Abstract
Cys2-His2 zinc finger proteins (ZFPs) are the largest family of transcription factors in higher metazoans. They also represent the most diverse family with regards to the composition of their recognition sequences. Although there are a number of ZFPs with characterized DNA-binding preferences, the specificity of the vast majority of ZFPs is unknown and cannot be directly inferred by homology due to the diversity of recognition residues present within individual fingers. Given the large number of unique zinc fingers and assemblies present across eukaryotes, a comprehensive predictive recognition model that could accurately estimate the DNA-binding specificity of any ZFP based on its amino acid sequence would have great utility. Toward this goal, we have used the DNA-binding specificities of 678 two-finger modules from both natural and artificial sources to construct a random forest-based predictive model for ZFP recognition. We find that our recognition model outperforms previously described determinant-based recognition models for ZFPs, and can successfully estimate the specificity of naturally occurring ZFPs with previously defined specificities.
INTRODUCTION
Defining the grammar underlying the transcriptional regulatory elements within the human genome remains a critical step in understanding both developmental and disease processes (1). The advent of high-throughput sequencing technology has fueled the development of methodologies for the genome-wide characterization of regulatory features, such as global histone modifications (1–10). These data coupled with global analysis of RNA transcript levels (6,11), chromatin immunoprecipitation (ChIP)-based occupancy data for sequence-specific transcription factors (TFs) (7,12–14) and chromatin conformational capture techniques (15) provide a framework for deconvoluting regulatory networks directing gene expression patterns (16,17). Currently, only a small subset of human TFs has been characterized by ChIP-based approaches in any given cell line (7,13,14), although some sequence occupancy can be inferred from DNaseI (12,17) and MNase (18) data. In the absence of genome-wide binding data, knowledge of the DNA-binding specificities of the TFs within regulatory networks in concert with data sets on sequence conservation, chromatin accessibility and histone modifications can be exploited by computational algorithms to predict TF genomic occupancy, and thereby construct more elaborate transcriptional regulatory models (1,9,17,19–24). Given the difficulty in characterizing the diverse binding patterns of all expressed TFs in all possible temporal and spatial expression patterns in vertebrates, the ability to estimate the specificity of the constellation of TFs expressed at any given time in a given cell type provides a critical data set for constructing these regulatory models.
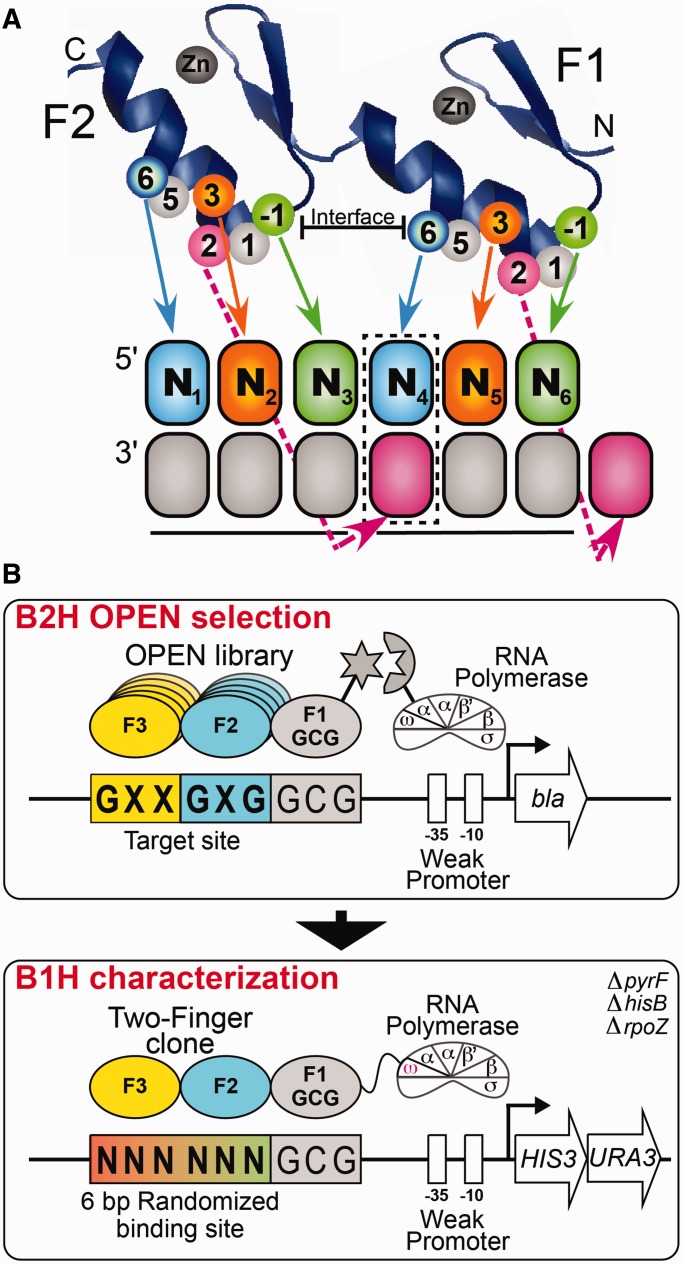
Cys2-His2 zinc finger proteins (ZFPs) are the largest class of TFs within most metazoans (25), with an estimated 675 members in the human genome (26) harboring an average of 8.5 finger units per gene (27). The majority of these ZFPs are believed to be involved in DNA-recognition, as many of the neighboring fingers are connected by a Krüppel-type TGE(K/R)P linker, which is a hallmark of DNA-binding fingers (28). The canonical DNA-recognition model for an individual finger is based on the ZFP-DNA co-crystal structure of Zif268 (29,30) and other naturally occurring and engineered ZFPs (31–35), wherein each finger potentially recognizes a 4-bp subsite that overlaps the recognition site of the neighboring N- and C-terminal fingers by 1 bp (Figure 1A). Amino acid residues at positions −1, +2, +3 and +6 of the recognition helix typically mediate the recognition preference of a finger within its subsite. The target site preference of a tandem array of fingers reflects a complex interaction between the individual finger modules, as the recognition properties of an individual finger can be influenced by its position within an array and the recognition determinants displayed by its immediate neighbors (36–41).
