Abstract
The emergence of Staphylococcus aureus strains that are resistant to glycopeptides has led to alarming scenarios where serious staphylococcal infections cannot be treated. The bacterium expresses many small regulatory RNAs (sRNAs) that have unknown biological functions for the most part. Here we show that an S. aureus sRNA, SprX (alias RsaOR), shapes bacterial resistance to glycopeptides, the invaluable treatments for Methicillin-resistant staphylococcal infections. Modifying SprX expression levels influences Vancomycin and Teicoplanin glycopeptide resistance. Comparative proteomic studies have identified that SprX specifically downregulates stage V sporulation protein G, SpoVG. SpoVG is produced from the yabJ-spoVG operon and contributes to S. aureus glycopeptide resistance. SprX negatively regulates SpoVG expression by direct antisense pairings at the internal translation initiation signals of the second operon gene, without modifying bicistronic mRNA expression levels or affecting YabJ translation. The SprX and yabJ-spoVG mRNA domains involved in the interaction have been identified, highlighting the importance of a CU-rich loop of SprX in the control of SpoVG expression. We have shown that SpoVG might not be the unique SprX target involved in the glycopeptide resistance and demonstrated that the regulation of glycopeptide sensitivity involves the CU-rich domain of SprX. Here we report the case of a sRNA influencing antibiotic resistance of a major human pathogen.
INTRODUCTION
Staphylococcus aureus is a human and animal pathogen that has spectacular adaptive capacities to various antimicrobial agents. Staphylococcus aureus rapidly acquires multiple antibiotic resistances, causing a wide spectrum of nosocomial and community-associated infections, and is thus a major worldwide health problem (1). Since the spread of staphylococcal resistance to β-lactams (such as Methicillin), glycopeptide antibiotics have been the most efficient weapons against Gram-positive infections, including the problematic Methicillin-resistant S. aureus (MRSA). Vancomycin and Teicoplanin glycopeptide antibiotics have a similar mode of action on cell wall synthesis (2,3). Unlike β-lactams, glycopeptides do not block enzymes involved in cell wall synthesis, but instead sequester the substrates required for peptidoglycan formation. In 1997, the first S. aureus with reduced susceptibility to glycopeptides was reported in Japan (1,4). Since then, a number of cases have been reported worldwide, increasing the threat of incurable staphylococcal infections. Three classes of S. aureus Vancomycin-resistance have emerged that differ in Vancomycin susceptibilities: high-level Vancomycin-resistant strains, also called VRSA (Minimum Inhibitory Concentration, MIC ≥32 µg/ml), Vancomycin-intermediate S. aureus (VISA; MIC, 8–16 µg/ml) and heterogeneous Vancomycin-intermediate S. aureus (hVISA), which are strains containing subpopulations of Vancomycin-intermediate daughter cells. High level VRSA appeared as a result of a horizontal transfer of Tn1546 encoding the multiprotein VanA complex from Enterococcus faecalis to MRSA clinical isolates (5). Intermediate glycopeptide resistance of S. aureus is much more prevalent and currently, the mechanism of such resistance is unknown. It is thought to result from stepwise accumulations of mutations that confer advantage in the face of drug encounters (6,7). One of the most consistent characteristic of S. aureus intermediate-resistant strains (hVISA and VISA) is a cell wall thickening, which might reduce the ability of the Vancomycin to diffuse into the division septum, although the mechanisms underlying the assembly of a thicker cell wall are unknown (5–7). Therefore, there is an urgent need to increase our knowledge of the molecular events involved in antibiotic resistance of this serious pathogen, with a necessary focus on the precious glycopeptides used for MRSA infections.
In S. aureus, Methicillin and glycopeptide resistance is adjusted by the yabJ-spoVG operon, which is in turn controlled by a nucleotide sequence recognized by the alternative sigma B factor (σB) (8,9). The yabJ-spoVG mRNA codes for two proteins: YabJ, which has unknown functions; and stage V sporulation protein G (SpoVG). SpoVG was initially identified in Bacillus subtilis and thought to be involved in an unknown way in sporulation (10). However, in non-sporulating bacteria, its mode of action and the molecular mechanisms involved are not known even though it was shown recently that SpoVG is a site-specific DNA-binding protein (11). SpoVG (but not YabJ) was shown to be the major regulator of the yabJ-spoVG operon (12). This yabJ-spoVG operon has been proposed as the σB-dependent secondary regulator (8). In addition to the control of Methicillin and glycopeptide resistance, deletion of the yabJ-spoVG operon was shown to alter the expression of extracellular nuclease, lipase and protease expressions (12) as well as affecting capsule formation and the transcriptional control of cap and esxA (8,13). Micro-arrays performed on yabJ-spoVG deletion in the Newman strain showed that yabJ-spoVG antagonizes the effects of σB on the expression levels of several proteins (12).
The coordinated expression of pathogenicity determinants is tightly controlled by a complex network of elements, including two component systems, transcription factors, small metabolites and small regulatory RNAs (sRNAs). Recently, bacterial sRNAs were shown to play a major role in a variety of regulatory processes (14). Staphylococcus aureus expresses ∼250 sRNAs, most of which have unknown biological functions (15–22). sRNAs control gene expression through various mechanisms, generally at the posttranscriptional level by direct pairing with mRNA targets (23). These interactions positively or negatively regulate translation and/or the stability of the mRNAs. Several S. aureus sRNAs have been shown to be involved in pathogenicity, e.g. RNAIII, which regulates the expression of numerous virulence factors (16,24–26). RNAIII is produced at the end of the exponential phase and allows for the transition between synthesis of surface-associated proteins and secreted factors (27). Moreover, the sRNAs SprD and Ssr42 have been shown to be involved in S. aureus virulence in animal models of infection (28,29).
Herein we report that the recently identified S. aureus sRNA SprX (small pathogenicity island RNA X) (20) shapes bacterial resistance to glycopeptide antibiotics and controls the SpoVG expression. In fact, SprX inhibits SpoVG expression through the direct interaction between a C-rich loop of SprX and spoVG ribosomal binding site of yabJ-spoVG mRNA. This complex prevents ribosomal loading onto spoVG, and specifically inhibits translation of the second downstream gene within the yabJ-spoVG operon without altering the stability of the yabJ-spoVG mRNA.
MATERIALS AND METHODS
Bacterial strains, culture conditions and susceptibility testing
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The bacteria were grown at 37°C in Brain–Heart Infusion broth (BHI, Oxoid) or in Tryptic Soy Broth (TSB, Oxoid). When necessary, the media were supplemented with either 10 µg of chloramphenicol or erythromycin for S. aureus, or 50 µg ampicillin per ml for Escherichia coli. For the spot assays, overnight cultures of the tested strains were diluted to obtain an OD600 of 8. From these cultures, seven consecutive 10-fold dilutions were prepared. Three microliters of each dilution was dropped on Muëller–Hinton (MH) or on MH supplemented by 0.75 or 0.85 µg/ml of Teicoplanin or Vancomycin antibiotics, then incubated for 24 h at 37°C. The first dilution corresponds to 107 bacteria. For susceptibility testing, plates containing an antibiotic gradient were prepared as described before (9) except that we used BHI agar plate instead of MH agar plate. Teicoplanin susceptibilities were determined by using Etest strips (BioMerieux, France) on MH agar plates according to the manufacturer’s instructions.
Plasmid and strain construction
In pCN38-sprX, sprX2 was expressed from its own promoter. The sprX2 sequence (containing sprX sequence, the 207 bp upstream and 258 bp downstream from sprX) was amplified from HG001 genomic DNA as a 615-bp fragment using primers listed in Supplementary Table S2. The pCN38-sprX_mutL3 was produced using the mutagenized oligonucleotides ‘mutfor’ and ‘mutrev’ (Supplementary Table S2). The fragments were flanked with PstI and EcoRI restriction sites. The polymerase chain reaction (PCR) products were cloned in pCN38 (30).
