hypothesized that the production of short rRNA transcripts induced by ActD treatment might stimulate ARTD2 activity. To test this hypothesis, we investigated the in vitro ARTD2 automodification in the presence of in vitro synthesized rRNA transcripts (corresponding to rDNA sequences from −16 to +130 bp) using P32-labeled NAD+ to monitor ADP-ribosylation (Figure 4C). Densitrometric analysis of the incorporated radiolabeled ADP-ribose revealed that the automodification of ARTD2 was stimulated by rRNA 3.4-fold stronger in comparison with double-stranded DNA, while rRNA stimulated ARTD1 only 0.7-fold compared with the stimulation with double-stranded DNA (Figure 4C). This effect did not seem to be mediated by specific rRNA sequences, as other nonnucleolar RNA transcripts were able to stimulate ARTD2 activity to a similar extent (Supplementary Figure S5C). In contrast to ARTD2, ARTD1 was strongly activated by double-stranded DNA and, as previously described (36), only exhibited 60% of its activity in the presence of RNA as compared with DNA (Figure 4C).
Because the ActD effect was only observed in the presence of genotoxic stress, we tested whether the activation of ARTD2 by RNA depends on double-stranded DNA. Addition of DNA did not enhance the stimulatory effect of RNA on ARTD2, confirming that RNA stimulates ARTD2 independent of DNA (Figure 4D).
ARTD2 binds to RNA through its SAP domain
To define which domain of ARTD2 is responsible for the RNA-mediated activation, we compared the effect of RNA on the activities of human ARTD2FL (full length) and ARTD2ΔSAP (aa 95–583), a mutant that lacks the SAP domain and displays similar basal activity as ARTD2FL. Although ARTD2FL was stimulated by RNA, the deletion of the SAP domain impaired RNA-mediated activation (Figure 4E). In addition to the loss of the stimulation by RNA, the additional deletion of the WGR domain (aa 231–583, ARTD2 ΔSΔW), or removal of the WGR domain alone (deletion of aa 116–193, ARTD2 ΔWGR) resulted in a strong reduction of the general ARTD2 activity (Figure 4E). RNA-binding experiments with these proteins confirmed that the SAP domain is responsible for the binding of ARTD2 to RNA. Together, these results suggest that RNA is a potent activator of ARTD2 enzymatic activity predominantly through the SAP domain, and that the WGR domain is an important structural element for the overall activity of the enzyme.
DISCUSSION
ADP-ribosylation has been implicated in several nucleolar processes such as ribosome biogenesis, formation of rDNA heterochromatin and stress sensing (32,34,36,42–44). Here, we show that H2O2 or MNNG induces PAR formation in the nucleoli of both mouse and human cells. This is in agreement with previous studies that found ARTD1 and ARTD2 to localize to nucleoli (34). The combination of H2O2 or MNNG treatment with low doses of ActD revealed a synergistic effect on PAR formation that is mediated by ARTD2. It was well-known before that ActD treatment leads to the accumulation of short rRNA transcripts, which might be responsible for the enhancement of PAR formation. Short rRNA transcripts were able to strongly stimulate ARTD2 activity via the SAP/WGR domain, while double-stranded DNA did not exhibit this effect. Our findings thus reveal a new activator for ARTD2-dependent ADP-ribosylation, which has important implications for the future analysis of the biological role of ARTD2 in the nucleus.
ADP-ribosylation and in particular the homologous enzymes ARTD1 and ARTD2 have been traditionally implicated in the response to DNA damage. An important cornerstone for this model is the strong in vitro activation of ARTD1 by double-stranded or nicked DNA, as well as the detection of PAR on treatment of cells with genotoxic compounds. In these DNA damage-dependent processes, ARTD2 displays much less activity than ARTD1, raising the question whether molecules other than activators of ARTD1 activate ARTD2. Our results strongly suggest that instead of binding to sites of DNA damage, ARTD2 associates with RNA in the nucleolus, providing an alternative cue to identify and respond to DNA damage. In support of this, recent data have implicated site-specific Dicer and Drosha RNA moieties in the control of DNA damage (45). Furthermore, the activation of ARTD2 by RNA may also be part of an intricate network of RNA surveillance and repair mechanisms to preserve RNA quality (46–48). The identification of ARTD2 activation by RNA is a new and unexpected result that may indicate an additional mean for monitoring not only genome integrity, but also other processes specific for ARTD2. Genetic disruption of ARTD2, but not of ARTD1, affects various differentiation processes in mice, including spermatogenesis (49), adipogenesis (50) and the survival of thymocytes (51).