Figure 1.
(A) Schematic representation of the canonical recognition pattern of two zinc fingers recognizing a hexamer sequence. Each zinc finger unit spans ∼30 amino acids and folds into a ββα-motif around a tetrahedrally coordinated zinc ion (42,43). DNA-binding specificity is typically mediated by residues at positions −1, +2, +3 and +6 of the recognition helix, where the numbering scheme refers to the position of each residue relative to the start of the α-helix. The boxed base pair (N4) represents the position of potential recognition overlap in the canonical recognition model. (B) Schematic representation of the two-stage process used to identify two-finger modules with the desired sequence preference. In Stage 1, the B2H system is used to select two-finger modules from an OPEN-based library, where the finger pools used correspond to the finger 2 (F2) and finger 3 (F3) subsites in each target site (44,45). These two-finger libraries are selected in the context of a constant finger 1 (F1) module that recognizes GCG in the neighboring subsite. The DNA-binding specificity of active clones recovered from the B2H selection was determined using the B1H system using a 6-bp randomized library adjacent to the constant GCG F1 binding site. The recovered binding sites are determined by Illumina sequencing and then a binding site motif is calculated from these sequences (46).
DNA-binding specificities have been determined for only a small fraction of ZFPs in metazoan genomes (13,17,26,47–50). Unlike other TF families where the majority of the resident factors in diverse species share a high degree of homology (26,51–54), evolutionary analysis of ZFPs indicates that a substantial fraction of resident members do not have highly conserved homologs across metazoans. Instead, the number and composition of fingers within these ZFPs is dynamic between species (27,55,56) and can even vary within a species [e.g. the variation in human PRDM9 isoforms (57,58)]. The specificity determinants within these ZFPs are under strong positive selection, implying the rapid diversification of their recognition potential (27). Consequently, naturally occurring ZFPs can specify a wide variety of different DNA sequences based on both the number and composition of fingers within the array.
Although some principles that govern the recognition properties of zinc fingers have been established, the accurate prediction of their DNA-binding specificity remains challenging. Specificity determinants at individual recognition helix positions with defined base preferences have been extracted from the biochemical and structural characterization of naturally occurring ZFPs (42,47,49,50,59–61) and the selection and characterization of artificial ZFPs that recognize novel target sequences (37,38,41,44,62–74). These data provide a foundation for the construction of predictive recognition models that estimate DNA-binding specificity based on the sequence of the recognition helix of each incorporated finger. Initial models focused on using the amino acid identity at key determinant positions (−1, +2, +3 and +6) to estimate the base preference at their primary DNA contact positions within the DNA subsite bound by each individual finger (75–77). Recently, more advanced predictive models have been constructed with improved performance that incorporate context-dependent recognition, which allows determinants to influence more binding site positions than prescribed by the standard recognition model (76–82). However, the construction of these models has been hampered by the limited amount of existing quantitative specificity data for ZFPs that links individual fingers with recognition of particular subsites.
A comprehensive recognition model for canonically binding ZFPs should be achievable using the growing archive of quantitative specificity data from recent bacterial one-hybrid (B1H) analysis of a large number of artificial (41,62,71) and naturally occurring ZFPs (49,50), where the position of each finger within the recognition sequence is defined or can be inferred. This data set spans 678 two-finger modules, including the characterization of 95 two-finger modules generated using the Oligomerized Pool ENgineering (OPEN) system (44,45) described herein. A sizeable fraction of these data explicitly examines the impact of recognition residues at the finger–finger interface on the preferred specificity at the junction of the finger binding sites, which remains the most challenging recognition feature to model. These data permit an improved estimation of context-dependent effects requiring the use of predictive models [such as support vector machine (83) or random forests (RFs) (84)] that implicitly capture these complex properties. Building on our previous efforts using RF models to estimate the specificity of homeodomains (85), we have constructed an RF predictive model for ZFPs using our B1H data that are superior to existing predictive models and that can effectively estimate the DNA-binding specificity of a number of naturally occurring ZFPs.
