Abstract
Leucyl-tRNA (transfer RNA) synthetase (LeuRS) is a multi-domain enzyme, which is divided into bacterial and archaeal/eukaryotic types. In general, one specific LeuRS, the domains of which are of the same type, exists in a single cell compartment. However, some species, such as the haloalkaliphile Natrialba magadii, encode two cytoplasmic LeuRSs, NmLeuRS1 and NmLeuRS2, which are the first examples of naturally occurring chimeric enzymes with different domains of bacterial and archaeal types. Furthermore, N. magadii encodes typical archaeal tRNALeus. The tRNA recognition mode, aminoacylation and translational quality control activities of these two LeuRSs are interesting questions to be addressed. Herein, active NmLeuRS1 and NmLeuRS2 were successfully purified after gene expression in Escherichia coli. Under the optimized aminoacylation conditions, we discovered that they distinguished cognate NmtRNALeu in the archaeal mode, whereas the N-terminal region was of the bacterial type. However, NmLeuRS1 exhibited much higher aminoacylation and editing activity than NmLeuRS2, suggesting that NmLeuRS1 is more likely to generate Leu-tRNALeu for protein biosynthesis. Moreover, using NmLeuRS1 as a model, we demonstrated misactivation of several non-cognate amino acids, and accuracy of protein synthesis was maintained mainly via post-transfer editing. This comprehensive study of the NmLeuRS/tRNALeu system provides a detailed understanding of the coevolution of aminoacyl-tRNA synthetases and tRNA.
INTRODUCTION
Protein biosynthesis is an essential step in all three domains of life (1). This complicated process requires a subset of proteins and nucleic acids. Aminoacyl-tRNA (transfer RNA) synthetases (aaRSs) are responsible for providing aminoacyl-tRNAs (aa-tRNAs) as the building blocks for protein biosynthesis by catalyzing the esterification of specific amino acids to their cognate tRNAs (2). This reaction (aminoacylation) is performed in two steps by most aaRSs; the amino acid is first activated with adenosine triphosphate (ATP) to form aminoacyl-adenylate (aa-AMP), which is then transferred to the 3′-terminus of the cognate tRNA to generate aa-tRNA for delivery to the ribosomes for protein biosynthesis. The aaRSs are divided into two classes (class I and class II) on the basis of conserved sequences and characteristic structural motifs (3,4). Leucyl-tRNA synthetase (LeuRS) belongs to class I aaRSs, which share a structurally similar Rossmann fold with two characteristic ‘HIGH’ and ‘KMSKS’ motifs in the catalytic domain.
Aminoacylation of tRNA requires precise regulation of the velocity of the aa-tRNA production for the ribosome and tight control of aberrant aa-tRNA synthesis (5). Various identity determinants and/or antideterminants are harbored by different tRNAs to facilitate the selection of cognate tRNA from a large set of tRNA species by specific aaRSs. However, the specificity of cognate amino acid activation is greatly challenged by the presence of various non-cognate amino acids and amino acid metabolites, which differ only in the side chain. Taking the mistranslation frequency (1/3000) in protein biosynthesis as the threshold (6), a proofreading (editing) mechanism is necessary for half of aaRSs to maintain accuracy during aa-tRNA synthesis. The editing activity can be divided further into the pre- and post-transfer editing. Pre-transfer editing refers to the hydrolysis of the misactivated aa-AMP either in the absence of cognate tRNA (tRNA-independent pre-transfer editing) or in the presence of cognate tRNA (tRNA-dependent pre-transfer editing), whereas post-transfer editing is performed by the editing domain [such as the connective peptide 1 (CP1) domain of class Ia synthetases (7), the N2 domain of class II threonyl-tRNA synthetase (8), the freestanding editing domain homologs (9)] after a misactivated amino acid has been loaded onto the cognate tRNA. The editing activity of an aaRS is of crucial importance in ensuring overall translational quality control. The impairment or loss of editing activity can lead to mistranslation and further ambiguity of the proteome with serious negative influences on the cellular function of most organisms (10–12). This is directly illustrated by the development of neurodegeneration in a mouse model with partial loss of the editing activity of alanyl-tRNA synthetase (13).
LeuRSs catalyze aminoacylation and editing reactions to synthesize Leu-tRNALeu and to prevent production of mischarged tRNALeu. Based on the primary sequence, relative domain position and orientation, LeuRS can be divided into bacterial (existing in bacteria and organelles) and archaeal/eukaryotic types (existing in eukaryotes and most archaea) (14,15). Both types of LeuRSs contain a Rossmann-fold domain (for amino acid activation and tRNA charging), a CP1 domain (for editing), an α-helix bundle domain (for tRNA binding) and a C-terminal domain (CTD, for tRNA binding) (16). Moreover, LeuRSs also contain some specific insertion domains, such as the leucine-specific domain to modulate their aminoacylation and editing activities (17,18). To date, all the domains identified in a single LeuRS polypeptide are of the same type, indicating that no gene (fragment) rearrangement or fusion between two LeuRS types has occurred.
LeuRSs use different tRNALeu discrimination modes besides recognition of the common specific discriminator base (A73) of tRNALeu. Biochemical (19,20) and 3D structural (21) data have revealed that the T-loop, D-loop and bases (47f–47i) on one side of the long variable stem in tRNALeu from bacteria (e.g. Escherichia coli) are recognized by the CTD in bacterial EcLeuRS or Mycobacterium tuberculosis LeuRS. In the hyperthermophilic bacteria Aquifex aeolicus, which contains a heterodimeric LeuRS, the core structure of tRNA formed by the tertiary interactions (U8-A14, G18-U55 and G19-C56) and the orientation of the long variable stem are critical elements for tRNALeu aminoacylation, and the anticodon stem acts as an additional determinant for editing by LeuRS (22). In Homo sapiens, three base pairs (C3:G70, A4:U69 and G5:C68) in the acceptor stem, C20a in the D-loop, the length of long variable stem and the number of the unpaired residues in long variable loop are the identity elements of tRNALeu (23). However, Saccharomyces cerevisiae uses the anticodon loop of tRNALeu (A35 and G37) as the major identity determinant (24). The recognition mode of tRNALeu in archaea is highly conserved and differs from those of the previously mentioned species. The most obvious feature is the critical dependence on the two absolutely conserved bases (A47c, G47d) at the tip of the long variable loop in tRNALeu, as reported in halophilic Haloferax volcanii and thermophilic Pyrococcus horikoshii (25,26). The 3D structure of the P. horikoshii LeuRS (PhLeuRS) showed that its CTD, which contains five α-helices, antiparallel β-sheets and many flexible loops, is responsible for tRNA recognition, especially of Asp845, Ile849 and the last few conserved residues (Pro962, Ile964, Ile966 and Glu967) (26). Deletion of more than one residue in the CTD led to the total loss of aminoacylation activity of PhLeuRS. In contrast, deletion of the entire CTD had no effect on its editing activity (14).
The distribution of LeuRSs is diverse in the three domains of life. In bacteria (e.g. E. coli), only a bacterial LeuRS is encoded. Eukaryotes (e.g. H. sapiens) generally contain two LeuRSs: an archaeal/eukaryotic cytoplasmic form and a bacterial form in mitochondria and/or chloroplasts. A majority of archaeal (e.g. P. horikoshii) encode a LeuRS, which is evolutionarily close to the cytoplasmic form of the enzyme in eukaryotes. However, in a limited number of cases, such as the halobacteria Natrialba magadii, which thrives in alkaline and hypersaline conditions (pH 9.5, 3.5 M NaCl) (2 g/l KCl, 0.1 g/l MgSO4·7H2O and microscale of FeSO4 and MnCl2 are also needed in the culture medium) (27,28), the LeuRS/tRNALeu system exhibits several striking features. First, the N. magadii genome encodes two LeuRS genes [designated as NmLeuRS1 (accession No. YP_003479843.1, 904-aa) and NmLeuRS2 (accession No. YP_003481407.1, 971-aa)] displaying 46% identity. Second, these two LeuRSs exhibit both bacterial and archaeal characteristics. Specifically, the N-terminal region (including the Rossmann fold, the CP1 domain, the CP2 domain and the α-helix bundle domain) shares high homology with bacterial LeuRSs (Supplementary Figure S1A); however, the CTD is highly homologous to its archaeal LeuRS counterpart (Supplementary Figure S1B). Moreover, N. magadii expresses typical archaeal tRNALeus in which the A47c and G47d bases in the long variable loop are absolutely conserved, whereas no bacterial tRNALeus are expressed (Supplementary Figure S2). This raises the interesting questions of the mechanisms underlying the aminoacylation and editing activities, the active sites of which reside in the bacterial and archaeal portion of two NmLeuRSs as well as the recognition mode for archaeal tRNALeu.
In the present study, genes encoding NmLeuRS1 and NmLeuRS2 were cloned and overexpressed in E. coli. Following successful purification, the aminoacylation, amino acid activation, tRNA recognition mode and editing activity of these NmLeuRSs were characterized. Our results provide an improved understanding of the activity and evolutionary pathway of tRNA synthetases from halobacteria.
MATERIALS AND METHODS
Materials
l-leucine (Leu), l-norvaline (Nva), α-amino butyric acid (ABA), l-isoleucine (Ile), l-methionine (Met), tetrasodium pyrophosphate, Tris–HCl, MgCl2, NaCl, KCl and activated charcoal were purchased from Sigma (St Louis, MO, USA). [3H]Leu, [32P]tetrasodium pyrophosphate and [α-32P]ATP were obtained from PerkinElmer (Waltham, MA, USA). Nucleoside triphophate (NTP) and deoxynucleoside triphosphate (dNTP) mixtures were purchased from Sangon Biotech (China). The Pfu DNA polymerase, the DNA fragment rapid purification kit and plasmid extraction kit were purchased from Tiangen Biotech (China). The KOD-plus mutagenesis kit was obtained from TOYOBO (Japan). T4 ligase, inorganic pyrophosphatase, protein ladder (#26614) and restriction endonucleases were obtained from Thermo Scientific (Waltham, MA, USA). The Ni2+-NTA (Ni2+-nitrilotriacetate) Superflow was purchased from Qiagen (Germany). Polyethyleneimine cellulose plates, Amicon Ultra-15 filter and nitrocellulose membranes (0.22 μm) were purchased from Merck (Germany). Oligonucleotide primers were synthesized by Life Technologies (Carlsbad, CA, USA). Competent E. coli BL21 (DE3) and Top10 cells were prepared in our laboratory. T7 RNA polymerase (29) and E. coli CCA-adding enzyme (30) were purified from an overproduction strain in our laboratory.
Sequence alignment of LeuRSs
For phylogenetic analysis of LeuRSs covering three domains, 63 protein sequences of various LeuRSs from representative species were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/protein/). Specific sequence alignments were carried out using the Clustal X2 program (31) and subsequent phylogenetic trees were constructed with the MEGA 5.2 program (32) using the minimal evolution algorithm. Bootstrap analysis was performed with 1000 replicates.
