Nucleic Acids Res. 2013; 41, 4307–4323. doi: 10.1093/nar/gkt101.
The authors would like to introduce the following changes to the original version of the article published in Nucleic Acids Research.
We corrected an error that affected our assessment of absolute and relative performance of the PETfold method. In the original version of the manuscript, results were reported for the use of unaligned RNA sequences as input for PETfold pre2.0. Jan Gorodkin and Stefan Seemann, the authors of this method (University of Copenhagen, Denmark), brought to our attention that this was incorrect, as the intended input for PETfold pre2.0 were aligned RNA sequences. Regrettably, PETfold pre2.0 did not validate the correct type of input data, and for unaligned data sets, it generated RNA secondary structure predictions, which scored poorly in our benchmark. We recalculated all predictions for PETfold pre2.0 with aligned RNA sequences (the same data sets as used for other methods that required aligned sequences). As a result, the performance of PETfold pre2.0 has significantly improved and according to the corrected rankings, this method has been re-evaluated as one of the best. Owing to the modification of relative scores calculated for PETfold pre2.0 with respect to other methods, other slight changes in the rankings occurred, which are now reflected in the corrected rankings presented on the CompaRNA Web site and in corrected Figure 3 and Table 6. We apologize to the authors of PETfold as well as to the readership of Nucleic Acids Research for erroneously reporting the performance of PETfold in our original rankings. Importantly, the authors of PETfold have subsequently released a new version PETfold 2.0 that checks whether input sequences are aligned. Currently, both PETfold pre2.0 and PETfold 2.0 are tested in CompaRNA.
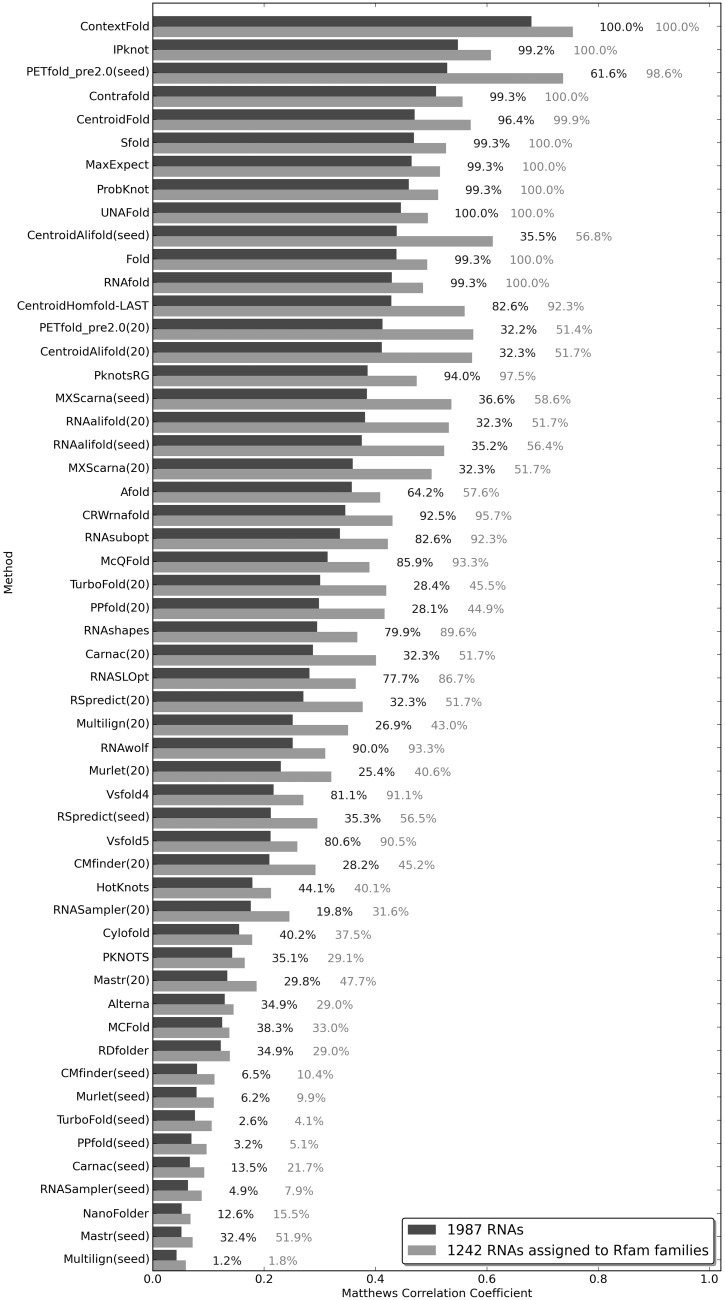
Figure 3.