To inactivate the HG001 (31) sprX genes, DNA fragments upstream and downstream of sprX were amplified by PCR from genomic DNA by primers listed in Supplementary Table S2. For the sprX1, the upstream fragment was 889 bp long and the downstream fragment was 949 bp long; for sprX2 the upstream was 988 bp long and the downstream was 949 bp long. A second PCR amplification was done by combining these fragments (using primers sprXD1 and sprXD6 for sprX1, and primers 2sprXD1 and 2sprXD4 for sprX2). These were then cloned using the PstI-BamHI sites in the temperature-sensitive plasmid pBT2 (32). To achieve gene disruption in the genome by homologous recombination, the resulting plasmids pBT2ΔsprX1 and pBT2ΔsprX2 were transformed into S. aureus strain RN4220 and then into S. aureus HG001. Mutants were enriched by cultivation at 42°C. Cells from the stationary phase culture were plated on Tryptic Soy Agar (TSA, Oxoid) plates and incubated at 37°C. Colonies were imprinted on plates that were supplemented with 10 µg/ml chloramphenicol. Chloramphenicol-sensitive colonies were tested by PCR for deletion of sprX1 and sprX2. The deletions were confirmed by northern blot assays. The primers used for constructing pBT2ΔsprX are shown in Supplementary Table S2. The S. aureus HG001ΔyabJ-spoVG::erm mutant was constructed by transducing the ΔyabJ-spoVG::erm mutation of strain RN4220 (9) into strain HG001. The deletion of the yabJ-spoVG locus was confirmed by PCR, northern blot and western blot analysis.
SpoVG-His6 expression and antibody preparation
The pSTM33 vector containing SpoVG-His6 (9) was electroporated into BL21 strain (DE3, Novagen). Cells were grown in Luria Broth (LB, MO BIO) broth to a 600 nm optical density of 0.5, and SpoVG-His6 expression was induced with 0.3 mM isopropyl-d-thiogalactopyranoside (IPTG, Eurobio). After 3 h, cells were collected by centrifugation and suspended in 30 ml phosphate-buffered saline (pH 7.4) supplemented with a complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail tablet (Roche) and 0.1 mg/ml DNase. The cells were disrupted and the debris separated by centrifugation at 13 000g for 10 min. Purification of the His-tagged protein was performed on nickel Sepharose High Performance columns (HisTrap HP; GE Healthcare) using an ÄKTA fast-performance liquid chromatography system. The correct molecular weight of the purified protein was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. One milligram of the purified SpoVG-His6 was used to raise polyclonal mouse antibodies (Eurogentec, Seraing, Belgium).
Protein isolation and western blots
For the total protein extractions, culture pellets corresponding to 2 ml of culture at an OD600 of 1 were suspended in 0.2 ml of lysis buffer [10 mM Tris–HCl, pH 7.5, 20 mM NaCl, 1 mM EDTA, 5 mM MgCl2 and complete EDTA-free protease inhibitor cocktail tablet (Roche) containing 0.1 mg/ml lysostaphin]. Following incubation at 37°C for 10 min, Laemmli sample buffer was added (33). Samples were then boiled for 5 min, separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred onto a hybond-P PolyVinyliDene Fluoride (PVDF) membrane (Amersham). To visualize SpoVG, antibodies were used at a dilution of 1:5000. The anti-rabbit antibodies were used at a dilution of 1:20 000. Western blots were revealed using the Amersham ECL Plus detection Kit. Signals were visualized using a Typhoon FLA 9500 and quantified using Image-QuantTL 7.0 (GE Healthcare).
Bidimensional gel electrophoresis
Overnight cultures of bacteria were diluted 1:100 in BHI and grown at 37°C to the exponential phase, then the cells were pelleted for 10 min at 4°C (8000g). Pellets of 2-ml culture were suspended in the same lysis buffer already described with the addition of 2 UI of DNase amp grade and 2 UI of RNase A. Following incubation at 37°C for 30 min, 1 ml of Tri-Reagent was added, then the samples were sonicated (3 × 30 s 20% active cycle). Hundred microliters of chloroform was added to each sample and incubated for 5 min at room temperature (RT). Next, 300 µl of ethanol was added and samples were incubated for 3 min at RT and precipitated overnight at −20°C using acetone. The proteins were centrifuged for 15 min at 4°C (4000g) and washed with 1 ml of 80% acetone, then pelleted for 5 min at 4°C (4000g). The pellets were dried at RT. Duplicate pellets were dissolved in 200 µl of urea 8 M. 2D-DIGE and mass spectrometry identification of proteins of interest were performed by the Cochin Institute (Paris). Spots corresponding to proteins with expression modified between HG001 wt and ΔsprX were subjected to in-gel trypsin digestion, peptide extraction and desalting, followed by MALDI-ToF/ToF analysis. Peptides were analyzed using MASCOT and the NCBInr database to identify the selected protein spots. The average ratio of protein levels was calculated using DeCyder 2D software (GE Healthcare) to determine the change in normalized spot volume between HG001 wt and HG001ΔsprX samples. The average ratios are shown in Supplementary Table S3, although the statistical analysis was based on the log of the true ratio measurements.
RNA isolation, northern blots, transcription and RNA labeling
Total RNAs were prepared as previously described (20). For SprX, northern blots were performed with 10 µg of total RNA (15). Specific 32P-labeled probes (Supplementary Table S2) were hybridized with ExpressHyb solution (Clontech) for 90 min, washed, exposed and then scanned with a Typhoon FLA 9500 scanner (GE Healthcare). For the northern blots of yabJ-spoVG mRNA, 10 µg of total RNAs were separated by 5% polyacrylamide/8M urea gel. The primer pair oSTM29/oSTM30 (Supplementary Table S2) was used to generate α32P-dCTP-labeled spoVG-specific probes by PCR labeling. For in vitro experiments, RNAs were transcribed from PCR fragments generated from genomic DNA using the primers described in Supplementary Table S2. To produce the template-encoding SprX_mutL3, mutagenized oligonucleotides were used (Supplementary Table S2). RNA was produced by in vitro transcription using MEGAscript (Ambion). 5′-RNA γ32P-dATP labeling was performed (34). The RNA was purified by 8% polyacrylamide/8M urea gel, eluted, ethanol precipitated, then stored at −80°C.
RNA structure probing
Structural assays were performed as previously described (34). SprX was prepared by incubating 14 pmol of unlabeled RNA in a buffer (10 mM Tris–HCl, pH 7.5, 60 mM NaCl, 1 mM EDTA) for 10 min at 25°C. The yabJ-spoVG167 mRNA was prepared by incubating 1 pmol of labeled RNA in the aforementioned buffer. MgCl2 was added to obtain a final concentration of 2.5 mM and this was then incubated for 10 min at 25°C. Preparation for structural analysis of the duplexes between SprX and yabJ-spoVG167 mRNA was done by incubating 0.2 pmol of labeled RNA with 10 pmol of unlabeled RNA for 15 min at 25°C. Cleavages with S1 nuclease (0.063 U/µl),V1 RNase (6.25 × 10−7 U/µl) and 1.25 mM lead acetate were carried out for 10 min at 25°C in the presence of 1 μg of total yeast tRNA. For sequencing, T1 at 0.038 U/µl and U2 at 0.0015 U/µl were used. The reactions were precipitated, and the pellets dissolved in loading buffer II (Ambion). The samples were denatured for 10 min at 65°C before separation on 8% polyacrylamide/8M urea gels. Gels were dried and visualized (Typhoon FLA 9500).
Toeprint assays
The toeprint assays were performed as previously described (20), but with modifications. Annealing mixtures containing 13 nM of yabJ-spoVG mRNA with either 67 nM of labeled Toeprint_spoVG or Toeprint_yabJ primers were incubated with a buffer (20 mM Tris–HCl, pH 7.5, 60 mM NH4Cl) for 2 min at 90°C followed by 1 min at RT. Renaturation was realized in the presence of 10 mM MgCl2 at 25°C for 20 min. For the assays in the presence of SprX, various concentrations of SprX were added before the purified E. coli 70S ribosomes. The ribosomes were reactivated for 15 min at 37°C and diluted in the reaction buffer in the presence of 1 mM MgCl2. 33 nM 70S were added in each assay, incubated for 5 min and the MgCl2 was adjusted to 10 mM. After 5 min, 0.83 µM uncharged tRNAfMet was added and this was incubated for 15 min. cDNAs were synthesized with 4 U of AMV RT (Biolabs) for 15 min. Reactions were ended by the addition of 15 µl of loading buffer II (Ambion). The cDNAs were loaded and separated onto 8% polyacrylamide/8M urea gels. Sequencing ladders were generated with the same 5′-end-labeled primer.