Interestingly, the stimulation of ARTD2 in vivo was dependent on the co-stimulation with ActD and H2O2 or MNNG, indicating that additional signals induced by H2O2 or MNNG are required for the activation of ARTD2 by RNA. These signals might include the damage of RNA, as oxidative damage in RNA is usually higher than in DNA (52). However, our experiments with H2O2-pretreated RNA did not strengthen this hypothesis (data not shown). Alternatively, the treatment of cells with H2O2 or MNNG might induce signaling cascades that lead to the posttranslational modification of ARTD2, which is required for the stimulation with RNA, besides damaging the DNA. Future studies of the nucleolar ADP-ribose acceptors and the effect of ADP-ribosylation on the response to RNA damage are needed to reveal and define these functions in detail. Interestingly, the ActD effect was observed in both human and mouse cells, suggesting that the stimulation of ARTD2 is a biologically conserved effect.
Under the study conditions described here, cellular PAR formation on H2O2/MNNG treatment occurred to a large portion within nucleoli. Growing evidence indicates that the nucleolus plays a key role in monitoring and responding to cellular stress. After exposure to extra- or intracellular stress, signal pathways induce rapid downregulation of 45S pre-rRNA synthesis that is followed by perturbation of nucleolar structure, cell cycle arrest and stabilization of p53 (53). The formation of high levels of PAR in nucleoli under genotoxic stress conditions might be part of this nucleolus-dependent signaling. Whether PAR formation is due to a high accessibility of nucleolar DNA and RNA to chemical agents and how this occurs remains to be investigated.
Previous studies have shown that RNA is a key regulator of ARTD1 in the nucleolus [(36) and Figure 4D/F]. Nucleolar localization of ARTD1 is dependent on RNA and ARTD1 binds in vivo and in vitro to the nucleolar noncoding pRNA, an intergenic transcript implicated in the establishment of rDNA heterochromatin (54). pRNA associates with the zinc finger DNA binding domain of ARTD1 and stimulates ARTD1 activity, although to a lesser degree than double-stranded DNA [(36) and Figure 4D].
In this work, we provide evidence that RNA is a key regulator of ARTD2 enzymatic activity. The nucleolar localization of overexpressed ARTD2 has already been described (34) and the currently available antibodies do not permit a direct immunofluorescent localization of endogenous ARTD2 or determination of how RNA affects nucleolar occupancy of ARTD2. However, we could demonstrate that RNA strongly activates ARTD2 activity. In contrast to ARTD1, double-stranded DNA did not activate ARTD2. These findings suggest that the structure and nature of nucleic acid is an important determinant for the regulation of ARTD2 function. Consistent with this, recent analyses with several DNA structures mimicking intermediates of different DNA metabolizing processes revealed that ARTD2 activation efficiency did not correlate with K(d) values for DNA (55). ARTD2 displayed the highest affinity for flap-containing DNA, but was more efficiently activated by 5′-overhang DNA, suggesting that single-stranded nucleic acids might be a general stimulator of ARTD2. The stimulation did not seem to be sequence-specific, as other RNAs tested were also able to strongly stimulate ARTD2 (36). We have identified SAP to be the main domain responsible for the stimulation by RNA. The SAP motif (after SAF-A/B, Acinus and PIAS) is a putative DNA/RNA binding domain found in diverse nuclear and cytoplasmic proteins (56,57). Based on the presented findings, it is thus possible that other proteins with a SAP motif similarly may bind RNA and be regulated by this binding. Interestingly, deleting the WGR domain only abolished the stimulation by RNA, but also the overall activity of ARTD2, indicating that the structural arrangement of the SAP and the CAT motif is functionally relevant.