Gene cloning and protein purification
NmLeuRS1 and NmLeuRS2 genes were amplified from N. magadii ATCC 43099 genomic DNA and cloned into pET28a (pre-cleaved with NdeI/BamHI) to produce pET28a-NmLeuRS1 and pET28a-NmLeuRS2. The primers for gene amplification were as follows: NmLeuRS1 forward (5′ ccatatggaaagttatacag3′), NmLeuRS1 reverse (5′ cggatcctcagtcctcgatc3′), NmLeuRS2 forward (5′ ccatatgacaaaccagtacgatc3′) and NmLeuRS2 reverse (5′ cggatcctcactgaatccgaat3′). Haloarcula hispanica LeuRS1 and LeuRS2 genes were amplified from H. hispanica ATCC 33960 genomic DNA and cloned into pET28a (pre-cleaved with NdeI/BamHI or NheI/BamHI) to produce pET28a-HhLeuRS1 and pET28a-HhLeuRS2. The primers for gene amplification were as follows: HhLeuRS1 forward (5′ ccatatgaccacgaccggtg3′), HhLeuRS1 reverse (5′ cggatccctactcgtcgatgt3′), HhLeuRS2 forward (5′ agctagcatgtcgcgccgatat3′) and HhLeuRS2 reverse (5′ tggatcctcaggagatgtgga3′). Escherichia coli BL21 (DE3) cells were transformed with these constructs. A single transformant was selected and cultured in 1 l of 2× Yeast extract and Tryptone (YT) medium (1% yeast extract, 1.6% tryptone and 0.5% NaCl) at 37°C. Cells were cultured to mid-log phase (OD600, 0.6) and protein expression was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside (final concentration, 200 μM). Cells were cultured at 22°C for a further 7 h before centrifugation at 3000g for 10 min at 4°C followed by washing with water. Purification was performed by Ni2+-NTA Superflow (Qiagen) chromatography according to the manufacturer’s protocol. The wet cells (∼3.5 g) were lysed by ultrasonication on ice in 15 ml of buffer A [20 mM, pH 9.0, Tris–HCl, 1.2 M NaCl, 2 mM imidazole, 10% glycerol and 10 mM phenylmethanesulfonyl fluoride]. The lysates were centrifuged at 40 000g for 60 min to remove the debris and insoluble fractions. The supernatant was applied to 2 ml of Ni2+-NTA resin mixed gently for 30 min at 4°C and then washed with 20 ml of buffer A and 20 ml of buffer B (20 mM, pH 9.0, Tris–HCl, 1.2 M NaCl, 25 mM imidazole and 10% glycerol) to remove nonspecific binding proteins. Binding proteins were eluted in 10 ml of buffer C (20 mM, pH 9.0, Tris–HCl, 1.2 M NaCl, 250 mM imidazole and 10% glycerol), and the eluted fractions were concentrated and dialyzed with buffer D (20 mM, pH 8.5, Tris–HCl, 3 M KCl, 0.1 M MgAc2 and 5 mM β-mercaptoethanol) (33) using Amicon Ultra-15 filters (Millipore; cutoff 30 kDa). The resulting protein solution was concentrated to ∼300 μl, mixed with glycerol to a final concentration of 40% (v/v) and stored at −20°C. Protein concentration was determined by active site titration (34).
Preparation of tRNALeu transcript and [32P] labeling at 3′ terminus of tRNA
The DNA sequence of the T7 promoter and the NmtRNALeu(GAG) gene was obtained by ligating three chemically synthetized DNA fragments for each strand, which were then ligated into pTrc99b plasmid (pre-cleaved with EcoRI/PstI) to construct pTrc99b-T7-NmtRNALeu(GAG). The forward primer (5′ ctaatacgactcactatagcgt 3′) and reverse primer (5′ tggtgcgtgggaccggattc 3′) were used to amplify the template for in vitro transcription by Pfu polymerase (T7 promoter underlined). The PCR product was extracted by phenol/chloroform and precipitated in three volumes of ethanol and 0.3 M Sodium acetate (NaAc), pH 5.2. The in vitro transcription of NmtRNALeu(GAG) was carried out in a reaction mixture containing 40 mM Tris–HCl, pH 8.0, 22 mM MgCl2, 1 mM spermidine, 5 mM dithiothreitol (DTT), 0.5% Triton X-100, 60 ng/µl tDNA template, 5 mM NTP (each), 1 U/μl ribonuclease inhibitor, 20 mM GMP, 500 U/µl T7 RNA polymerase and 1 U/ml pyrophosphatase for 3 h. Subsequently, 5 U/ml DNase I was added to digest the tDNA template for 1 h. The transcript was separated by 8 M urea −15% PAGE gel electrophoresis. The tRNA was excised from the gel, eluted with 0.5 M NaAc, pH 5.2, at room temperature three times, precipitated with three volumes of ethanol at −20°C and dissolved in 5 mM MgCl2 after centrifugation (15 000g, 30 min, 4°C) and drying. The tRNA was annealed at 85°C for 10 min and cooled naturally in the bath of 1 l water to room temperature for correct folding. NmtRNALeu(CAA), PhtRNALeu(GAG) and the corresponding mutants were prepared in the same way.
NmtRNALeu(GAG) was labeled with [α-32P]ATP as reported previously (35). In detail, 750 pmol tRNA was added in a 50 µl of reaction mixture containing 60 mM Tris–HCl, pH 8.0, 12 mM MgCl2, 1 mM DTT, 50 µM Na4PPi, 15 µM ATP, 0.666 µM [α-32P]ATP and 10 µM E. coli CCA-adding enzyme at 37°C for 5 min. Subsequently, 0.1 U pyrophosphatase was added for a further 5 min. The solution was extracted twice using phenol/chloroform and then precipitated in three volumes of ethanol. The ratio of [32P]-labeled tRNA was determined by liquid scintillation counting of the sample washed with and without 5% trichloroacetic acid.
Aminoacylation, misacylation and deacylation
The aminoacylation assays of NmLeuRSs were carried out in a reaction mixture containing 20 mM Tris–HCl, pH 9.0, 3.5 M KCl, 30 mM MgCl2, 1 mM DTT, 4 mM ATP, 20 µM [3H]Leu (11 Ci/mM), various tRNAs and NmLeuRS1 (50–500 nM) or NmLeuRS2 (2 µM) unless otherwise stated at 40°C. Misacylation was performed under similar conditions with 8.80 μM NmtRNALeu containing 3.67 nM [32P]NmtRNALeu, 50 mM Nva or 170 mM ABA or 100 mM Ile or 100 mM Met, and 2 µM NmLeuRSs or their mutants. The misacylated Nva-[32P]NmtRNALeu was obtained by using PhLeuRS1-D332A. Deacylation of Nva-[32P]NmtRNALeu was carried out under similar conditions except pH 7.5 with 500 nM NmLeuRSs or mutants were used.
To determine the aminoacylation level of [32P]NmtRNALeu, the misacylation and deacylation samples were ethanol precipitated, digested by nuclease S1 and then thin-layer chromatography (TLC) was performed to separate Nva-[32P] adenosine monophosphate (AMP) (from Nva-[32P]NmtRNALeu), [32P]AMP (from [32P]NmtRNALeu) and [32P]ATP in 0.1 M NH4Ac and 5% acetic acid. The plates were visualized by autoradiography, and the data were analyzed using MultiGauge Version 3.0 software (FUJIFILM).
Measurement of equilibrium dissociation constants for tRNA by filter binding assays
Nitrocellulose filter binding assays were performed to detect the formation of NmLeuRSs/[32P]-NmtRNALeu(GAG) complexes at 4°C in a 50 μl of reaction mixture containing 20 mM Tris–HCl, pH 9.0, 3 M KCl, 30 mM MgCl2, 1 mM DTT, 75 nM NmtRNALeu(GAG) [∼13 000 counts per minute (CPM)/pmol, 1 nM [32P]NmtRNALeu(GAG)] and NmLeuRSs (1–15 μM) for 30 min (36). The mixtures were then filtered through the nitrocellulose membrane (0.22 μm) [preequilibrated in washing buffer (50 mM potassium phosphate, pH 5.5, 50 mM MgCl2) for at least 10 min] and washed twice with 0.3 ml of washing buffer. The membranes then dried before quantitation by radioactivity. Data were analyzed using GraphPad Prism 5 software.
ATP-PPi exchange and AMP formation
ATP-PPi exchange assays were performed at 40°C in a reaction mixture containing 20 mM Tris–HCl, pH 9.0, 3.5 M KCl, 30 mM MgCl2, 1 mM DTT, 4 mM ATP, 2 mM [32P]tetrasodium pyrophosphate, Leu (0.5 µM–10 mM) or Nva (0.05–50 mM) or ABA (0.5–170 mM) and 1 µM NmLeuRS1 or 3 µM NmLeuRS2. Samples at specific time-points were added to 200 µl of quenching solution containing 2% activated charcoal, 3.5% HClO4 and 50 mM tetrasodium pyrophosphate.
AMP formation assays were carried out at 40°C in a reaction mixture containing 20 mM Tris–HCl, pH 9.0, 3.4 M KCl, 30 mM MgCl2, 1 mM DTT, 15 mM Nva, 4 mM [α-32P]ATP, 2 µM NmLeuRS1 or NmLeuRS1-D354A mutant, in the presence or absence of 25 µM NmtRNALeu(GAG). Samples were quenched in 200 mM NaAc (pH 5.2) and then spotted onto polyethyleneimine cellulose plates. TLC was performed as described previously.
RESULTS
Natrialba magadii encodes two chimeric LeuRSs and archaeal tRNALeus
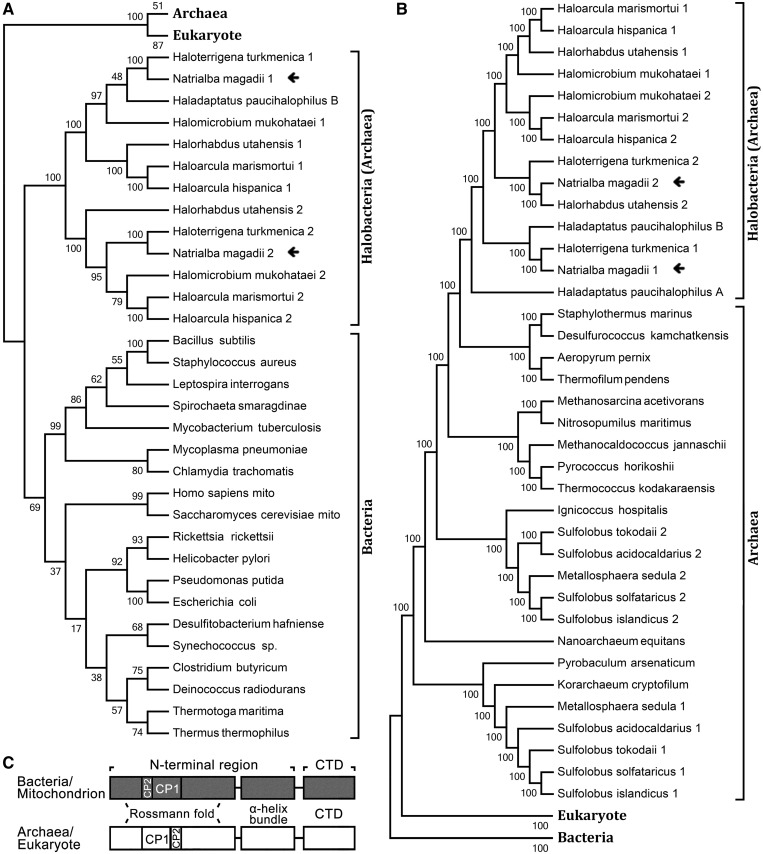
The expanded genome analysis of various species from all three domains of life significantly facilitated gene function investigations. Analysis of LeuRSs from bacteria, archaea and eukaryotes revealed that some halobacteria, such as N. magadii, H. hispanica, Halorhabdus utahensis, Haloarcula marismortui, Halomicrobium mukohataei, Haloterrigena turkmenica and Haladaptatus paucihalophilus, and some sulfolobus (e.g. Sulfolobus islandicus) harbor two copies of leuS, encoding LeuRS. Sequence alignment showed that the N-terminal region of both NmLeuRSs (Met1-Pro756 of NmLeuRS1 and Met1-Pro788 of NmLeuRS2) displayed a high level of homology with the corresponding region of bacterial LeuRSs (e.g. Met1-Pro788 of EcLeuRS) (Supplementary Figure S1A), whereas the CTDs of both NmLeuRSs (Thr757-Asp904 of NmLeuRS1 and Ala789-Gln971 of NmLeuRS2) were highly homologous to their archaeal LeuRS counterparts (e.g. Glu822-Glu967 of PhLeuRS) (Supplementary Figure S1B). This observation was further supported by phylogenetic analysis of either the N-terminal regions or CTDs of LeuRSs from the three domains of life (Figure 1). In particular, the homology or identity of two LeuRSs in a single species was not high. For instance, the protein products NmLeuRS1 and NmLeuRS2 displayed 46.6% identity and 56.9% homology. Consistently, NmLeuRS1 and NmLeuRS2 were in separate branches of the phylogenetic tree, indicating the possibility of horizontal gene transfer, or gene duplication and subsequent divergence.