RESULTS
SprX affects S. aureus resistance to glycopeptide antibiotics
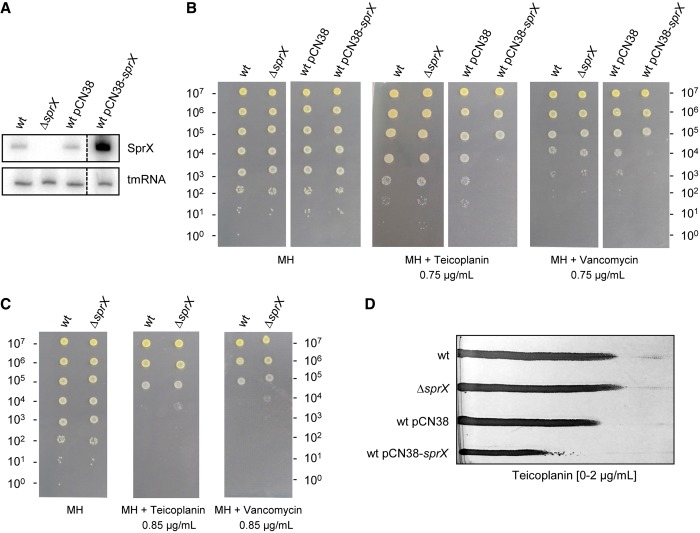
In a large-scale analysis of S. aureus N315 strain, we identified the small RNA SprX (alias RsaOR) (20). The SprX sequence is highly conserved among S. aureus strains. sprX is present in the majority of sequenced S. aureus strains and is encoded by genomes of converting phages. Most strains contain one copy of the sprX gene as for N315 (35), but some comprise two copies, as for example HG001 (36), or even three copies of sprX, as in the case of strain Newman (37). Its expression has been found to decrease during stationary growth phase and to increase on salt stress (20). To elucidate the functions of SprX, we analyzed the phenotypes of strains with different sRNA expression levels. For this purpose, we used HG001 agr+ S. aureus strain carrying two copies of sprX gene (Supplementary Figure S1A). Alignments of SprX1 and SprX2 showed that the RNA sequences are identical in each, except for 3 nt (Supplementary Figure S1B). We disrupted the two copies of sprX in strain HG001 (ΔsprX) by homologous recombination, thus abolishing SprX expression (Figure 1A). Overexpression of SprX was achieved using a multicopy plasmid expressing SprX from its endogenous promoter (pCN38-sprX, Figure 1A). Among the phenotypes tested of strains possessing different SprX expression levels, we measured their resistance to Teicoplanin, a glycopeptide antibiotic. Using Etest strips, we have detected that the deletion of sprX slightly increased the resistance to Teicoplanin (MIC 1.5 µg/ml) compared with parent wt HG001 strain (MIC 1 µg/ml). For a thorough analysis of the effect of SprX on glycopeptide susceptibility, we used a spot Population Analysis Profile assay (spot PAP), which allows rapid, sensitive and reproducible qualitative and quantitative testing of antibiotic resistance that is not limited by log2 dilutions used in the Etest strips (7). We have tested the effect of SprX on susceptibility to Teicoplanin and Vancomycin, two antibiotics from the glycopeptide family (Figure 1B).
Figure 1.
SprX sRNA modulates S. aureus resistance to two glycopeptide antibiotics. (A) Upper panel: northern blots showing SprX expression when grown until the late-exponential phase in the wt HG001 (wt) strain; HG001ΔsprX (ΔsprX); wt HG001 transformed by pCN38 (wt pCN38); and wt HG001 transformed by pCN38−sprX (wt pCN38-sprX). Lower panel: tmRNA used as a loading control. (B and C) Overnight cultures were prepared in BHI from strains wt HG001 (wt), HG001ΔsprX (ΔsprX) and wt HG001 transformed by either pCN38 (wt pCN38) or by pCN38-sprX (wt pCN38-sprX). Ten-fold serial dilutions of cultures were deposited from top (most concentrated: 107 bacteria) to bottom (100 bacteria) on MH plates and on MH plates supplemented with either Teicoplanin or Vancomycin at 0.75 µg/ml (B) and 0.85 µg/ml (C). They were then incubated for 24 h at 37°C. (D) Susceptibility testing of strains from the panel B on gradient plates containing Teicoplanin.
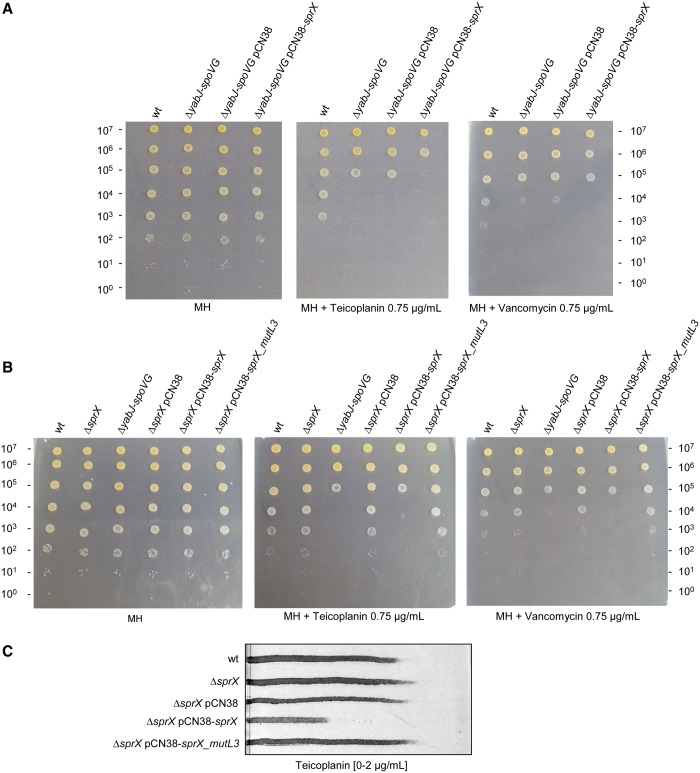
SprX deletion and overexpression had no effect on bacterial growth in MH medium. Interestingly, we observed and reproduced a 2-log10 diminution of colony formation with Teicoplanin and Vancomycin antibiotics for the strain overexpressing SprX when compared with wild-type (wt) strain (Figure 1B). We also observed that the sprX deletion strain was slightly more resistant than the wt strain when growing on MH with an increased concentration of antibiotics of 0.85 µg/ml (Figure 1C). Similar results were obtained using gradient plates, demonstrating that the deletion of SprX slightly increases while its overexpression reduces resistance to Teicoplanin (Figure 1D).
To assess if these results are relevant in a clinical isolate, we transformed VISA strain Mu50 (4), which carries a single copy of sprX in its genome (35), by both pCN38 and pCN38-sprX plasmids (Supplementary Figure S2A). Both, the spot PAP and the gradient plate tests showed that SprX overexpression in strain Mu50 resulted in a decrease in Teicoplanin resistance compared with the strain harboring pCN38 empty plasmid (Supplementary Figure S2B and C). Taken together, these results indicate that SprX could be involved in S. aureus resistance to Teicoplanin and Vancomycin. This interesting observation prompted us to identify the SprX target(s) involved in this antibiotic resistance, and to elucidate the mechanism of their regulation by SprX.
SprX reduces the expression of a protein involved in bacterial resistance to numerous antibiotics
To identify SprX targets, we analyzed whether SprX modulates the expression of S. aureus proteins. We thus compared the protein profiles of HG001 wt and ΔsprX strains by bidimensional gel electrophoresis (2D-DiGE). In total, five proteins were identified as being regulated by SprX (Supplementary Table S3) and all of these were upregulated in the ΔsprX strain. Interestingly, mass spectrometry analysis of these proteins identified SpoVG, which is involved in bacterial resistance to Methicillin and glycopeptide antibiotics (9).