The results of a robustness test on the RNAstrand data set. The numbers on the right to each bar correspond to the percent of RNAs for which a given method returned predictions (dark = 1987 RNAs from the RNAstrand data set; light = 1242 RNAs for which CompaRNA assigned an Rfam family). (20), refers to the test of a comparative method in which 20 representatives of an Rfam seed alignment were used; (seed), refers to the test in which all sequences from a given seed alignment were used.
Table 6.
Best methods according to rankings on the RNAstrand data set
| Ranking type | First rank | Second rank | Third rank | |
|---|---|---|---|---|
| All RNAs | Ext | TurboFold(seed) (W: 52, L: 1, NW: 0) | ContextFold & PETfold_pre2.0(seed) (W: 51, L: 2, NW: 0) | TurboFold(20) (W: 50, L: 2, NW: 1)a |
| Short RNAs (20–200 nt) | Ext | ContextFold (W: 53, L: 0, NW: 0) | TurboFold(20) (W: 51, L: 1, NW: 1) | CentroidAlifold(seed) (W: 49, L: 3, NW: 1) |
| Medium-sized RNAs (201–800 nt) | Ext | PETfold_pre2.0(seed) (W: 44, L: 2, NW: 7) | ContextFold (W: 44, L: 3 NW: 6) | TurboFold(20) (W: 39, L: 2, NW: 12) |
| Long RNAs (801–30 000 nt) | Ext | PETfold_pre2.0(seed) (W: 24, L: 0, NW: 29) | ContextFold (W: 22, L: 1, NW: 30) | CentroidAlifold(seed) (W: 21, L: 2, NW: 30) |
| All pseudoknotted RNAs | Ext | PETfold_pre2.0(seed) (W: 48, L: 3, NW: 2) | ContextFold (W: 45, L: 4, NW: 4) | CentroidAlifold(seed) (W: 44, L: 6, NW: 3) |
| Pseudoknotted short RNAs (20–200 nt) | Ext | Cylofold (W: 35, L: 0, NW: 18) | McQFold (W: 35, L: 1, NW: 17) | PKNOTS (W: 33, L: 2, NW: 18) |
| Pseudoknotted medium-sized RNAs (201–800 nt) | Ext | ContextFold (W: 42, L: 0, NW: 11) | PETfold_pre2.0(seed) (W: 41, L: 1, NW: 11) | TurboFold(20) (W: 38, L: 2, NW: 13) |
| Pseudoknotted long RNAs (801–30 000 nt) | Ext | PETfold_pre2.0(seed) (W: 24, L: 0, NW: 29) | PETfold_pre2.0(20) & ContextFold (W: 22, L: 2, NW: 29) | CentroidAlifold(seed) (W: 21, L: 2, NW: 30)a |
| Robustness test–1987 sequences | Ext | ContextFold (W: 53, L: 0, NW: 0) | IPknot (W: 52, L: 1, NW: 0) | PETfold_pre2.0(seed) (W: 51, L: 2, NW: 0) |
| Robustness test–1242 sequences with Rfam family assigned | Ext | ContextFold (W: 53, L: 0, NW: 0) | PETfold_pre2.0(seed) (W: 52, L: 1, NW: 0) | CentroidAlifold(seed) (W: 51, L: 2, NW: 0) |
aFourth place.
W, number of wins; L, number of defeats; NW, number of cases in which it was impossible to select winner; (20), refers to the test of a comparative method in which 20 representatives of an Rfam seed alignment were used; (seed), refers to the test in which all sequences from a given seed alignment were used.
Moreover, the following minor changes ought to be introduced to the original article:
We identified and corrected a minor error in the script used to parse predictions generated by RSpredict. We recalculated the results with a correctly parsed data, leading to a relatively modest change of RSpredict performance. The changes have been communicated with the authors of publication describing RSpredict, Juna Spirollari and Jason Wang (New Jersey Institute of Technology, USA). The corrected results are now reflected in the corrected rankings presented on the CompaRNA Web site and in corrected Figure 3.