Figure 1.
Phylogenetic analyses of LeuRSs from different species. (A) Phylogenetic analyses of the N-terminal regions (including Rossmann-fold, CP1, CP2 and α-helix bundle domains) of various LeuRSs. (B) Phylogenetic analyses of the C-terminal domains (CTDs) of various LeuRSs. (C) Domain scheme of LeuRSs from bacteria/mitochondrion and archaea/eukaryote, respectively. N-terminal regions included Rossmann-fold, CP1, CP2 and α-helix bundle domains.
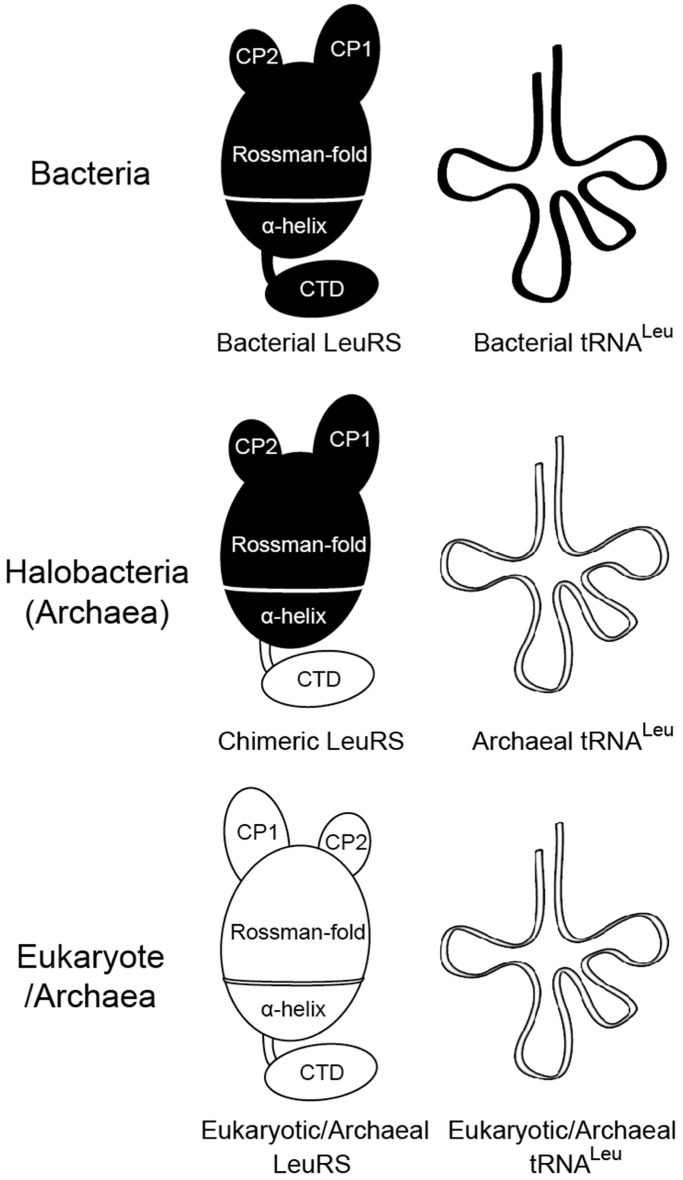
On the other hand, the N. magadii genome has five tRNALeu genes, including tRNALeu(GAG), tRNALeu(CAG), tRNALeu(UAG), tRNALeu(CAA) and tRNALeu(UAA) (37). Sequence analysis of tRNALeus revealed that these tRNAs were all archaeal type, with absolutely conserved A47c and G47d bases in the long variable loop (Supplementary Figure S2). Above all, in halophiles such as N. magadii, two LeuRSs and tRNALeu exhibit chimeric and archaeal features, respectively (Figure 2). To our knowledge, this is the first description of a naturally occurring chimeric LeuRS.
Figure 2.
Schema showing the different interaction modes of LeuRS/tRNALeu in bacteria, halobacteria (archaea) and eukaryote/archaea. The models of LeuRS and tRNALeu in black and white represent bacterial and archaeal/eukaryotic types, respectively. CP, connective peptide domains; CTD, C-terminal domain.
NmLeuRS activity requires high KCl and high pH
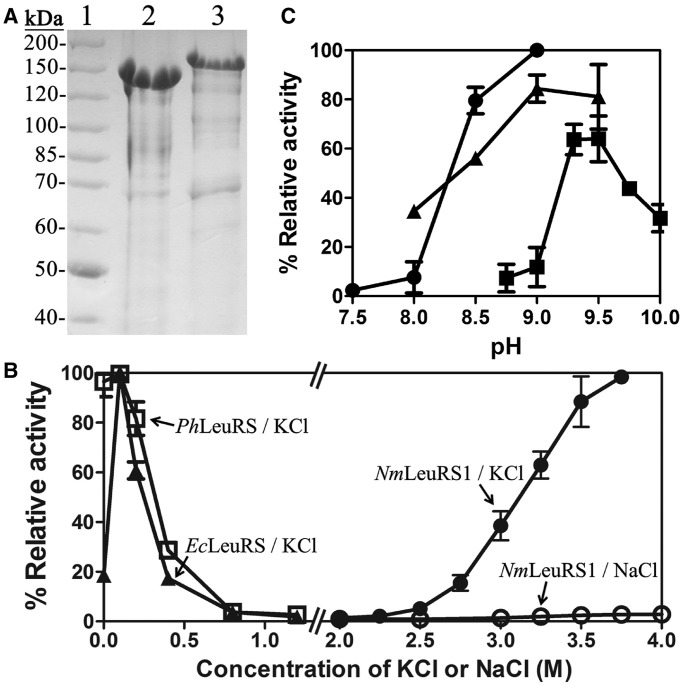
To characterize the function of both NmLeuRSs, the genes encoding two NmLeuRSs were overexpressed in E. coli. Both NmLeuRSs were purified in Tris–HCl buffer, pH 9.0, by Ni2+-NTA affinity chromatography (see ‘Materials and Methods’ section). In accordance with previous reports (38,39), NmLeuRS1 (102.2 kDa) and NmLeuRS2 (107.6 kDa) migrated slowly in 8% SDS-PAGE gel (Figure 3A) because of the presence of more negative charges with more acidic (Asp, Glu) and less basic (Lys, Arg) amino acid residues in halophilic proteins compared with their mesophilic homologs.
Figure 3.
Purification of NmLeuRSs and optimization of aminoacylation conditions. (A) A total of 8% SDS-PAGE analysis of the purified NmLeuRSs from E. coli. Lanes: 1, 2 and 3 are molecular markers (Thermo Scientific, #26614), NmLeuRS1 and NmLeuRS2, respectively. (B) Relative aminoacylation activity of NmLeuRS1, EcLeuRS and PhLeuRS under different KCl or NaCl concentrations. The activities of EcLeuRS in 0.1 M KCl, PhLeuRS in 0 M KCl and NmLeuRS1 in 3.75 M KCl were defined as 100%. (C) Relative aminoacylation activities of NmLeuRS1 under different pH conditions in Tris–HCl (black circle), Bis-Tris-propane-HCl (black up-pointing triangle) and CHES-KOH (black square). The activity of NmLeuRS1 in Tris–HCl, pH 9.0, was defined as 100%.
To investigate the aminoacylation reaction, NmLeuRS1 was used initially to determine an appropriate reaction system, followed by investigation of NmLeuRS2. As a halophilic protein, NmLeuRS1 showed a linear increase in aminoacylation activity with increasing KCl concentration above the threshold of 2.5 M. It displayed the highest activity in the presence of 3.75 M KCl (higher KCl concentrations were not evaluated) (Figure 3B). To facilitate preparation of the reaction system, 3.5 M KCl was chosen for use in further assays. Surprisingly, no activity of NmLeuRS1 was detected in NaCl solutions, even at concentrations as high as 4 M (Figure 3B). A similar phenomenon was reported for H. volcanii DNA ligase, in that K+ mediated stabilization and modulation of the enzyme activity, whereas Na+ did not (40). In contrast, the activity of mesophilic homologs such as EcLeuRS and PhLeuRS was obviously different from that of NmLeuRS1. Although the relative activity of EcLeuRS increased from 20 to 100% after the addition of 0.1 M KCl, that lost in the presence of >0.8 M KCl (Figure 3B). The activity of PhLeuRS was totally inversely correlated with the concentration of KCl, in >0.8 M KCl solution PhLeuRS had no activity (Figure 3B). Similar to NmLeuRS1, the activity of NmLeuRS2 increased significantly from 2.0–3.1 M KCl; no obvious further stimulation of activity occurred at higher KCl concentrations (Supplementary Figure S3A).
The pH optimization for the amino acylation activities of NmLeuRS1 was then studied. NmLeuRS1 exhibited the highest activity at pH 9.0 in Tris–HCl or Bis-Tris-propane-HCl buffer or at pH 9.5 in CHES-KOH buffer (Figure 3C). The activity decreased obviously at pH 8.0 or below. Similar results were obtained with NmLeuRS2 (Supplementary Figure S3B). High pH is optimal for the activity of many extracellular proteins of alkaliphiles (41,42). However, an almost neutral cytoplasmic pH is maintained by Na+/H+ antiporters (43,44) or cell wall macromolecules, which serve as a specific barrier to the flux of relevant ions (45). Our results suggest that the cytoplasmic proteins in N. magadii perform their physiological function in an alkaline solution through another mechanism. The genes necessary for survival in alkaline conditions have not been identified in the genome of N. magadii (46). The in vitro characteristics of recombinant NmLeuRSs may differ in the in vivo situation.