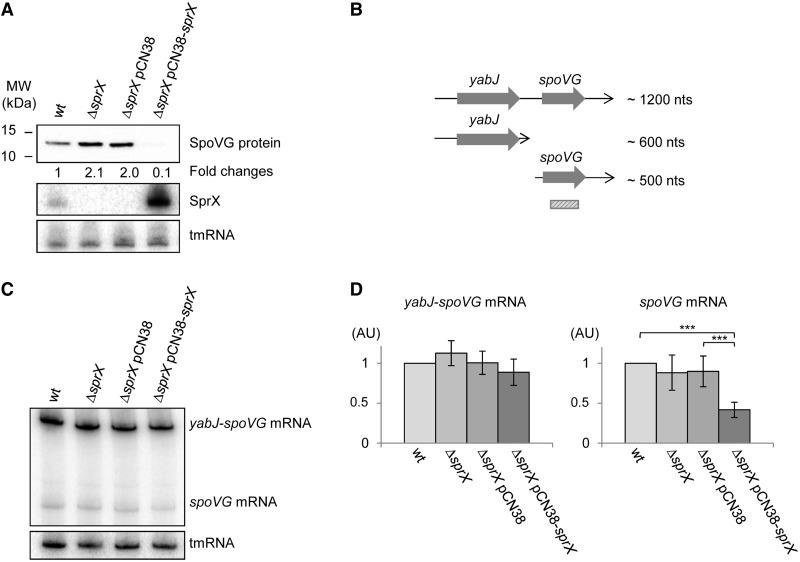
To confirm the downregulation of SpoVG expression by SprX, SpoVG protein levels were monitored by western blots using polyclonal antibodies raised against the SpoVG protein (Figure 2A). In agreement with the 2D-DiGE data, we observed an increase of SpoVG protein levels in the ΔsprX strain (Figure 2A). Furthermore, the effect of sprX inactivation resulting in an increase in SpoVG protein levels was detected at different phases of growth (Supplementary Figure S4). To confirm that the enhanced expression of SpoVG was linked to SprX inactivation, we performed complementation experiments. Introducing pCN38-sprX into the ΔsprX strain decreased SpoVG protein levels as compared with the ΔsprX strain and also compared with those in the wt strain (Figure 2A). These results are explained by the overexpression of SprX from pCN38 compared with its endogenous expression levels in the wt strain. Altogether, these data demonstrate that the SprX sRNA lowers SpoVG expression.
Figure 2.
SprX inhibits SpoVG protein expression at the translational level. (A) Upper panel: western blot of SpoVG protein levels when grown until the late-exponential phase in the wt HG001 strain (wt), HG001ΔsprX (ΔsprX) and in HG001ΔsprX transformed by either pCN38 (ΔsprX pCN38) or pCN38-sprX (ΔsprX pCN38-sprX). Coomassie staining was used as a loading control (Supplementary Figure S3A). SpoVG protein expression levels were calculated relative to the wt strain. The value of the SpoVG protein in wt strain was normalized to 1. Middle panel: northern blots showing SprX expression. Lower panel: tmRNA used as a loading control. (B) Schematic representation of yabJ-spoVG mRNA. ORFs, mRNA lengths and the probe used for detection by nothern blots of spoVG and yabJ-spoVG mRNAs are indicated. nts, nucleotides. (C) Northern blots detecting the expression levels of yabJ-spoVG and spoVG mRNAs in wt HG001 strain (wt), HG001ΔsprX (ΔsprX) and in HG001ΔsprX transformed by either pCN38 (ΔsprX pCN38) or pCN38-sprX (ΔsprX pCN38-sprX) grown until late-exponential (OD600 9) phases of growth. Probing for the tmRNA was used as a loading control (probes are listed in Supplementary Table S2). (D) Northern blot quantification of the levels of spoVG and yabJ-spoVG mRNAs of the strains from panel C. mRNA expression levels were calculated relative to the value measured for the wt strain. The error bars are the mean values derived from four independent experiments with independent RNA purifications. The tmRNA was used for normalization. The value of the mRNA levels in wild-type strain was normalized to 1. AU, arbitrary units. Difference in expression was measured by student’s T-test, ***P < 0.001.
Because SpoVG protein expression is downregulated by SprX, we monitored mRNA levels to determine whether this occurs at transcriptional and/or translational levels. SpoVG is translated from an ∼1200-nt long bicistronic yabJ-spoVG mRNA that is cleaved by an unknown mechanism into two transcripts of ∼600 and ∼500 nt, which correspond, respectively, to yabJ and spoVG mRNAs [(8); Figure 2B]. We used a labeled DNA probe of 300 bp encompassing SpoVG open reading frame (ORF) to detect both the spoVG and yabJ-spoVG mRNAs (Figure 2B). We also used a labeled DNA probe of 370 bp encompassing the YabJ ORF, which allows the detection of both yabJ and yabJ-spoVG mRNAs (Supplementary Figure S5A). No significant changes were seen in levels of either mRNA in the ΔsprX strain when compared with the wt parental strain (Figure 2C and D and Supplementary Figure S5B and C). Overexpression of SprX did not induce significant changes in the expression levels of full-length yabJ-spoVG mRNA, but decreased the amount of both spoVG and yabJ mRNAs (Figure 2C and D and Supplementary Figure S5B and C). Why the yabJ and spoVG fragments are produced is currently unknown; however, it has been previously shown that SpoVG is translated from full-length 1200-nt long yabJ-spoVG transcript (9). Consequently, our results showing that SprX does not affect the level of yabJ-spoVG mRNA suggest that SprX might affect the translation of SpoVG protein.
Structural changes of the yabJ-spoVG mRNA induced by SprX
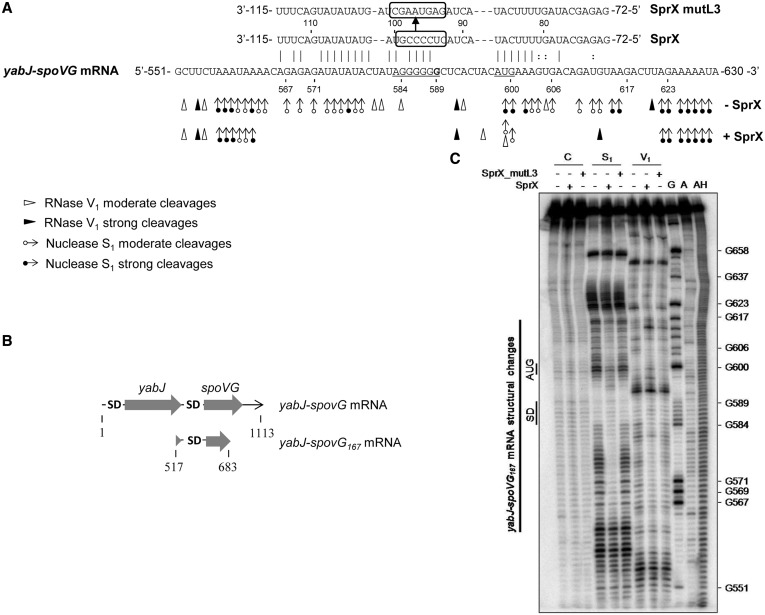
In vivo results prompted us to explore the interaction between SprX and yabJ-spoVG mRNA. Interestingly, we identified a putative pairing site between SprX and the translational initiation site of spoVG in silico using targetRNA2 (38) and sRNA TarBase (39). The predicted interaction between the two RNAs includes 26 bp and occurs between the U79-U115 nt from SprX and the A566-A606 nt from the yabJ-spoVG mRNA [a fragment that includes its Shine–Dalgarno (SD) sequence and the initiation codon of spoVG] (Figure 3A). To determine whether SprX interacts with the yabJ-spoVG mRNA in vitro, we used enzymatic probes to monitor the structural changes of yabJ-spoVG mRNA induced by SprX binding. For this purpose, we designed and produced a 167-nt-long mRNA construct, yabJ-spoVG167, which contains the ribosome binding site (RBS) of spoVG and the first 86 nt of the spoVG ORF (Figure 3B). We subjected the yabJ-spoVG167 alone or in complex with SprX to nuclease S1 (specific for single-stranded RNAs) and to RNase V1 cleavages (specific for double-stranded RNAs) (Figure 3C). The presence of many S1 cleavages in the region A558-A630 of yabJ-spoVG167 mRNA supports the existence of unpaired single-strand domains (Figure 3A and C). These data indicate that the proposed yabJ-spoVG mRNA region that binds with SprX is in fact accessible for interaction. SprX-induced structural changes on the yabJ-spoVG167 mRNA are located from A563 to U621 (Figure 3A and C). S1 cleavages disappeared at positions A563-A578 and A602-A616. Several weak V1 cleavages situated at A593, U605 and G580-G584 disappeared within the mRNA. Moreover, on duplex formation a strong V1 cut at position G613 and two weak ones at G596 and U599 appeared in the mRNA. Therefore, we can conclude that the binding of SprX induces structural changes encompassing the SD and AUG initiation codon of spoVG in yabJ-spoVG mRNA.