Together, the strong activation of ARTD2 by RNA through the SAP domain is a likely underlying cause for the distinct and complementary functions mediated by this homolog as compared with ARTD1. The fact that RNA stimulates PAR formation by ARTD2 has not only mechanistic implications but also sheds new light on how ARTD2 and poly-ADP-ribosylation function during the genotoxic stress response.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Forschungskredit of the University of Zurich (to K.L. and N.S.); Swiss National Science Foundation [310003A-135801 to R.S., 310030B_138667 and PDMFP3_127315 to M.O.H.]; Kanton of Zurich (to M.O.H.). Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
P.O. Hassa is acknowledged for providing the 10H antibody and M. Stucki (University of Zurich, Switzerland) is acknowledged for providing the T24 cell line. Confocal microscopy was performed at the Center for Microscopy and Image Analysis of the University of Zurich. M. Fey is acknowledged for technical support. Florian Freimoser and Stephan Christen (University of Zurich, Switzerland) provided editorial assistance and critical input during the writing. K.L. designed and performed experiments; D.B. performed Northwestern analysis; N.S. produced synthetic RNA; R.S. and M.O.H. supervised the work and M.O.H. wrote the manuscript.
REFERENCES
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 3.Dianov GL, Hubscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassa PO, Haenni S, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Bio.l Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber V, Dantzer F, Ame J-C, De Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 8.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPs, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 9.Kraus WL, Hottiger MO. PARP-1 and gene regulation: Progress and puzzles. Mol. Aspects Med. 2013;34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 10.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front. Biosci. 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee S, Kaye S. PARP inhibitors in BRCA gene-mutated ovarian cancer and beyond. Curr. Oncol. Rep. 2011;13:442–449. doi: 10.1007/s11912-011-0193-9. [DOI] [PubMed] [Google Scholar]
- 12.Bryant H, Schultz N, Thomas H, Parker K, Flower D, Lopez E, Kyle S, Meuth M, Curtin N, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 13.Telli ML. PARP inhibitors in cancer: moving beyond BRCA. Lancet Oncol. 2011;12:827–828. doi: 10.1016/S1470-2045(11)70236-4. [DOI] [PubMed] [Google Scholar]
- 14.Underhill C, Toulmonde M, Bonnefoi H. A review of PARP inhibitors: from bench to bedside. Ann. Oncol. 2010;22:268–279. doi: 10.1093/annonc/mdq322. [DOI] [PubMed] [Google Scholar]
- 15.Amé J, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Höger T, Ménissier-de Murcia J, de Murcia G. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- 16.Yelamos J, Schreiber V, Dantzer F. Toward specific functions of poly(ADP-ribose) polymerase-2. Trends Mol. Med. 2008;14:169–178. doi: 10.1016/j.molmed.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Oliver AW, Ame JC, Roe SM, Good V, de Murcia G, Pearl LH. Crystal structure of the catalytic fragment of murine poly(ADP-ribose) polymerase-2. Nucleic Acids Res. 2004;32:456–464. doi: 10.1093/nar/gkh215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yelamos J, Farres J, Llacuna L, Ampurdanes C, Martin-Caballero J. PARP-1 and PARP-2: New players in tumour development. Am. J. Cancer Res. 2011;1:328–346. [PMC free article] [PubMed] [Google Scholar]
- 19.Troiani S, Lupi R, Perego R, Depaolini SR, Thieffine S, Bosotti R, Rusconi L. Identification of candidate substrates for poly(ADP-ribose) polymerase-2 (PARP2) in the absence of DNA damage using high-density protein microarrays. FEBS J. 2011;278:3676–3687. doi: 10.1111/j.1742-4658.2011.08286.x. [DOI] [PubMed] [Google Scholar]
- 20.Brown DD, Gurdon JB. Absence of ribosomal RNA synthesis in the anucleolate mutant of Xenopus laevis. Proc. Natl Acad. Sci. USA. 1964;51:139–146. doi: 10.1073/pnas.51.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadjiolov AA. In: Cell Biology Monographs. Beerman AM, Goldstein L, Portrer KR, Sitte P, editors. New York: Springer; 1985. pp. 1–263. [Google Scholar]
- 22.Santoro R. The silence of the ribosomal RNA genes. Cell Mol. Life Sci. 2005;62:2067–2079. doi: 10.1007/s00018-005-5110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santoro R. In: The Nucleolus. Olson MJ, editor. Basel, Switzerland: Springer; 2011. pp. 57–82. [Google Scholar]
- 24.Haaf T, Hayman DL, Schmid M. Quantitative determination of rDNA transcription units in vertebrate cells. Exp. Cell Res. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-b. [DOI] [PubMed] [Google Scholar]