Distinct aminoacylation activities of the NmLeuRSs
NmLeuRSs have similar domain architecture, display 46% identity and coexist in a single cell compartment. It is possible that either one or both NmLeuRSs play an essential aminoacylation role in protein biosynthesis. The activities of the two NmLeuRSs were initially compared in ATP-PPi exchange assays. NmLeuRS1 displayed a much higher amino acid activation activity than NmLeuRS2, the Km value of NmLeuRS2 was nearly 105-fold greater than that of NmLeuRS1 (Km: 891 ± 2 μM versus 8.43 ± 1.30 μM), and the activation rate of NmLeuRS1 was 58-fold greater than that of NmLeuRS2 [kcat: (5.63 ± 0.12) × 10−1 s−1 versus (9.61 ± 0.47) × 10−3 s−1] (Table 1).
Table 1.
Leucine activation kinetics constants of NmLeuRSs at 40°Ca
| Km (μM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Activity ratio | |
|---|---|---|---|---|
| NmLeuRS1 | 8.43 ± 1.30 | (5.63 ± 0.12) × 10−1 | 66.8 | 1b |
| NmLeuRS2 | 891 ± 2 | (9.61 ± 0.47) × 10−3 | 0.0108 | 1/6185 |
aThe results are the average of three independent repeats with standard deviations indicated.
bThe activity of NmLeuRS1 for Leu is defined as 1.
The aminoacylation assays showed that both NmLeuRSs were able to charge NmtRNALeu(GAG), NmtRNALeu(CAA) and PhtRNALeu(GAG) (the Leu accepting activity of the aforementioned tRNALeu transcripts was 1350, 1468 and 1225 pmol/A260), although the catalytic efficiencies (kcat/Km) of NmLeuRS2 for different tRNALeus were only 1/130 to 1/40 of those of NmLeuRS1 (Table 2). The data showed a distinct difference in the catalytic velocity (kcat) but not the Km value for tRNALeu during aminoacylation, which may be due to the much lower amino acid activation activity of NmLeuRS2. To further define the affinity between NmLeuRSs and NmtRNALeu(GAG), filter binding assays were performed as reported previously (36). The equilibrium dissociation constants (kd) of NmLeuRS1 and NmLeuRS2 for NmtRNALeu(GAG) were 6.36 ± 1.17 μM and 3.36 ± 0.89 μM, respectively (Table 3). These results showed that NmLeuRS2 had obviously decreased aminoacylation activity compared with NmLeuRS1, whereas the affinity for cognate NmtRNALeu was retained.
Table 2.
Aminoacylation kinetics of NmLeuRSs at 40°Ca
| tRNALeu | LeuRS | Km (μM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Activity ratio |
|---|---|---|---|---|---|
| NmtRNALeu(GAG) | NmLeuRS1 | 7.73 ± 0.22 | (6.40 ± 0.19) × 10−2 | 8.28 | 1b |
| NmLeuRS2 | 3.35 ± 0.21 | (6.68 ± 0.22) × 10−4 | 0.199 | 1/42 | |
| NmtRNALeu(CAA) | NmLeuRS1 | 11.01 ± 1.36 | (4.64 ± 0.34) × 10−2 | 4.21 | 1/2 |
| NmLeuRS2 | 13.13 ± 1.57 | (4.18 ± 0.50) × 10−4 | 0.0318 | 1/260 | |
| PhtRNALeu(GAG) | NmLeuRS1 | 12.51 ± 2.16 | (4.18 ± 0.60) × 10−2 | 3.34 | 1/2.5 |
| NmLeuRS2 | 16.43 ± 0.62 | (1.19 ± 0.08) × 10−3 | 0.0724 | 1/114 |
aThe results are the average of three independent repeats with standard deviations indicated.
bThe activity of NmLeuRS1 for NmtRNALeu(GAG) is defined as 1.
Table 3.
kd of NmtRNALeu(GAG) in filter binding reaction at 4°Ca
| LeuRS | kd (μM) |
|---|---|
| NmLeuRS1 | 6.36 ± 1.17 |
| NmLeuRS2 | 3.36 ± 0.89 |
aThe results are the average of three independent repeats with standard deviations indicated.
NmLeuRS1 showed the highest catalytic activity for NmtRNALeu(GAG), which was used predominantly (4.93%) among all NmtRNALeu isoacceptors. The catalytic activity for NmtRNALeu(CAA) was only half of that for NmtRNALeu(GAG). In addition, the catalytic activity for the heterologous PhtRNALeu(GAG) was slightly lower than that for the homologous NmtRNALeu (Table 2), implying that NmLeuRSs, despite the presence of the bacterial aminoacylation and editing domains, effectively distinguished archaeal tRNALeu.
To further explore the aminoacylation activities of other coexisted LeuRSs present in halobacteria, we then purified the protein of two leuS genes of H. hispanica after being overexpressed in E. coli [designated as HhLeuRS1 (accession No. YP_004794828.1, 895-aa) and HhLeuRS2 (accession No. YP_004797622.1, 893-aa)] (Supplementary Figure S4A) (47). Similar results with NmLeuRSs were obtained, that HhLeuRS2 had only little amino acid activation and aminoacylation activities, whereas HhLeuRS1 retained the normal activities (Supplementary Figure S4B and C). These results suggested that the coexistence of an active LeuRS and an inactive one might be a widespread phenomenon in halobacteria.
NmLeuRSs distinguish NmtRNALeu in the archaeal mode
With both bacterial N-terminal regions and an archaeal C-terminal tRNA binding domain, the mechanism by which NmLeuRSs distinguish NmtRNALeu presents an interesting question. NmLeuRS1 was chosen as a model for this investigation because it exhibited higher activity than NmLeuRS2. Previous investigation of the archaeal PhLeuRS/tRNALeu system showed that A47c and G47d in the long variable loop of PhtRNALeu are critical elements, which are recognized by Asp845, Ile849 and the last few conserved residues (Pro962, Ile964, Ile966 and Glu967) of PhLeuRS (26).
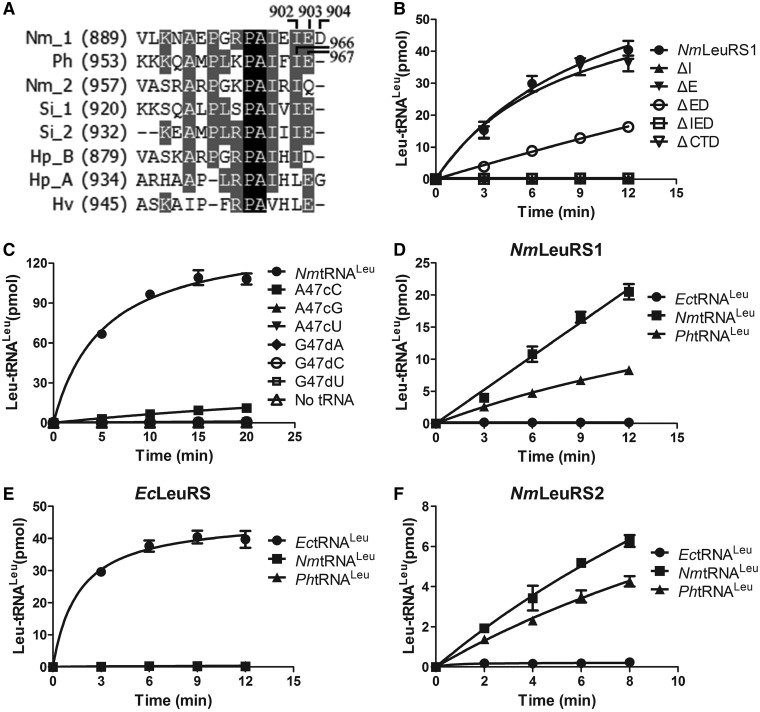
We first studied the recognition mode based on analysis of LeuRS. Owing to the high homology between the CTDs of NmLeuRS1 and PhLeuRS, we prepared several deletion mutants at the C-terminus of NmLeuRS1, including NmLeuRS1-ΔE (Glu903 deletion), NmLeuRS1-ΔED (Glu903 and Asp904 deletions), NmLeuRS1-ΔI (Ile902 deletion) and NmLeuRS1-ΔIED (Ile902, Glu903 and Asp904 deletions) (Figure 4A). NmLeuRS1-ΔE showed similar activity compared with the wild-type enzyme, whereas NmLeuRS1-ΔED exhibited an ∼60% loss, and NmLeuRS1-ΔIED and NmLeuRS1-ΔI lost their activity completely (Figure 4B), implicating the conserved Ile902 of NmLeuRS1 as the most important residue in recognition. These data suggested that the mode of NmtRNALeus distinguished by NmLeuRS1 was similar to that of PhLeuRS, both depending on the conserved Ile residue (Ile902 and Ile966 for NmLeuRS1 and PhLeuRS, respectively) (Figure 4A) (26). Moreover, complete deletion of the CTD (NmLeuRS1-ΔCTD) led to the total loss of aminoacylation activity (Figure 4B), indicating the importance of the archaeal CTD in the NmLeuRS1 aminoacylation reaction.
Figure 4.
NmLeuRSs discriminate NmtRNALeu in archaeal mode. (A) Sequence alignment of the C-terminus of different LeuRSs. Nm_1 and Nm_2, N. magadii LeuRS1 and LeuRS2; Ph, P. horikoshii; Si_1 and Si_2, S. islandicus LeuRS [LeuRS1 (accession No. YP_002829648.1) and LeuRS2 (accession No. YP_002829589.1)]; Hp_B and Hp_A, H. paucihalophilus LeuRS [bacterial type (accession No. WP_007982262.1) and archaeal type (accession No. WP_007982263.1)]; Hv, H. volcanii. (B) The aminoacylation activities of 500 nM NmLeuRS1 (black circle), 500 nM NmLeuRS1-ΔI (black up-pointing triangle), 500 nM NmLeuRS1-ΔE (black down-pointing triangle), 500 nM NmLeuRS1-ΔED (white circle), 500 nM NmLeuRS1-ΔIED (white square) and 500 nM NmLeuRS1-ΔCTD (white down-pointing triangle) for 5.6 μM NmtRNALeu(GAG). (C) The aminoacylation activities of 500 nM NmLeuRS1 for 12 μM (0.3 μg/μl) NmtRNALeu(GAG) (black circle), 0.3 μg/μl NmtRNALeu-A47cC (black square), 0.3 μg/μl NmtRNALeu-A47cG (black up-pointing triangle), 0.3 μg/μl NmtRNALeu-A47cU (black down-pointing triangle), 0.3 μg/μl NmtRNALeu-G47dA (black diamond), 0.3 μg/μl NmtRNALeu-G47dC (white circle) and 0.3 μg/μl NmtRNALeu-G47dU (white square); the cross-species aminoacylation activities of 200 nM NmLeuRS1 (D), 10 nM EcLeuRS (E) and 3 μM NmLeuRS2 (F) for 4 μM EctRNALeu (black circle), 4 μM NmtRNALeu (black square) and 4 μM PhtRNALeu (black up-pointing triangle), respectively.