Figure 3.
SprX interacts directly with the yabJ-spoVG mRNA by base-pairing at the spoVG RBS. (A) SprX base pairings with the yabJ-spoVG mRNA. Binding of SprX encompasses the spoVG translational initiation site within the yabJ-spoVG mRNA. The SD sequence (5′-AGGGGG-3′) and initiation codon are underlined. The cleavage site of yabJ-spoVG mRNA corresponding to G at position 589 (9) is indicated in bold. We outline the mutated nucleotides in SprX_mutL3 for 5′-GAGUAAGC-3′. Probing data in the absence (−SprX) and in the presence of SprX (+SprX) are indicated. Enzymatic cleavages are as follows: moderate (white circle with rightward arrow) and strong (black circle with rightward arrow) S1 Nuclease cleavages; moderate (white right-pointing pointer) and strong (black right-pointing pointer) RNase V1 cleavages. (B) Schematic representation of yabJ-spoVG167 mRNA. The two ORFs and the two SD sequences are indicated. (C) Structural probing of the yabJ-spoVG167 mRNA in the presence and absence of SprX. Enzymatic hydrolysis (RNases S1 and V1) of 5′-end-labeled yabJ-spoVG167 mRNA free (−) or with an excess of either SprX or SprX_mutL3. Lanes are as follows: C, control; S1, nuclease S1; V1, RNase V1; G, RNase T2; A, RNase U2; and AH, alkaline ladder. The bar denotes the localization of the main reactivity changes that are induced by complex formation with SprX but not with SprX_mutL3.
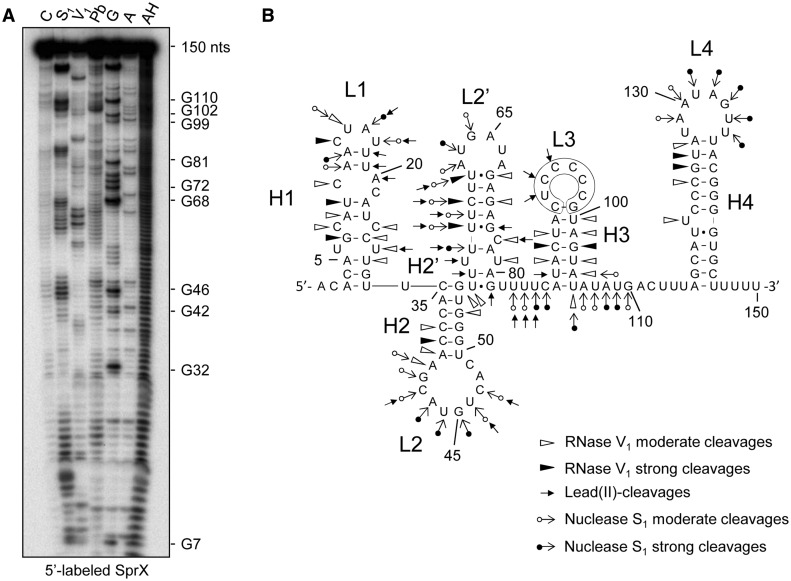
We investigated the structure of free SprX (nts 1–150) using chemical and enzymatic probes. To probe the SprX solution structure, we used lead (II) and nuclease S1, which cleave accessible single-stranded RNA, and RNase V1, which specifically cleaves double-stranded RNA (Figure 4A). The data support a SprX structure presented in Figure 4B. The simultaneous presence of V1 and S1 cleavages at the same nucleotide position within H1-L1 and H2’-L2’ may be explained by the coexistence of two alternating structures at the SprX 5′-end (Figure 4B and Supplementary Figure S6). Hairpins H3-L3 and H4-L4 in the 3′ region of SprX are, however, well-supported by the probing patterns. Moreover, a C-rich nucleotide sequence situated within the SprX L3 loop has been proposed to interact through base pairings with the spoVG SD sequence (Figure 3A). We tested the importance of the SprX L3 loop in the interaction between SprX and yabJ-spoVG mRNA by mutating the 5′-CUCCCCG-3′ sequence of the SprX L3 loop termed SprX_mutL3 (Figure 3A). The probing pattern of the yabJ-spoVG mRNA was identical in the presence of SprX_mutL3 and in the absence of SprX (Figure 3C), implying that the mutated SprX failed to induce structural changes in the spoVG RBS of the yabJ-spoVG167 mRNA. These results emphasize the importance of that loop in the interaction with the yabJ-spoVG target. Taken together, these mutational data support the proposed model of SprX-yabJ-spoVG mRNA interaction (Figure 3A), which involves pairings between the SprX L3 loop and the spoVG RBS of the yabJ-spoVG mRNA.
Figure 4.
Structural probe monitoring of SprX conformation in solution. (A) 5′-labeled SprX was probed by RNase V1, which cleaves double-strands or stacked nucleotides; and by nuclease S1 and lead acetate, both of which cleave accessible single-strands. Lanes are as follows: C, control; S1, Nuclease S1; V1, RNase V1; Pb, lead(II)-induced cleavages; G, RNase T2; A, RNase U2; and AH, alkaline ladder. SprX numbering is on the right. (B) Secondary structure of SprX from strain N315 based on probing data. The nucleotide numbering corresponds to SprX2 from HG001 (Supplementary Figure S1). The mutated nucleotides in SprX_mutL3 are outlined on the SprX secondary structure model. Enzymatic cleavages are as follows: moderate (white circle with rightward arrow) and strong (black circle with rightward arrow) S1 Nuclease cleavages; (rightward arrow) lead(II)-induced cleavages; moderate (white right-pointing pointer) and strong (black right-pointing pointer) RNase V1 cleavages.
SprX specifically reduces spoVG translation initiation of the yabJ-spoVG operon mRNA
Because SprX interacts with the spoVG RBS of yabJ-spoVG mRNA covered by the ribosomes during translation initiation, we conjectured that SprX could prevent ribosome loading on the spoVG RBS. We tested this hypothesis by performing toeprint analysis. We formed a ternary initiation complex consisting 70S ribosomes, initiator tRNAfMet and yabJ-spoVG mRNA. The ribosome blocked the elongation of reverse transcription, and produced a toeprint 16 nt downstream from the AUG initiation codon of spoVG (Figure 5A). SprX significantly reduced this toeprint in a concentration-dependent manner, indicating that in vitro SprX inhibits ribosome binding onto the spoVG RBS of yabJ-spoVG mRNA. These results are in agreement with our in vivo data, which show that SprX reduces SpoVG protein expression (Figure 2A). Interestingly, SprX_mutL3 failed to prevent ribosome loading onto the spoVG of yabJ-spoVG mRNA, indicating the essential role of the SprX L3 loop in regulating SpoVG expression (Figure 5A). Because yabJ-spoVG is a bicistronic mRNA, we next tested whether SprX might also prevent ribosome loading onto the yabJ RBS of the yabJ-spoVG mRNA (Figure 5B). One ribosome toeprint was detected 16 nt downstream from the predicted yabJ initiation codon. The addition of increasing concentrations of SprX did not alter the ribosome binding. These results show that in vitro, SprX specifically inhibits SpoVG translation initiation by antisense pairings with the spoVG RBS of the yabJ-spoVG mRNA, but it has no effect on YabJ translation initiation.