- 25.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 26.Olson MO. Sensing cellular stress: another new function for the nucleolus? Sci. STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 27.Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J. Cell Physiol. 1970;76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- 28.Sobell HM. Actinomycin and DNA transcription. Proc. Natl Acad. Sci. USA. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trask DK, Muller MT. Stabilization of type I topoisomerase-DNA covalent complexes by actinomycin D. Proc. Natl Acad. Sci. USA. 1988;85:1417–1421. doi: 10.1073/pnas.85.5.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sentenac A, Simon EJ, Fromageot P. Initiation of chains by RNA polymerase and the effects of inhibitors studied by a direct filtration technique. Biochim. Biophys. Acta. 1968;161:299–308. doi: 10.1016/0005-2787(68)90108-1. [DOI] [PubMed] [Google Scholar]
- 31.Hadjiolova KV, Hadjiolov AA, Bachellerie JP. Actinomycin D stimulates the transcription of rRNA minigenes transfected into mouse cells. Implications for the in vivo hypersensitivity of rRNA gene transcription. Eur. J. Biochem. 1995;228:605–615. doi: 10.1111/j.1432-1033.1995.0605m.x. [DOI] [PubMed] [Google Scholar]
- 32.Boamah EK, Kotova E, Garabedian M, Jarnik M, Tulin AV. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 2012;8:e1002442. doi: 10.1371/journal.pgen.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leitinger N, Wesierska-Gadek J. ADP-ribosylation of nucleolar proteins in HeLa tumor cells. J. Cell Biochem. 1993;52:153–158. doi: 10.1002/jcb.240520207. [DOI] [PubMed] [Google Scholar]
- 34.Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J. Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 35.Guetg C, Santoro R. Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics. 2012;7:811–814. doi: 10.4161/epi.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell. 2012;45:790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 37.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 40.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 41.Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat. Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 42.Tulin A, Stewart D, Spradling A. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guerrero PA, Maggert KA. The CCCTC-binding factor (CTCF) of Drosophila contributes to the regulation of the ribosomal DNA and nucleolar stability. PLoS One. 2011;6:e16401. doi: 10.1371/journal.pone.0016401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rancourt A, Satoh MS. Delocalization of nucleolar poly(ADP-ribose) polymerase-1 to the nucleoplasm and its novel link to cellular sensitivity to DNA damage. DNA Repair (Amst.) 2009;8:286–297. doi: 10.1016/j.dnarep.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 45.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wurtmann EJ, Wolin SL. RNA under attack: cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Wu J, Deleo CJ. RNA damage and surveillance under oxidative stress. IUBMB Life. 2006;58:581–588. doi: 10.1080/15216540600946456. [DOI] [PubMed] [Google Scholar]
- 48.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 49.Dantzer F, Mark M, Quenet D, Scherthan H, Huber A, Liebe B, Monaco L, Chicheportiche A, Sassone-Corsi P, de Murcia G, et al. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc. Natl Acad. Sci. USA. 2006;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Menissier-de Murcia J. Poly(ADP-ribose) polymerase-2 [corrected] controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-gamma [corrected] heterodimer. J. Biol. Chem. 2007;282:37738–37746. doi: 10.1074/jbc.M701021200. [DOI] [PubMed] [Google Scholar]
- 51.Yelamos J, Monreal Y, Saenz L, Aguado E, Schreiber V, Mota R, Fuente T, Minguela A, Parrilla P, de Murcia G, et al. PARP-2 deficiency affects the survival of CD4+CD8+ double-positive thymocytes. EMBO J. 2006;25:4350–4360. doi: 10.1038/sj.emboj.7601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hofer T, Badouard C, Bajak E, Ravanat JL, Mattsson A, Cotgreave IA. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 2005;386:333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 53.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 55.Kutuzov MM, Khodyreva SN, Ame JC, Ilina ES, Sukhanova MV, Schreiber V, Lavrik OI. Interaction of PARP-2 with DNA structures mimicking DNA repair intermediates and consequences on activity of base excision repair proteins. Biochimie. 2013;95:1208–1215. doi: 10.1016/j.biochi.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 57.Iida T, Kawaguchi R, Nakayama J. Conserved ribonuclease, Eri1, negatively regulates heterochromatin assembly in fission yeast. Curr. Biol. 2006;16:1459–1464. doi: 10.1016/j.cub.2006.05.061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.