We then explored the recognition mode based on analysis of tRNALeu. Sequence alignment of tRNALeus showed that A47c and G47d at the tip of the long variable loop were absolutely conserved in archaea (Supplementary Figure S2). These residues have been identified as the sequence-specific determinants in archaeal H. volcanii tRNALeu (HvtRNALeu) and PhtRNALeu (25,26). Mutations of A47c and G47d to the other three nucleotides in NmtRNALeu(GAG) were performed to obtain NmtRNALeu(GAG)-A47cC, -A47cG, -A47cU, -G47dA, -G47dC and -G47dU, respectively. None of the mutants were efficiently aminoacylated by NmLeuRS1 except NmtRNALeu(GAG)-A47cC, which retained 7% of the catalytic activity of the wild-type tRNALeu (Figure 4C, Supplementary Table S1). Therefore, like archaeal PhtRNALeu and HvtRNALeu, the A47c and G47d bases located in the tip of the long variable loop were of great importance in the recognition process.
These results showed that NmLeuRS1 distinguished NmtRNALeu via a similar mechanism to that used by archaeal PhLeuRS with the last few residues of enzyme (especially conserved Ile902) and the conserved A47c and G47d residues of tRNALeu being crucial elements for recognition. As NmLeuRS1 also possesses bacterial aminoacylation and editing domains, the question of whether these two domains conferred the ability to recognize bacterial tRNALeus on NmLeuRS1 remained unresolved. To address this issue, the aminoacylation activity of NmLeuRS1 for bacterial EctRNALeu (the Leu accepting activity was 1600 pmol/A260) and archaeal PhtRNALeu was determined. NmLeuRS1 was able to charge archaeal PhtRNALeu but not bacterial EctRNALeu (Table 2, Figure 4D), implying that the bacterial aminoacylation and editing domains contributed little to tRNA species specificity. On the other hand, EcLeuRS was only able to charge its cognate EctRNALeu but not NmtRNALeu and PhtRNALeu (Figure 4E). Cross-activation of these tRNALeus by NmLeuRS2 was also evaluated, and similar results were obtained (Figure 4F), suggesting that both NmLeuRSs distinguished tRNALeus in the archaeal mode. In addition, we replaced the CTD (Thr759-Asp904) of NmLeuRS1 with its EcLeuRS counterpart (Asp791-Gly860) to generate NmLeuRS1-EcCTD to mimic a complete bacterial LeuRS. However, NmLeuRS1-EcCTD failed to aminoacylate EctRNALeu, PhtRNALeu and NmtRNALeu (Supplementary Figure S5), indicating that the archaeal CTD plays a key role in archaeal tRNALeu recognition and that fusion of a bacterial CTD with the basis of bacterial region of NmLeuRS1 failed to reinstate bacterial tRNALeu aminoacylation capacity.
Taken together, these lines of evidence suggested that NmLeuRS1 distinguished NmtRNALeu in the archaeal mode and indicated that the archaeal CTD, but not the bacterial aminoacylation and editing domains of NmLeuRS1, contributed its archaeal tRNA recognition capacity.
The N-terminal region of NmLeuRS1 is of bacterial type
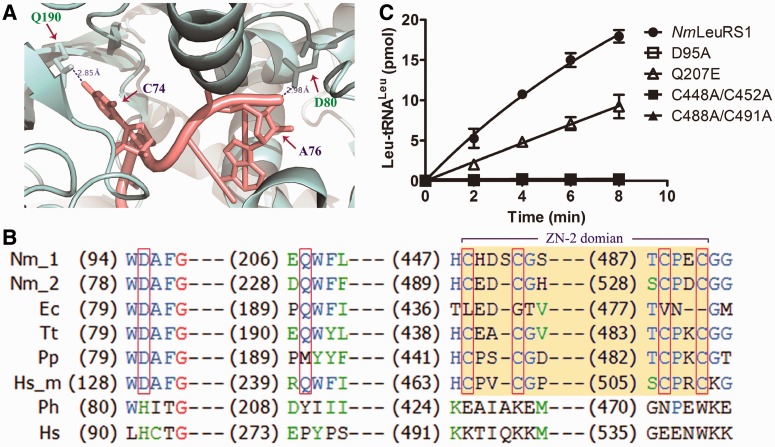
To explore whether the N-terminal region of NmLeuRSs was of bacterial type, we constructed several mutations in amino acid residues of NmLeuRS1, only conserved in bacterial but not archaeal/eukaryotic LeuRSs. Asp95 and Gln207 of NmLeuRS1 were equivalent to Asp80 and Gln190 in EcLeuRS, which, respectively, interacted with A76 and C74 of EctRNALeu during aminoacylation reaction, as showed in its 3D structure (PDB: 4AQ7) (Figure 5A) (21), and conserved in only bacterial but not archaeal/eukaryotic LeuRSs (Figure 5B), suggesting they might play a key role in the aminoacylation activity of bacterial LeuRS. We obtained NmLeuRS1-D95A and -Q207E mutants, which would disturb the interaction of LeuRS and tRNALeu.
Figure 5.
The N-terminal region of NmLeuRS1 is of bacterial type. (A) The structure of EcLeuRS complex with tRNALeu in aminoacylation conformation [PDB code 4AQ7, (21)]. EcLeuRS is gray colored and the tRNA molecule is in salmon. The residues D80, Q190 in EcLeuRS (equivalent to D95, Q207 in NmLeuRS1) and the respectively interacted nucleotides A76, C74 are indicated. (B) Sequence alignment of the N-terminal region of different LeuRSs. The red boxes and the yellow block indicate the specific residues and the ZN-2 domain investigated in this study. Nm_1 and Nm_2, N. magadii LeuRS1 and LeuRS2; Ec, E. coli; Tt, Thermus thermophilus; Pp, Pseudomonas putida; Hs_m, H. sapiens mitochondrial LeuRS; Ph, P. horikoshii; Hs, H. sapiens. (C) The aminoacylation activities of 200 nM NmLeuRS1 (black circle), 200 nM NmLeuRS1-D95A (white square), 200 nM NmLeuRS1-Q207E (white up-pointing triangle), 200 nM NmLeuRS1-C448A/C452A (black square) and 200 nM NmLeuRS1-C488A/C491A (black up-pointing triangle) for 4 μM NmtRNALeu (GAG).
The results showed that the mutant NmLeuRS1-D95A abolished all the aminoacylation activity and NmLeuRS1-Q207E only retained half of the aminoacylation activity compared with NmLeuRS1 (Figure 5C), suggesting the bacterial type of the N-terminal region of NmLeuRS1.
In addition, zinc binding domains (ZN domain) were conserved in many class I tRNA synthetase. However, besides the universal ZN-1 domain, only bacterial/mitochondrial but not archaeal/eukaryotic LeuRSs contained the ZN-2 domain (48). It has shown that mutations of conserved Cys residues in ZN domain obviously reduced the aaRS activities and the growth of cells (49,50). Sequence analysis showed the ZN-2 domain existed in both NmLeuRSs as most bacterial LeuRSs (Figure 5B). Therefore, we performed the mutations of Cys to Ala at ZN-2 domain of NmLeuRS1 to produce double mutants NmLeuRS1-C448A/C452A and -C488A/C491A. The results showed both mutants totally lost their aminoacylation activities (Figure 5C), indicating the crucial role of bacterial LeuRS-specific ZN-2 domain and further supporting our suggestion that the N-terminal region of NmLeuRS1 was of bacterial type.
NmLeuRS1 misactivates several non-cognate amino acids
LeuRSs are characterized by the capacity to activate a wide range of amino acids that are structurally similar to cognate Leu (30,35). We determined the amino acid activation activity of NmLeuRS1 toward non-cognate Ile and Met as well as the non-proteinogenic amino acids Nva and ABA. Nva and ABA were misactivated by NmLeuRS1, whereas the kinetic constants of Ile and Met could not be determined because kinetic analysis at high concentrations of Ile and Met was not realized. The kcat of NmLeuRS1 for Nva (0.364 ± 0.008 s−1) and ABA (0.518 ± 0.028 s−1) activation were ∼60 and 90% of that for cognate Leu (0.563 ± 0.012 s−1), although the Km values for Nva (955 ± 22 μM) and ABA (37 820 ± 1630 μM) were much higher compared with that of cognate Leu (8.43 ± 1.3 μM) (Table 4). For ABA, the discrimination factor (DF) was 1/4912, which was lower than the mistranslation frequency (1/3000) of protein biosynthesis (6). In contrast, NmLeuRS1 has to clear the misactivated Nva (DF: 1/175) by its editing activity. Because of the extremely low amino acid activation activity of NmLeuRS2 even toward cognate Leu, accurate evaluation of the kinetics of misactivation for non-cognate amino acids was not achieved.
Table 4.
Amino acid activation kinetics of NmLeuRS1 for Leu, Nva and ABAa
| Amino acids | Km (μM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Discriminate factor (DF)b |
|---|---|---|---|---|
| Leu | 8.43 ± 1.3 | 0.563 ± 0.012 | 66.8 | 1 |
| Nva | 955 ± 22 | 0.364 ± 0.008 | 0.381 | 1/175 |
| ABA | 37 820 ± 1630 | 0.518 ± 0.028 | 0.0136 | 1/4912 |
aThe results are the average of three independent repeats with standard deviations indicated.
bDF is defined as the relative ratio of kcat/Km for non-cognate amino acids compared with that for Leu.
NmLeuRS1 exhibits obvious post-transfer editing activity to prevent generation of mischarged tRNALeu
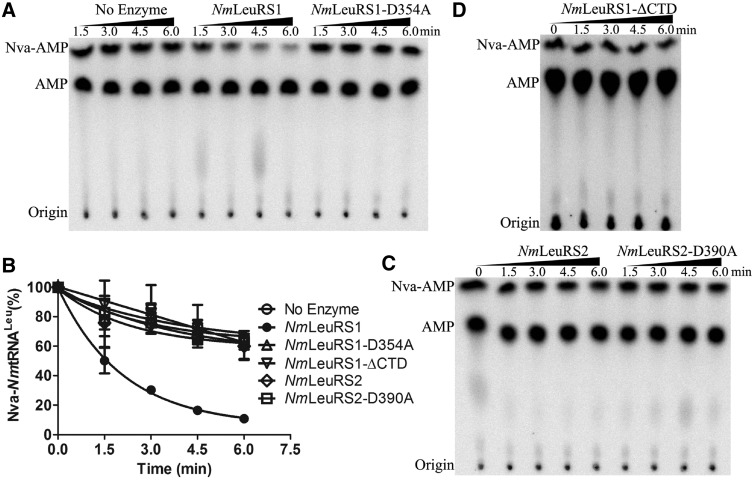
NmLeuRS1 has the bacterial Rossmann fold aminoacylation and editing domains, which means that the orientation and the insertion point of the CP1 editing domain into the Rossmann fold is totally different from its archaeal counterparts, such as the CP1 domain of PhLeuRS (14). However, it has the normal archaeal CTD for tRNALeu binding. Therefore, whether the editing activity of the bacterial CP1 domain and the tRNALeu binding capacity of the archaeal CTD coordinate to mediate post-transfer editing is an interesting question. To confirm the editing activity of NmLeuRS1, the post-transfer editing essential Asp residue (Asp354 of NmLeuRS1), as revealed in various LeuRS systems (14,30,35,51,52), was mutated to Ala residue to generate NmLeuRS1-D354A. To monitor the post-transfer editing directly, misacylated Nva-[32P]NmtRNALeu was produced by PhLeuRS1-D332A (14). The hydrolysis of Nva-NmtRNALeu showed that only NmLeuRS1 was able to deacylate the misacylated product (Figure 6). In contrast, NmLeuRS1-D354A lost its editing ability (Figure 6A and B). We also analyzed the editing activity of NmLeuRS2 in a similar manner; however, both wild-type NmLeuRS2 and the mutant of the conserved Asp390 (NmLeuRS2-D390A) exhibited no editing capacity (Figure 6B and C), suggesting the post-transfer editing of NmLeuRS2 was lost or too weak to be detected in deacylation assays.