Figure 5.
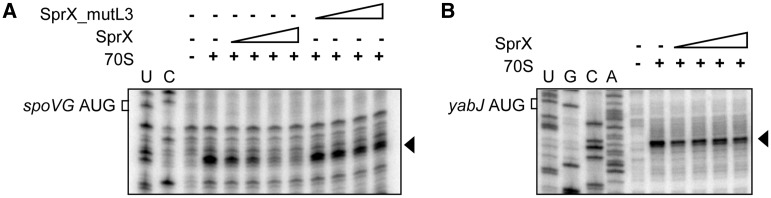
SprX specifically prevents ribosome loading on the spoVG translational initiation site within the 1200 nt long yabJ-spoVG mRNA. Here we show toeprint assays on (A) spoVG or (B) yabJ within the mRNA in the presence of increasing concentrations of SprX or SprX_mutL3 (33, 67, 133 and 267 nM). ‘+’ indicates the presence of purified 70S ribosomes. An arrow indicates the location of the experimentally determined toeprint. U, A, G and C refer to the yabJ-spoVG mRNA sequencing ladders.
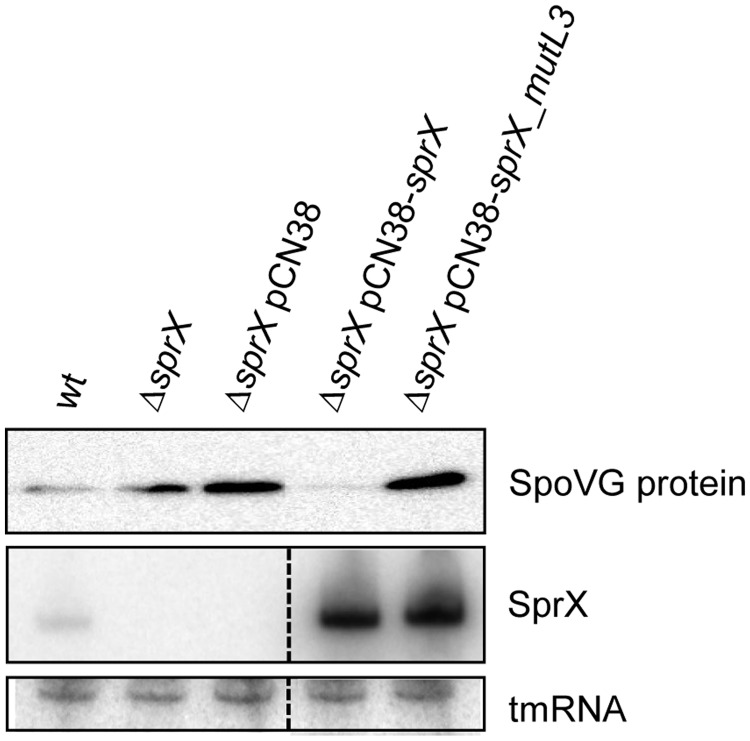
To assess the functional importance of the SprX L3 loop in vivo, we tested whether SprX_mutL3 is able to regulate SpoVG protein expression in vivo. To address this, we transformed ΔsprX strain by pCN38− sprX_mutL3 and monitored SpoVG protein levels using western blots. Overexpressing SprX strongly reduces SpoVG protein levels (Figure 6). In contrast, overexpressing SprX_mutL3 was unable to inhibit SpoVG protein expression, resulting in SpoVG protein levels identical to those in the strain ΔsprX transformed with pCN38 (Figure 6). SprX_mutL3 thus failed to regulate SpoVG expression, demonstrating the importance of the SprX loop in regulating SpoVG in vivo. Northern blots indicated that the SprX_mutL3 and wt SprX are expressed at similar levels from pCN38 (Figure 6), indicating that the absence of SpoVG regulation by SprX_mutL3 is not due to its in vivo instability. Taken altogether, our in vitro and in vivo results show that SprX inhibits SpoVG expression at the translational level by antisense pairings occurring between a C-rich loop from SprX and the spoVG RBS from yabJ-spoVG mRNA.
Figure 6.
A C-rich loop from SprX is essential in reducing SpoVG expression in vivo. Upper panel: western blot analysis of the SpoVG protein levels in wt HG001 strain (wt), HG001ΔsprX (ΔsprX) and then HG001ΔsprX transformed by pCN38 (ΔsprX pCN38), pCN38-sprX (ΔsprX pCN38-sprX) or by pCN38-sprX_mutL3 (ΔsprX pCN38-sprX_mutL3). Proteins were prepared at the late-exponential phase of growth (OD600 nm of 8). Coomassie staining gel was used as a loading control (Supplementary Figure S3B). Middle panel: northern blot analysis of SprX expression at late-exponential growth phase. Lower panel: tmRNA used as an internal loading control.
SprX influences glycopeptide antibiotic resistance through its C-rich loop
The deletion of the yabJ-spoVG operon reduces the resistance to glycopeptide antibiotics in glycopeptide-intermediate-resistant S. aureus (9). Complementation by a vector allowing the expression of only the SpoVG protein was sufficient to restore the antibiotic resistance (9). We verified whether the deletion of yabJ-spoVG would affect resistance to Teicoplanin and Vancomycin in the susceptible S. aureus strain HG001. In agreement with a previous report (9), deletion of the yabJ-spoVG operon reduces bacterial resistance to both antibiotics (Figure 7A). To assess if the action of SprX in the S. aureus resistance to antibiotics involves other SprX targets in addition to SpoVG, we tested whether SprX is able to modify glycopeptide susceptibility in the absence of SpoVG. For this purpose, we transformed pCN38-sprX in strain HG001ΔyabJ-spoVG. Overexpression of SprX in this strain slightly reduced bacterial resistance to Teicoplanin and Vancomycin compared with the one transformed with pCN38 (Figure 7B), indicating that, in addition to SpoVG, SprX might control other targets involved in glycopeptide resistance. However, the difference in the Teicoplanin and Vancomycin resistance between HG001ΔyabJ-spoVG transformed by pCN38-sprX or pCN38 is small, suggesting that the contribution of putative additional SprX targets involved in glycopeptide sensibility is limited compared with SpoVG.
Figure 7.
SprX modulates S. aureus glycopeptide resistance by its C-rich sequence. (A) Ten-fold serial dilutions of overnight cultures were deposited from top (most concentrated: 107 bacteria) to bottom (100 bacteria) on MH plates or on MH plates supplemented or not with Teicoplanin or Vancomycin (0.75 µg/ml), then incubated at 37°C. Strain are HG001 (wt), HG001ΔyabJ-spoVG (ΔyabJ-spoVG) and HG001ΔyabJ-spoVG transformed by pCN38 (ΔyabJ-spoVG pCN38) or pCN38-sprX (ΔyabJ-spoVG pCN38-sprX). (B) Ten-fold serial dilutions of strains: wt HG001 strain (wt), HG001ΔsprX (ΔsprX) and then HG001ΔsprX transformed by pCN38 (ΔsprX pCN38), pCN38-sprX (ΔsprX pCN38-sprX) or pCN38-sprX_mutL3 (ΔsprX pCN38-sprX_mutL3) and HG001ΔyabJ-spoVG (ΔyabJ-spoVG) were performed and incubated on MH plated supplemented with Teicoplanin or Vancomycin (0.75 µg/ml) at 37°C. (C) Level of resistance of strains from panel B on gradient plates containing Teicoplanin.
To see if SprX modulates bacterial resistance to glycopeptide antibiotics through its C-rich domain, we tested whether SprX_mutL3 (already shown to not influence SpoVG expression) could affect this susceptibility. We have observed that the effect on antibiotics resistance of sprX overexpression is similar to yabJ-spoVG deletion (Figures 1B and 7B), and more pronounced than the one observed for sprX deletion (Figures 1C and 7B). This could be explained by the moderate effect of sprX deletion on SpoVG expression levels (about a 2-fold effect, Figure 2A), while sprX overexpression induces a severe decrease in SpoVG expression levels, resembling the yabJ-spoVG deletion (∼10-fold). However, the strain transformed by pCN38−sprX_mutL3 exhibits the same resistance levels as the one transformed with pCN38 (Figure 7B), indicating that the overexpression of SprX_mutL3 failed to modify their antibiotic susceptibility. Similar results were obtained using gradient plates, illustrating that SprX_mutL3 was unable to modify Teicoplanin resistance (Figure 7C). Collectively, our results emphasize the importance of the C-rich sequence within the third loop of SprX in regulation of SpoVG and the S. aureus susceptibility to two glycopeptide antibiotics.