The description of the PknotsRG program in the original version of our article suggested incorrectly that PKNOTS does not use the Turner energy rules, nor does it find the minimum free energy structure. In fact, PKNOTS does use the Turner rules when applicable, and finds the minimum free energy structure with exact dynamic programming, and PknotsRG uses a similar model, but instead of finding the optimal minimum free energy, it applies a heuristic approach to find a structure that is not guaranteed to be necessarily the minimum free energy structure. This correction has been communicated with Elena Rivas (Howard Hughes Medical Institute, USA), the author of PKNOTS.
The two data sets consisting of pseudoknotted RNAs described in Table 4, which were used for testing methods predicting RNA secondary structure, contained a different number of RNAs than originally stated in our article. A data set created by taking into account standard base-pair definition contained 31 RNAs, instead of 33 as stated in the original article, and the data set based on extended base-pair definition contained 58 RNAs, instead of 62 as stated in the original article. This had a small impact on the relative performance of a few methods, which is reflected in the corrected version of Table 5. This inconsistency was brought to our attention by Cong Zeng (Laboratoire de Recherche en Informatique at Université Paris-Sud, France). We have also corrected the number of short RNAs from the RNAstrand dataset, which is 882, not 869 (Table 4). This was a typographical error, and its correction has no effect on the rankings.
Table 4.
Data sets used for benchmarking methods predicting RNA secondary structure
| Source | Data set name | Type of RNAs | Sequence length | Number of sequences |
|---|---|---|---|---|
| PDB | All RNAs, standard base-pair definition | All | ≥20 | 121 |
| All RNAs, extended base-pair definition | All | ≥20 | 121 | |
| Only pseudoknotted RNAs, standard base-pair definition | Pseudoknotted | ≥20 | 31 | |
| Only pseudoknotted RNAs, extended base-pair definition | Pseudoknotted | ≥20 | 58 | |
| RNAstrand | All RNAs | All | ≥20 | 1987 |
| All short RNAs | All | 21–200 | 882 | |
| All medium-sized RNAs | All | 201–800 | 818 | |
| All long RNAs | All | >800 | 287 | |
| Pseudoknotted RNAs | Pseudoknotted | ≥20 | 919 | |
| Pseudoknotted short RNAs | Pseudoknotted | 21–200 | 53 | |
| Pseudoknotted medium-sized RNAs | Pseudoknotted | 201–800 | 610 | |
| Pseudoknotted long RNAs | Pseudoknotted | >800 | 256 |
Table 5.
Best methods according to rankings on the PDB data set
| Ranking type | First rank | Second rank | Third rank | |
|---|---|---|---|---|
| All RNAs | Std | MXScarna(seed) (W: 38, L: 3, NW: 12) | CentroidAlifold(20) (W: 36, L: 0, NW: 17) | CentroidFold (W: 36, L: 8, NW: 9) |
| Ext | MXScarna(seed) (W: 38, L: 2, NW: 13) | CentroidFold (W: 37, L: 7, NW: 9) | CentroidAlifold(20) (W: 36, L: 0, NW: 17) | |
| Pseudoknotted RNAs | Std | CentroidAlifold(20) (W: 33, L: 0, NW: 20) | CentroidAlifold(seed) (W: 33, L: 1, NW: 19) | RNAalifold(20) (W: 31, L: 2, NW: 20) |
| Ext | MXScarna(seed) (W: 36, L: 1, NW: 16) | CentroidAlifold(20) (W: 35, L: 0, NW: 18) | RNAalifold(20) (W: 33, L: 2, NW: 18) | |
Std, standard base-pair definition; Ext, extended base-pair definition (see Materials and Methods section); W, number of wins; L, number of defeats; NW, number of cases in which it was impossible to select winner; (20), refers to the test of a comparative method in which 20 representatives of an Rfam seed alignment were used; (seed), refers to the test in which all sequences from a given seed alignment were used.
Per suggestion by the authors of PETfold (Stefan Seemann, Jan Gorodkin and Rolf Backofen), we would like to update the reference to their method and to replace a citation of their article describing the PETfold Web server (reference 53) by a citation of the following article: Seemann,S.E., Gorodkin,J. and Backofen,R. (2008) Unifying evolutionary and thermodynamic information for RNA folding of multiple alignments. Nucleic Acids Res, 36, 6355–6362.
We also thank all researchers mentioned in this corrigendum for their feedback that helped us improve the article as well as the CompaRNA Web server.
We have provided corrected Figure 3 and Tables 4–6, which differ with respect to the original article and incorporate the aforementioned changes.