Figure 6.
NmLeuRS1 but not NmLeuRS2 deacylates Nva-NmtRNALeu. (A) Representative graph showing the post-transfer editing activity of 500 nM NmLeuRS1 and 500 nM NmLeuRS1-D354A based on TLC assay. (B) Quantification of the deacylation activities of NmLeuRS (black circle), NmLeuRS1-D354A (white up-pointing triangle), NmLeuRS1-ΔCTD (white down-pointing triangle), NmLeuRS2 (white diamond) and NmLeuRS2-D390A (white square) in A, C and D. Representative graphs showing the post-transfer editing activity of 500 nM NmLeuRS2 and 500 nM NmLeuRS2-D390A (C), 500 nM NmLeuRS1-ΔCTD (D) based on TLC assay.
Whether HhLeuRSs exhibited similar results with NmLeuRSs in deacylation activities was then explored. Our data revealed that although both HhLeuRSs could deacylate mischarged Nva-HhtRNALeu, but HhLeuRS2 showed weaker activity than HhLeuRS1. These further confirmed the phenomena that an active LeuRS and an inactive (or slightly active) one coexisted in halobacteria (Supplementary Figure S4D and E).
Previous work also showed that the editing activity of PhLeuRS was not altered by deletion of the CTD (14). Therefore, to investigate the editing function of the archaeal CTD of NmLeuRS1, the NmLeuRS1-ΔCTD mutant was generated and shown to exhibit a complete loss of deacylation activity (Figure 6B and D). This suggested that the archaeal CTD of NmLeuRS1, unlike that of PhLeuRS, coordinated with its CP1 domain to play a key role in post-transfer editing.
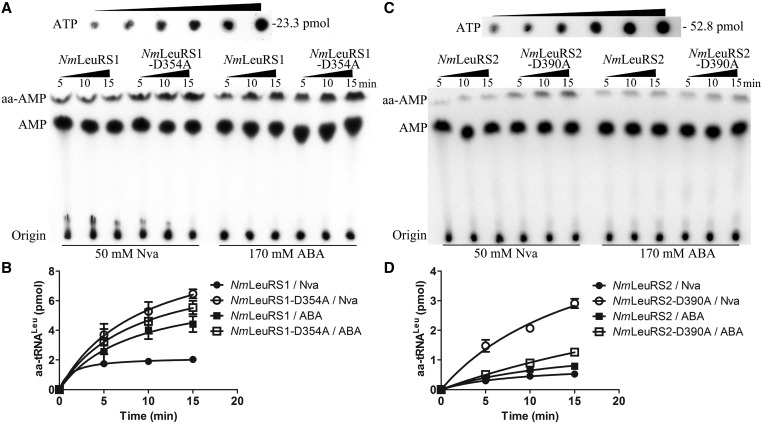
We then determined the misacylation activity of NmLeuRSs. The result showed no Ile and little Met could be misloaded to NmtRNALeu by NmLeuRSs (Supplementary Figure S6). In addition, NmLeuRS1-D354A produced significantly more (∼3-fold) Nva-NmtRNALeu and 30% more ABA-NmtRNALeu compared with wild-type NmLeuRS1 (Figure 7A and B), suggesting the essential editing role of NmLeuRS1 in preventing the synthesis of mischarged tRNALeu. However, the amount of generated Nva-NmtRNALeu and ABA-NmtRNALeu was comparable with wild-type NmLeuRS2 and mutant NmLeuRS2-D390A (Figure 7C and D), which is consistent with the observation that post-transfer editing was lost or significantly impaired in both enzymes (Figure 6B and C). In addition, it is notable that wild-type NmLeuRSs were able to synthesize trace amount of Nva-tRNALeu and an obvious amount of ABA-tRNALeu. This is also the case for Candida albicans LeuRS (CaLeuRS) (30), suggesting that editing capacity is insufficient for proofreading non-cognate amino acids at saturated concentrations. It can be speculated that the paradox between ABA misaminoacylation and charging accuracy can be accounted for by fine discrimination against ABA at the aminoacylation active site (1/4912) as proposed for CaLeuRS (30).
Figure 7.
NmLeuRSs misacylate NmtRNALeu with Nva and ABA. (A) Representative image showing the misacylation activities of 500 nM NmLeuRS1 and 500 nM NmLeuRS1-D354A for Nva and ABA based on TLC assay. A 2-fold diluted [α-32P]ATP for quantification (initial, 52.8 pmol) is also shown. (B) Quantification of the misacylation activities of NmLeuRS1 [for Nva (black circle), for ABA (black square)] and NmLeuRS1-D354A [for Nva (white circle), for ABA (white square)] in A. (C) Representative image showing the misacylation activities of 2 μM NmLeuRS2 and 2 μM NmLeuRS2-D390A for Nva and ABA based on TLC assay. (D) Quantification of the misacylation activities of NmLeuRS2 [for Nva (black circle), for ABA (black square)] and NmLeuRS2-D390A [for Nva (black square), for ABA (white square)] in C.
Editing characteristics of NmLeuRS1
On the basis of the efficient editing activity of NmLeuRS1, we further investigated the editing characteristics of NmLeuRS1 in TLC-based AMP formation assays on the basis that a single AMP molecule is released by either the hydrolysis of misactivated aa-AMP by pre-transfer editing or in the cycle of misacylation and post-transfer editing; therefore, measurement of the released AMP in the TLC assay can be used to quantify the editing capacity (53).
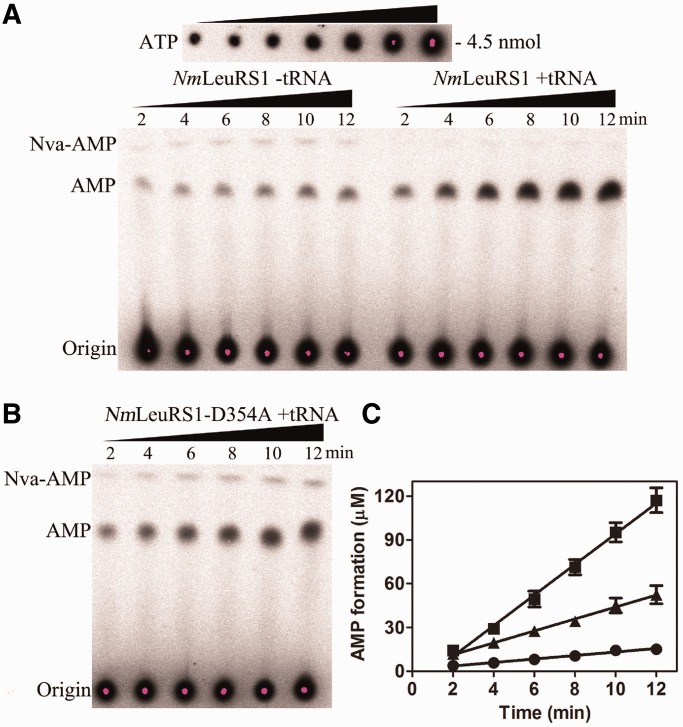
In the presence of NmtRNALeu, the observed rate constant (kobs) of AMP formation by NmLeuRS1 was (8.74 ± 0.36) × 10−2 s−1 (Figure 8A, Table 5), which represented the global editing activity. In the absence of NmtRNALeu, the AMP formation rate was (1.00 ± 0.13) × 10−2 s−1, ∼11.4% of that in the presence of NmtRNALeu (1.00/8.74). This result suggested that tRNA-independent pre-transfer editing contributed little to the total editing activity. Theoretically, the enhanced AMP formation activity, as a result of tRNA addition, may be derived from tRNA-dependent pre-transfer editing and/or post-transfer editing. To distinguish these two pathways, we analyzed AMP formation using NmLeuRS1-D354A. The kobs of NmLeuRS1-D354A in the presence of tRNA was (3.40 ± 0.19) × 10−2 s−1 (Figure 8B, Table 5), indicating that tRNA-dependent pre-transfer editing contributed ∼27.5% to the total editing activity [(3.40 − 1.00)/8.74]. Taken together, these results demonstrate that post-transfer editing contributed more than half (61.1%) of the Nva editing activity [(8.74 − 3.40)/8.74].
Figure 8.
The editing properties for Nva by NmLeuRS1. (A) A representative image showing the formation of [32P]AMP by 2 μM NmLeuRS1 in the absence (− tRNA) or presence (+ tRNA) of 25 μM NmtRNALeu. A 2-fold diluted [α-32P]ATP for quantification (initial concentration, 4.5 nmol) is also shown. (B) A representative image showing the formation of [32P]AMP by NmLeuRS1-D354A in the presence of NmtRNALeu. (C) Quantification of the formation of [32P]AMP by NmLeuRS1without tRNA (black circle) and with tRNA (black square) and by NmLeuRS1-D354A with tRNA (black up-pointing triangle).
Table 5.
The kobs of NmLeuRS1 and NmLeuRS1-D354A in AMP formation with Nvaa
| NmtRNALeu(GAG) | kobs (s−1) | |
|---|---|---|
| NmLeuRS1 | − | (1.00 ± 0.13) × 10−2 |
| + | (8.74 ± 0.36) × 10−2 | |
| NmLeuRS1-D354A | + | (3.40 ± 0.19) × 10−2 |
aThe results are the average of three independent repeats with standard deviations indicated.
In addition, if the editing activities failed to clear all the misproducts, Nva-AMP should be detected in the TLC assays. In the presence of NmtRNALeu, the NmLeuRS1-D354A mutant still failed to clear all the misproducts (with obvious accumulation of Nva-AMP) (Figure 8B) in contrast to NmLeuRS1 with tRNALeu (without obvious accumulation of Nva-AMP), suggesting that pre-transfer editing is not sufficient for maintaining the catalytic fidelity. Only the wild-type NmLeuRS1, which has post-transfer editing in the presence of NmtRNALeu, efficiently prevented the accumulation of Nva-AMP (Figure 8A).
DISCUSSION
Unique LeuRS models identified to date
There are several interesting examples of LeuRSs with unique features with regard to primary or tertiary structure. For example, in A. aeolicus, LeuRS is no longer a single polypeptide but rather a heterodimeric enzyme (αβ-LeuRS), although no advantage associated with this structure has been identified (34). However, both α and β subunits are of bacterial origin. In Mycoplasma mobile, LeuRS is naturally devoid of the CP1 editing domain (54). This is the first example of a truncated LeuRS, leading to an error-prone synthetase. Consequently, M. mobile exhibits genetic code ambiguity in its proteome, which might be advantageous for avoiding host immune defense (55). In addition, human mitochondrial LeuRS contains a naturally degenerated CP1 domain (56), which suggests a degenerate editing activity. However, mitochondrial LeuRS exhibited a fine discrimination capacity toward cognate Leu and non-cognate amino acids in the amino activation step. The reason for this degeneration has not been definitely identified. This work describes a unique example of a LeuRS/tRNALeu system, in which two chimeric LeuRSs coexist in a single cell compartment. Furthermore, we show that NmLeuRSs distinguish NmtRNALeu in the archaeal mode.