DISCUSSION
Here, we report on the function of SprX (alias RsaOR), a recently identified small RNA expressed from an S. aureus pathogenicity island. We provide evidence that SprX shapes S. aureus resistance to Vancomycin and Teicoplanin glycopeptides, two invaluable antibiotics for treatment of Methicillin-resistant staphylococcal infections. To investigate the mechanism underlying this modification in antibiotic resistance, we searched for SprX targets. 2D-DiGE analysis of wild-type (wt) and isogenic sprX deletion strains allowed us to identify the SpoVG protein, whose expression is reduced by SprX. SpoVG is translated from bicistronic yabJ-spoVG mRNA transcribed from a σB-dependent promoter that is responsible for yabJ-spoVG mRNA accumulation during bacterial growth (8). The yabJ-spoVG operon is involved in capsule formation; controls the expression of extracellular lipase, nuclease and protease; and is also involved in bacterial resistance to Methicillin and glycopeptides (8,9,12).
In fact, SpoVG (not YabJ) is the major regulator of the yabJ-spoVG operon (9,12) and on its own it can rescue the phenotypes related to the deletion of the yabJ-spoVG operon. However, the molecular mechanisms underlying SpoVG action in these biological pathways remain unknown. Recently, it was reported that SpoVG is a DNA-binding protein supporting the hypothesis that it could act as a transcriptional factor (11). In this report, we found that by reducing SpoVG expression levels, SprX affects S. aureus resistance to two glycopeptide antibiotics. Moreover, among other yabJ-spoVG–related phenotypes, we tested the effect of SprX on the resistance to Oxacillin and showed that SprX modify the susceptibility to this antibiotic (Supplementary Figure S7). Further studies on the regulation of the additional phenotypes related to yabJ-spoVG will be necessary to fully understand the importance of SprX in the regulation of other bacterial processes. We have shown that SpoVG might not be the unique target of SprX involved in the glycopeptide resistance. sRNAs are known to control the expression of multiple mRNA targets, and such mechanism allows a coordinated regulation of factors, involved in joint cellular processes (40).
The effect of SprX overexpression on SpoVG expression and antibiotic susceptibility is more pronounced compared with sprX deletion, suggesting that in the wt strain grown under laboratory conditions, SprX could be present in low amounts for the regulation of SpoVG. The effect of sprX deletion on SpoVG accumulation is only 2-fold and has a small reproducible effect on antibiotic resistance. Reversely, sprX overexpression induces a severe decrease in SpoVG expression (∼10 times), which modulates the antibiotic resistance. Because sRNAs are known to allow the bacteria to respond to different environmental changes, some growth conditions could increase SprX expression above the limiting amount to decrease SpoVG expression. On the other hand, the smaller effect of sprX deletion compared with overexpression of SprX could be explained by the existence of other SpoVG inhibitors, which could decrease the SpoVG amount in the absence of SprX. Further studies on the control of SprX expression and on other SprX targets will be necessary to fully understand the network of SprX regulation.
We have uncovered the mechanism by which SprX lowers SpoVG expression. SprX interacts by antisense pairings with the spoVG ribosomal binding site (including its SD and AUG initiation codon) of the yabJ-spoVG mRNA. By mutational analysis, we identified the functional sequence within SprX that interacts with spoVG RBS: a C-rich sequence situated in an accessible loop (L3 loop) within the SprX structure. This functional sequence contains an UCCC motif, a specific conserved signature that has been detected in several previously studied S. aureus mRNAs (16). Moreover, as SprX_mutL3 does not influence the glycopeptide susceptibility (Figure 7B and C), the C-rich region of SprX is likely responsible for the regulation of other SprX targets involved in antibiotic resistance. This reinforces the notion that gene expression regulation by these sRNAs in S. aureus occurs through a shared mechanism. In vitro, SprX-mediated downregulation of SpoVG translation does not require any additional factors. This result is in agreement with previously reported S. aureus sRNAs that overcome the Hfq protein requirement for sRNA–mRNA duplex stabilization in enterobacteria (41).
The SprX-yabJ-spoVG mRNA interaction inhibits SpoVG translation initiation by preventing the binding of ribosomes to the spoVG RBS. The translation initiation inhibition is a strategy commonly used by bacterial sRNAs to control gene expression. sRNA pairing with mRNA targets can enhance or repress targeted gene translation (23,42,43). Generally, duplex formation between the mRNA target and sRNAs induces mRNA target degradation through the recruitment of bacterial ribonucleases (44). However, the strategy of translational inhibition without target mRNA degradation was shown to be sufficient to silence gene expression in both Gram-negative and -positive bacteria (28,45). Here, we show that SprX inhibits SpoVG translational initiation without inducing degradation of bicistronic yabJ-spoVG mRNA from which SpoVG is translated. Furthermore, the yabJ-spoVG mRNA is cleaved by an unknown mechanism into two separate transcripts that correspond to the yabJ and the spoVG mRNAs (8). Interestingly, SprX interacts with yabJ-spoVG mRNA in the region of the cleavage site (Figure 3A). We have shown that SprX does not affect the levels of full-length yabJ-spoVG; however, the overexpression of SprX decreases the spoVG and yabJ mRNAs levels (Figure 2C and D and Supplementary Figure S5B and C). This suggests that because SprX interacts within the region of internal processing of yabJ-spoVG mRNA, its binding influence the mRNA cleavage. It has been shown that yabJ-spoVG mRNA cleavage occurs downstream from spoVG SD nucleotide sequence (9), which makes the translation of SpoVG impossible from spoVG transcript. Moreover, SpoVG is translated from full-length yabJ-spoVG mRNA (9). The physiological functions as well as the mechanism of the yabJ-spoVG mRNA processing remain to be identified, but this cleavage could serve to irreversibly inhibit spoVG expression.
Our results raised an important question concerning the combination of sRNA-mediated translational inhibition with mRNA degradation in S. aureus. mRNA–sRNA interactions can result in mRNA target degradations; therefore, gene silencing becomes irreversible, leading to the elimination of target mRNAs and sRNAs. In contrast, in the absence of sRNA-triggered mRNA degradation, mRNA target translation would rapidly resume once the sRNAs released target repression. Moreover, as SpoVG is translated from bicistronic yabJ-spoVG mRNA, sRNA-triggered operon mRNA degradation would also affect the expression of the operon’s first protein, YabJ. We hypothesize that the strategy wherein SprX inhibits translation of SpoVG but without promotion of mRNA degradation, may allow for discoordinate gene expression in the yabJ-spoVG operon. Such a strategy of specific translational repression of a targeted gene within an operon while not modifying other operon gene expressions was described in E. coli, where Spot42 specifically inhibits galK of galETKM operon (45). We showed in vitro that SprX does not influence YabJ translation. Because the conditions of YabJ protein expression are not known (12), further studies will be necessary to investigate the conditions of YabJ expression in vivo and to show whether SprX sRNA is involved in its regulation in vivo.
In fact, sRNA-mediated operon control adds an additional layer to gene expression regulation. Diverse sRNA mechanisms for the adjustment of operon expression have been discovered. In addition to the aforementioned mechanism of specific translational inhibition of target operon genes, sRNA could also influence operon mRNA stability. sRNAs could trigger degradation of the entire operon mRNA (34), or of just a part of the operon, releasing a translationally active mRNA fragment (46). Furthermore, sRNA could also induce operon-mRNA cleavages (47). We presume that further studies on sRNA-mediated regulation of operon expression will help us to understand the complexity of their regulation, and will reveal new mechanisms of action.
Until now, sRNAs were described as being involved in the control of diverse cellular processes, including pathogenicity control. Two S. aureus regulatory sRNAs, SprD and Ssr42, have been shown to be essential for the virulence in an animal model of infection (28,29). An S. aureus paradigm for this emerging and expanding class of regulatory RNAs is RNAIII, the effector of the global agr regulon that controls the synthesis of multiple virulence factors (16,24–26). Moreover, a number of studies have linked alteration of the agr system function with Vancomycin tolerance [reviewed in (6)]. Although the mechanisms are not yet understood, the effect of agr on Vancomycin tolerance was proposed to be mediated by the expression of RNAIII (48). Recently, a case of antibiotic resistance regulation by a riboswitch was described (49). In that report, a riboswitch (a 5′ leader sequence within mRNA) was shown to control the translation of the mRNA-encoding aminoglycoside adenyl-transferase enzymes that confer resistance to aminoglycoside antibiotics. Drug binding to the mRNA leader releases the translation repression imposed by the riboswitch and induces bacterial resistance to aminoglycosides. Here, we report on a sRNA involved in bacterial resistance to antibiotics in a major human pathogen, S. aureus.
ACCESSION NUMBERS
NCTC8325 (accession no. NC_007795)
SpoVG NCTC8325 SAOUHSC_00469
GeneID:3920329
YabJ NCTC8325 SAOUHSC_00468
GeneID:3920328
RNAIII NCTC 8325 SAOUHSC_02260
GeneID:3919680
(Gene accession number: X52543)
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut National de la Santé et de la Recherche Médicale (INSERM), the Rennes University (Défis Scientifiques Emergents), the Région Bretagne (‘allocation doctorale’, PhD thesis) and the Fondation pour la Recherche Médicale [FRM; FDT20130928407 to A.E.]; Agence Nationale de la Recherche [ANR-09-MIEN-030-01 to B.F.]. Funding for open access charge: Inserm.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr B. Berger-Bächi (Zurich, Switzerland) for the plasmid pSTM33 and the RN4220ΔyabJ-spoVG strain. We thank Dr M. Hallier and Y. Le Pétillon from the lab for the technical assistance and Pr. P.Y. Donnio (Rennes University) for helpful discussions and advices.
REFERENCES
- 1.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 2.Boger DL. Vancomycin, teicoplanin, and ramoplanin: synthetic and mechanistic studies. Med. Res. Rev. 2001;21:356–381. doi: 10.1002/med.1014. [DOI] [PubMed] [Google Scholar]
- 3.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 2006;42(Suppl. 1):S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 4.Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 1997;40:135–136. doi: 10.1093/jac/40.1.135. [DOI] [PubMed] [Google Scholar]
- 5.Liu C, Chambers HF. Staphylococcus aureus with heterogeneous resistance to vancomycin: epidemiology, clinical significance, and critical assessment of diagnostic methods. Antimicrob. Agents Chemother. 2003;47:3040–3045. doi: 10.1128/AAC.47.10.3040-3045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010;23:99–139. doi: 10.1128/CMR.00042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renzoni A, Andrey DO, Jousselin A, Barras C, Monod A, Vaudaux P, Lew D, Kelley WL. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS One. 2011;6:e21577. doi: 10.1371/journal.pone.0021577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier S, Goerke C, Wolz C, Seidl K, Homerova D, Schulthess B, Kormanec J, Berger-Bachi B, Bischoff M. sigmaB and the sigmaB-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus. Infect. Immun. 2007;75:4562–4571. doi: 10.1128/IAI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulthess B, Meier S, Homerova D, Goerke C, Wolz C, Kormanec J, Berger-Bachi B, Bischoff M. Functional characterization of the sigmaB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob. Agents Chemother. 2009;53:1832–1839. doi: 10.1128/AAC.01255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuno K, Sonenshein AL. Role of SpoVG in asymmetric septation in Bacillus subtilis. J. Bacteriol. 1999;181:3392–3401. doi: 10.1128/jb.181.11.3392-3401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jutras BL, Chenail AM, Rowland CL, Carroll D, Miller MC, Bykowski T, Stevenson B. Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins. PLoS One. 2013;8:e66683. doi: 10.1371/journal.pone.0066683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulthess B, Bloes DA, Francois P, Girard M, Schrenzel J, Bischoff M, Berger-Bachi B. The sigmaB-dependent yabJ-spoVG operon is involved in the regulation of extracellular nuclease, lipase, and protease expression in Staphylococcus aureus. J. Bacteriol. 2011;193:4954–4962. doi: 10.1128/JB.05362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulthess B, Bloes DA, Berger-Bachi B. Opposing roles of sigmaB and sigmaB-controlled SpoVG in the global regulation of esxA in Staphylococcus aureus. BMC Microbiol. 2012;12:17. doi: 10.1186/1471-2180-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 2010;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abu-Qatouseh LF, Chinni SV, Seggewiss J, Proctor RA, Brosius J, Rozhdestvensky TS, Peters G, von Eiff C, Becker K. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J. Mol. Med. 2010;88:565–575. doi: 10.1007/s00109-010-0597-2. [DOI] [PubMed] [Google Scholar]
- 19.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One. 2010;5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezee-Durant E, Barbet R, Jacquet E, Jacq A, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillet J, Hallier M, Felden B. Emerging functions for the Staphylococcus aureus RNome. PLoS Pathog. 2013;9:e1003767. doi: 10.1371/journal.ppat.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomasini A, Francois P, Howden BP, Fechter P, Romby P, Caldelari I. The importance of regulatory RNAs in Staphylococcus aureus. Infect. Genet. Evol. 2013;21:616–626. doi: 10.1016/j.meegid.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevalier C, Boisset S, Romilly C, Masquida B, Fechter P, Geissmann T, Vandenesch F, Romby P. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 2010;6:e1000809. doi: 10.1371/journal.ppat.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chabelskaya S, Gaillot O, Felden B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010;6:e1000927. doi: 10.1371/journal.ppat.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison JM, Miller EW, Benson MA, Alonzo F, III, Yoong P, Torres VJ, Hinrichs SH, Dunman PM. Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative. J. Bacteriol. 2012;194:2924–2938. doi: 10.1128/JB.06708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herbert S, Ziebandt AK, Ohlsen K, Schafer T, Hecker M, Albrecht D, Novick R, Gotz F. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 2010;78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 35.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 36.Gillaspy AF, Worrell V, Orvis J, Roe BA, Dyer DW, Iandolo JJ. In: Gram-positive Pathogens. Fischetti V, Ferretti RNJ, Portnoy D, Rood J, editors. Washington, DC: ASM Press; 2006. pp. 381–412. [Google Scholar]
- 37.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tjaden B. Computational identification of sRNA targets. Methods Mol. Biol. 2012;905:227–234. doi: 10.1007/978-1-61779-949-5_14. [DOI] [PubMed] [Google Scholar]
- 39.Cao Y, Wu J, Liu Q, Zhao Y, Ying X, Cha L, Wang L, Li W. sRNATarBase: a comprehensive database of bacterial sRNA targets verified by experiments. RNA. 2010;16:2051–2057. doi: 10.1261/rna.2193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Jousselin A, Metzinger L, Felden B. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol. 2009;17:399–405. doi: 10.1016/j.tim.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Frohlich KS, Vogel J. Activation of gene expression by small RNA. Curr. Opin. Microbiol. 2009;12:674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 43.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lalaouna D, Simoneau-Roy M, Lafontaine D, Masse E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim. Biophys. Acta. 2013;1829:742–747. doi: 10.1016/j.bbagrm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desnoyers G, Morissette A, Prevost K, Masse E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 2009;28:1551–1561. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Opdyke JA, Fozo EM, Hemm MR, Storz G. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J. Mol. Biol. 2011;406:29–43. doi: 10.1016/j.jmb.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulander W, Nissen Varming A, Baek KT, Haaber J, Frees D, Ingmer H. Antibiotic-mediated selection of quorum-sensing-negative Staphylococcus aureus. MBio. 2013;3:e00459–e00412. doi: 10.1128/mBio.00459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia X, Zhang J, Sun W, He W, Jiang H, Chen D, Murchie AI. Riboswitch control of aminoglycoside antibiotic resistance. Cell. 2013;152:68–81. doi: 10.1016/j.cell.2012.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.