Coexistence of two aaRSs in a single compartment
There are sporadic examples of the existence of dual aaRS activities in a single cell compartment. In human cytoplasm, two arginyl-tRNA synthetases (ArgRSs) are derived from two translational initiation sites by a single messenger RNA (57). The short ArgRS is proposed to participate in modification at the N-terminus of proteins targeted for degradation by providing the substrate for argninyl-tRNA transferase (58). Moreover, higher eukaryotes encode two threonyl-tRNA synthetases. Sequence analysis and genetic studies have shown that the short form generates Thr-tRNAThr for human cytoplasmic protein biosynthesis, whereas the significance of the longer isoform remains to be clarified (53). The coexistence of two types of glycyl-tRNA synthetase (GlyRS, ScGlyRS1 and ScGlyRS2) has been reported in S. cerevisiae. ScGlyRS1 is responsible for both cytoplasmic and mitochondrial activities, whereas GlyRS2 is dysfunctional and not essential for growth (59). Many bacterial species, such as Helicobacter pylori, also contain two duplicated glutamyl-tRNA synthetase (GluRS, HpGluRS1 and HpGluRS2). It has been revealed that HpGluRS1 acylated only tRNAGlu; whereas HpGluRS2 was specific solely for tRNAGln (60). In addition, two types of cytoplasmic LeuRSs have been reported in Agrobacterium radiobacter K84 (61), in which a second non-essential LeuRS (AgnB2) is encoded by the pAgK84 plasmid, in addition to the essential genomic form. The activity of the genomic form can be inhibited by TM 84, which is synthetized to kill Agrobacterium tumefaciens, which causes crown gall tumors in plants by specific inhibition of LeuRS activity. AgnB2 LeuRS, which is non-sensitive to TM 84, works as a self-protective copy.
Considering the obviously high aminoacylation and editing activity of NmLeuRS1, we propose that NmLeuRS1 is responsible for Leu-tRNALeu generation. However, both the aminoacylation and editing activities of NmLeuRS2 were significantly lower than those of NmLeuRS1; therefore, NmLeuRS2 may be not essential for protein biosynthesis. Compared with NmLeuRS1 and other bacterial LeuRSs, there are several substitutions at key amino acids (such as Ser631 instead of Lys in the catalysis-essential KMSKS motif) or insertion in the Rossmann-fold amino acid activation domain in NmLeuRS2 [such as a 38-aa insertion (Glu174-Thr211), Supplementary Figure S1A]. These changes may, individually or collectively, have some direct or indirect influence on the activity of NmLeuRS2. Unfortunately, substitutions of these conserved residues with the corresponding sites in EcLeuRS (such as Ser631 to Lys631 to regenerate the KMSKS motif) or deletion of the 38-aa insertion in NmLeuRS2 failed to restore its amino acid activation and aminoacylation activities (data not shown).Therefore, the detailed mechanism underlying the loss of NmLeuRS2 activity requires further investigation.
In contrast, NmLeuRS2 retained the affinity for NmtRNALeu, indicating NmLeuRS2 mediates other functions in a tRNA-dependent manner. Many aaRS-like proteins are responsible for tRNA-dependent trans-editing activities rather than the cis-editing domains, such as human ProX for Ala-tRNAPro, AlaXp for Ser-tRNAAla and Gly-tRNAAla, ThrXp for Ser-tRNAThr and Ybak for Cys-tRNAPro (9). However, the undetectable editing activity of NmLeuRS2 may exclude such a possibility. It is also possible that the editing function of NmLeuRS2 relies on other cofactors. Other studies revealed that a lysyl-tRNA synthetase-like protein PoxA modifies elongation factor-P with (R)-β-lysine and that a SerRS-like insect mitochondrial protein is also a tRNA-binding protein without aminoacylation activity but with an essential mitochondrial function (62,63). Therefore, it can be speculated that NmLeuRS2 functions in other pathways besides tRNA aminoacylation.
Chimeric characteristics of NmLeuRSs
Most archaea, such as P. horikoshii, harbor only one archaeal LeuRS gene. However, N. magadii, H. paucihalophilus and S. islandicus are extremophilic archaea, all of which contain two leuS genes. Sequence alignment revealed that the CTD in these LeuRSs are all of the archaeal type (Supplemental Figure S1B), suggesting the same archaeal recognition mode. However, the N-terminal region (including the Rossmann-fold, the CP1 domain and the α-helix bundle domain) is different. Specifically, the N-terminal regions of both NmLeuRSs are bacterial type (Supplemental Figure S1A), but the counterparts [Met1-Pro781 in the LeuRS (accession No. YP_002829648.1), Met1-Pro792 in another LeuRS (accession No. YP_002829589.1)] of both S. islandicus LeuRSs are archaeal type. In contrast, the N-terminal region [Met1-Pro746 in the LeuRS (accession No. WP_007982262.1)] of one H. paucihalophilus LeuRS (HpLeuRS) is bacterial type, whereas the other [Met1-Pro790 in another HpLeuRS (accession No. WP_007982263.1)] is archaeal type. This phenomenon indicates the complex distribution pattern of LeuRSs in the archaeal world. The functional detail of both S. islandicus LeuRSs and HpLeuRSs requires investigation.
Based on the complicated distribution of LeuRS in these species, we propose that the ancestor of most archaeal species encoded only one archaeal LeuRS. However, an ancient horizontal gene transfer of bacterial LeuRS occurred from an organism related to the bacterial ancestor to the haloarchaea. In some cases, such as in N. magadii, gene fusion may have occurred between the original archaeal LeuRS and the transferred bacterial LeuRS, leading to the presence of a chimeric LeuRS with the N-terminal region replaced by the bacterial counterpart. This new isoform coexists with the original archaeal copy. The bacterial form of LeuRS (or the newly generated chimeric LeuRS) further underwent gene duplications (64) and/or horizontal gene transfers (65) within the haloarchaea. It is possible that this new bacterial or chimeric LeuRS provided advantages in participating in the translational machinery in those species and thus, the original archaeal LeuRS copy was lost subsequently.
Despite the divergence of the N-terminal region of archaeal LeuRSs, all archaeal LeuRSs contain a similar archaeal CTD, suggesting the essential role of this domain in recognizing archaeal tRNALeus. The specific NmLeuRS/tRNALeu interaction mode strongly suggests that tRNA synthetases are able to evolve to adapt to the tRNAs and thus provides a striking example of coevolution between tRNA synthetase/tRNA. We showed that the CTD of NmLeuRS1 was essential for both aminoacylation and editing reactions, confirming its crucial role in Leu-tRNALeu synthesis and translational quality control. Furthermore, substitution of archaeal CTD with bacterial CTD totally abolished aminoacylation (Supplementary Figure S5) and editing activities (data not shown), which is consistent with its maintenance of the archaeal CTD. Considering that EcLeuRS lost its aminoacylation activity in high KCl solution (Figure 3B), we cannot rule out the potential possibility that the bacterial CTD in swapping mutant could not fold correctly in reaction buffer. For NmLeuRS1, it is also possible that the archaeal CTD prevents misacylation of tRNAIle as revealed in the PhLeuRS system (66).
In summary, our data reveal the haloalkaliphilic nature of NmLeuRSs and further elucidate the aminoacylation and editing activities of NmLeuRSs as well as the function of CTD in both the recognition and editing of NmtRNALeu. Thus, this study provides a comprehensive characterization of the aaRS/tRNA system in halobacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Basic Research Foundation of China [2012CB911000]; Natural Science Foundation of China [31130064, 31270852]. Funding for open access charge: [2012CB911000].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Profs. Li Huang and Hua Xiang and Drs Li Guo and Da-He Zhao (Institute of Microbiology, Beijing, China) for providing us N. magadii ATCC 43099 and H. hispanica ATCC 33960 genomic DNA. They are also grateful to Dr Yuchen Liu (Department of Molecular Biophysics and Biochemistry, Yale University) for valuable suggestions. The authors gratefully acknowledge the support of SA-SIBS scholarship program.
REFERENCES
- 1.Lucas-Lenard J, Lipmann F. Protein biosynthesis. Annu. Rev. Biochem. 1971;40:409–448. doi: 10.1146/annurev.bi.40.070171.002205. [DOI] [PubMed] [Google Scholar]
- 2.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 3.Cusack S, Berthet-Colominas C, Hartlein M, Nassar N, Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature. 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 4.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Wang E. Transfer RNA: a dancer between charging and mis-charging for protein biosynthesis. Sci. China Life Sci. 2013;56:921–932. doi: 10.1007/s11427-013-4542-9. [DOI] [PubMed] [Google Scholar]
- 6.Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem. J. 1972;128:1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JF, Guo NN, Li T, Wang ED, Wang YL. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry. 2000;39:6726–6731. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- 8.Dock-Bregeon A, Sankaranarayanan R, Romby P, Caillet J, Springer M, Rees B, Francklyn CS, Ehresmann C, Moras D. Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell. 2000;103:877–884. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- 9.Ruan LL, Zhou XL, Tan M, Wang ED. Human cytoplasmic ProX edits mischarged tRNAPro with amino acid but not tRNA specificity. Biochem. J. 2013;450:243–252. doi: 10.1042/BJ20121493. [DOI] [PubMed] [Google Scholar]
- 10.Nangle LA, De Crecy Lagard V, Doring V, Schimmel P. Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J. Biol. Chem. 2002;277:45729–45733. doi: 10.1074/jbc.M208093200. [DOI] [PubMed] [Google Scholar]
- 11.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem. Biol. 2006;13:1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Zhou XL, Du DH, Tan M, Lei HY, Ruan LL, Eriani G, Wang ED. Role of tRNA amino acid-accepting end in aminoacylation and its quality control. Nucleic Acids Res. 2011;39:8857–8868. doi: 10.1093/nar/gkr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 14.Fukunaga R, Yokoyama S. Crystal structure of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii reveals a novel editing domain orientation. J. Mol. Biol. 2005;346:57–71. doi: 10.1016/j.jmb.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 15.Zhou XL, Zhu B, Wang ED. The CP2 domain of leucyl-tRNA synthetase is crucial for amino acid activation and post-transfer editing. J. Biol. Chem. 2008;283:36608–36616. doi: 10.1074/jbc.M806745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lincecum TL, Jr, Tukalo M, Yaremchuk A, Mursinna RS, Williams AM, Sproat BS, Van Den Eynde W, Link A, Van Calenbergh S, Grotli M, et al. Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase. Mol. Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhou XL, Wang M, Tan M, Huang Q, Eriani G, Wang ED. Functional characterization of leucine-specific domain 1 from eukaryal and archaeal leucyl-tRNA synthetases. Biochem. J. 2010;429:505–513. doi: 10.1042/BJ20100235. [DOI] [PubMed] [Google Scholar]
- 18.Yan W, Tan M, Eriani G, Wang ED. Leucine-specific domain modulates the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nucleic Acids Res. 2013;41:4988–4998. doi: 10.1093/nar/gkt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du X, Wang ED. Tertiary structure base pairs between D- and TpsiC-loops of Escherichia coli tRNA(Leu) play important roles in both aminoacylation and editing. Nucleic Acids Res. 2003;31:2865–2872. doi: 10.1093/nar/gkg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu QH, Huang Q, Wang ED. Crucial role of the C-terminal domain of Mycobacterium tuberculosis leucyl-tRNA synthetase in aminoacylation and editing. Nucleic Acids Res. 2013;41:1859–1872. doi: 10.1093/nar/gks1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palencia A, Crépin T, Vu MT, Lincecum TL, Martinis SA, Cusack S. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2012;19:677–684. doi: 10.1038/nsmb.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao P, Zhu B, Jaeger S, Eriani G, Wang ED. Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps. Nucleic Acids Res. 2008;36:2728–2738. doi: 10.1093/nar/gkn028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breitschopf K, Achsel T, Busch K, Gross HJ. Identity elements of human tRNA(Leu): structural requirements for converting human tRNA(Ser) into a leucine acceptor in vitro. Nucleic Acids Res. 1995;23:3633–3637. doi: 10.1093/nar/23.18.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soma A, Kumagai R, Nishikawa K, Himeno H. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNA(Leu) J. Mol. Biol. 1996;263:707–714. doi: 10.1006/jmbi.1996.0610. [DOI] [PubMed] [Google Scholar]
- 25.Soma A, Uchiyama K, Sakamoto T, Maeda M, Himeno H. Unique recognition style of tRNALeu by Haloferax volcanii Leucyl-tRNA synthetase. J. Mol. Biol. 1999;293:1029–1038. doi: 10.1006/jmbi.1999.3219. [DOI] [PubMed] [Google Scholar]
- 26.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 27.Tindall BJ, Ross HNM, Grant WD. Natronobacterium gen. nov. and Natronococcus gen. nov., Two New Genera of Haloalkaliphilic Archaebacteria. Syst. Appl. Microbiol. 1984;5:41–57. [Google Scholar]
- 28.Kamekura M, Dyall-Smith ML, Upasani V, Ventosa A, Kates M. Diversity of alkaliphilic halobacteria: proposals for transfer of Natronobacterium vacuolatum, Natronobacterium magadii, and Natronobacterium pharaonis to Halorubrum, Natrialba, and Natronomonas gen. nov., respectively, as Halorubrum vacuolatum comb. nov., Natrialba magadii comb. nov., and Natronomonas pharaonis comb. nov., respectively. Int. J. Syst. Bacteriol. 1997;47:853–857. doi: 10.1099/00207713-47-3-853. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Wang E, Wang Y. A modified procedure for fast purification of T7 RNA polymerase. Protein Expr. Purif. 1999;16:355–358. doi: 10.1006/prep.1999.1083. [DOI] [PubMed] [Google Scholar]
- 30.Zhou XL, Fang ZP, Ruan ZR, Wang M, Liu RJ, Tan M, Anella FM, Wang ED. Aminoacylation and translational quality control strategy employed by leucyl-tRNA synthetase from a human pathogen with genetic code ambiguity. Nucleic Acids Res. 2013;41:9825–9838. doi: 10.1093/nar/gkt741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 1984;259:9461–9471. [PubMed] [Google Scholar]
- 34.Xu MG, Chen JF, Martin F, Zhao MW, Eriani G, Wang ED. Leucyl-tRNA synthetase consisting of two subunits from hyperthermophilic bacteria Aquifex aeolicus. J. Biol. Chem. 2002;277:41590–41596. doi: 10.1074/jbc.M205126200. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Ma JJ, Tan M, Yao P, Hu QH, Eriani G, Wang ED. Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase. Nucleic Acids Res. 2011;39:235–247. doi: 10.1093/nar/gkq763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, Zhou XL, Liu RJ, Fang ZP, Zhou M, Eriani G, Wang ED. Multilevel functional and structural defects induced by two pathogenic mitochondrial tRNA mutations. Biochem. J. 2013;453:455–465. doi: 10.1042/BJ20130294. [DOI] [PubMed] [Google Scholar]
- 37.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou S, Larsen RW, Boudko D, Riley CW, Karatan E, Zimmer M, Ordal GW, Alam M. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature. 2000;403:540–544. doi: 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- 39.Fukuchi S, Yoshimune K, Wakayama M, Moriguchi M, Nishikawa K. Unique amino acid composition of proteins in halophilic bacteria. J. Mol. Biol. 2003;327:347–357. doi: 10.1016/s0022-2836(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 40.Ortega G, Lain A, Tadeo X, Lopez-Mendez B, Castano D, Millet O. Halophilic enzyme activation induced by salts. Sci. Rep. 2011;1:6. doi: 10.1038/srep00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito S, Kobayashi T, Ara K, Ozaki K, Kawai S, Hatada Y. Alkaline detergent enzymes from alkaliphiles: enzymatic properties, genetics, and structures. Extremophiles. 1998;2:185–190. doi: 10.1007/s007920050059. [DOI] [PubMed] [Google Scholar]
- 42.De Castro RE, Ruiz DM, Gimenez MI, Silveyra MX, Paggi RA, Maupin-Furlow JA. Gene cloning and heterologous synthesis of a haloalkaliphilic extracellular protease of Natrialba magadii (Nep) Extremophiles. 2008;12:677–687. doi: 10.1007/s00792-008-0174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krulwich TA. Alkaliphiles: ‘basic' molecular problems of pH tolerance and bioenergetics. Mol. Microbiol. 1995;15:403–410. doi: 10.1111/j.1365-2958.1995.tb02253.x. [DOI] [PubMed] [Google Scholar]
- 44.Kitada M, Kosono S, Kudo T. The Na+/H+ antiporter of alkaliphilic Bacillus sp. Extremophiles. 2000;4:253–258. doi: 10.1007/s007920070010. [DOI] [PubMed] [Google Scholar]
- 45.Aono R, Ito M, Machida T. Contribution of the cell wall component teichuronopeptide to pH homeostasis and alkaliphily in the alkaliphile Bacillus lentus C-125. J. Bacteriol. 1999;181:6600–6606. doi: 10.1128/jb.181.21.6600-6606.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siddaramappa S, Challacombe JF, Decastro RE, Pfeiffer F, Sastre DE, Gimenez MI, Paggi RA, Detter JC, Davenport KW, Goodwin LA, et al. A comparative genomics perspective on the genetic content of the alkaliphilic haloarchaeon Natrialba magadii ATCC 43099T. BMC Genomics. 2012;13:165. doi: 10.1186/1471-2164-13-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H, Wu Z, Li M, Zhang F, Zheng H, Han J, Liu J, Zhou J, Wang S, Xiang H. Complete genome sequence of Haloarcula hispanica, a Model Haloarchaeon for studying genetics, metabolism, and virus-host interaction. J. Bacteriol. 2011;193:6086–6087. doi: 10.1128/JB.05953-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cusack S, Yaremchuk A, Tukalo M. The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 2000;19:2351–2361. doi: 10.1093/emboj/19.10.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landro JA, Schimmel P. Metal-binding site in a class I tRNA synthetase localized to a cysteine cluster inserted into nucleotide-binding fold. Proc. Natl Acad. Sci. USA. 1993;90:2261–2265. doi: 10.1073/pnas.90.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landro JA, Schimmel P. Zinc-dependent cell growth conferred by mutant tRNA synthetase. J. Biol. Chem. 1994;269:20217–20220. [PubMed] [Google Scholar]
- 51.Zhu B, Yao P, Tan M, Eriani G, Wang ED. tRNA-independent pretransfer editing by class I leucyl-tRNA synthetase. J. Biol. Chem. 2009;284:3418–3424. doi: 10.1074/jbc.M806717200. [DOI] [PubMed] [Google Scholar]
- 52.Zhou XL, Tan M, Wang M, Chen X, Wang ED. Post-transfer editing by a eukaryotic leucyl-tRNA synthetase resistant to the broad-spectrum drug AN2690. Biochem. J. 2010;430:325–333. doi: 10.1042/BJ20100474. [DOI] [PubMed] [Google Scholar]
- 53.Zhou XL, Ruan ZR, Huang Q, Tan M, Wang ED. Translational fidelity maintenance preventing Ser mis-incorporation at Thr codon in protein from eukaryote. Nucleic Acids Res. 2013;41:302–314. doi: 10.1093/nar/gks982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan M, Yan W, Liu RJ, Wang M, Chen X, Zhou XL, Wang ED. A naturally occurring nonapeptide functionally compensates the CP1 domain of leucyl-tRNA synthetase to modulate aminoacylation activity. Biochem. J. 2012;443:477–484. doi: 10.1042/BJ20111925. [DOI] [PubMed] [Google Scholar]
- 55.Li L, Palencia A, Lukk T, Li Z, Luthey-Schulten ZA, Cusack S, Martinis SA, Boniecki MT. Leucyl-tRNA synthetase editing domain functions as a molecular rheostat to control codon ambiguity in Mycoplasma pathogens. Proc. Natl Acad. Sci. USA. 2013;110:3817–3822. doi: 10.1073/pnas.1218374110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lue SW, Kelley SO. An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry. 2005;44:3010–3016. doi: 10.1021/bi047901v. [DOI] [PubMed] [Google Scholar]
- 57.Zheng YG, Wei H, Ling C, Xu MG, Wang ED. Two forms of human cytoplasmic arginyl-tRNA synthetase produced from two translation initiations by a single mRNA. Biochemistry. 2006;45:1338–1344. doi: 10.1021/bi051675n. [DOI] [PubMed] [Google Scholar]
- 58.Sivaram P, Deutscher MP. Existence of two forms of rat liver arginyl-tRNA synthetase suggests channeling of aminoacyl-tRNA for protein synthesis. Proc. Natl Acad. Sci. USA. 1990;87:3665–3669. doi: 10.1073/pnas.87.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu YH, Chang CP, Chien CI, Tseng YK, Wang CC. An insertion peptide of yeast glycyl-tRNA synthetase facilitates both productive docking and catalysis of cognate tRNAs. Mol. Cell Biol. 2013;33:3515–3523. doi: 10.1128/MCB.00122-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, Söll D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc. Natl Acad. Sci. USA. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reader JS, Ordoukhanian PT, Kim JG, de Crécy-Lagard V, Hwang I, Farrand S, Schimmel P. Major Biocontrol of Plant Tumors Targets tRNA Synthetase. Science. 2005;309:1533. doi: 10.1126/science.1116841. [DOI] [PubMed] [Google Scholar]
- 62.Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, Navarre WW, Ibba M. The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-beta-lysine. Nat. Chem. Biol. 2011;7:667–669. doi: 10.1038/nchembio.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guitart T, Leon Bernardo T, Sagales J, Stratmann T, Bernues J, Ribas de Pouplana L. New aminoacyl-tRNA synthetase-like protein in insecta with an essential mitochondrial function. J. Biol. Chem. 2010;285:38157–38166. doi: 10.1074/jbc.M110.167486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown JR, Doolittle WF. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl Acad. Sci. USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andam CP, Harlow TJ, Papke RT, Gogarten JP. Ancient origin of the divergent forms of leucyl-tRNA synthetases in Halobacteriales. BMC Evol. Biol. 2012;12:85. doi: 10.1186/1471-2148-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukunaga R, Yokoyama S. The C-terminal domain of the archaeal leucyl-tRNA synthetase prevents misediting of isoleucyl-tRNA(Ile) Biochemistry. 2007;46:4985–4996. doi: 10.1021/bi6024935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.