MATERIALS AND METHODS
OPEN finger selections
OPEN selections were performed to generate a set of two-finger modules that recognize all 64 possible GNNGNG-type sequences in the context of an N-terminal ‘GCG’ binding anchor zinc finger (recognition helix: RSDTLAR). All target sites used in the selection of novel recognition fingers were of the form GNNGNGGCG. Zinc finger libraries for each target site were assembled from the corresponding Finger 2 and Finger 3 OPEN pools as previously described but with a fixed Finger 1 module (44,45). OPEN selections were performed essentially as previously described (44,45) but using a beta-lactamase (bla) antibiotic-resistance gene instead of the HIS3 gene (70). For each of the 64 selections, we assayed the abilities of up to five clones to activate expression of a lacZ reporter gene in a bacterial two-hybrid (B2H) system as previously described (45) and determined the amino acid sequences of these clones. Fifty-eight of the 64 selections displayed active clones, from which we chose 95 clones that could activate expression of lacZ in the B2H system by ∼2.5-fold or more for further evaluation via B1H binding site selections (Supplementary Table S1).
CV-B1H method
To determine binding site specificities of OPEN-selected and other 2F-modules, the CV-B1H (Constrained Variation Bacterial one-Hybrid) assay was performed essentially as described previously (46). Two-finger modules were evaluated as fusions to the GCG anchor finger. Following transformation into the selection strain, 1 × 106 cells containing the zinc finger plasmid (1352-omega-UV2-ZFP) and the 6-bp randomized binding site library (in pH3U3) were plated on selective NM minimal medium plates (100 × 15 mm) containing 50 µM IPTG and 1 or 2 mM 3-AT and grown at 37°C for 22–30 h. All cells on the plate were pooled, and the pH3U3 plasmids containing the compatible binding sites were isolated for identification of the functional DNA sequences. The binding site region was PCR amplified, barcoded and sequenced via Illumina sequencing, and then binding specificities were determined from these data using GRaMS modeling and the log-odds method (46,71,86).
Construction of the RF ZFP regression model
Based on a pilot study and previous work with homeodomain recognition modeling (85), we developed a recognition modeler based on a RF regression approach (84) using the ‘randomForest’ module from the R package [http://www.r-project.org/(87)]. Two different ZFP RF regression models were trained based on the B1H specificity data: one-finger and two-finger models. The training data for the two-finger model consisted of 678 protein sequences for two fingers of ZFPs and the position frequency matrices (PFMs) obtained from the B1H experiments described above. The one-finger model was trained on the same set but contained 1209 individual fingers (redundancy removed, Supplementary Table S2). Preliminary analysis showed that including additional protein positions beyond the canonical −1, +2, +3 and +6 recognition positions in each finger did not improve the accuracy of the model, so all further training used only those positions. Of the 678 two-finger examples, there are 530 unique combinations of residues at positions −1, +2, +3 and +6; all of them are kept in the data set because the PFMs, while similar between repeats, are not identical and this maintains the inherent variability in the data. These models use the RF regression engine that was previously described (85). The modeler predicts the PFM for a zinc finger protein based on its sequence at the recognition positions, and the RF regression minimizes the mean-squared error (MSE) between the predicted and observed PFMs. MSE values for a single position can range from 0, if the two PFMs are identical, to 0.5 if they contain probabilities of 1.0 for different bases. A random position (probability of 0.25 for each base) would have a maximum MSE of 0.1875 compared with a position with probability of 1.0 for any base. This has the effect of generating PFMs that tend toward random at some positions instead of making high probability predictions that are frequently incorrect.
We used the default value of 500 trees while training the RF model. In this model, a single tree picks predictive variables, specific amino acids at specific positions, randomly and then applies regression to estimate their contribution to each PFM parameter. The set of individual trees are then weighted by regression to minimize the overall MSE between the observed and predicted PFMs. Accuracies were determined by 10-fold cross-validation, where the total data set was divided into 10 subsets and training was based on nine of them and the accuracy measured on the remaining subset. Each of the subsets was left out in turn, and the testing accuracy is reported as the means and medians on the test sets.
We chose to minimize MSE because we are specifically trying to find optimal PFMs that fit the entire distribution of binding site affinities. However, other objectives could be used instead. There have been a large number of different methods proposed to compare motifs with each other and determine a quantitative measure of similarity (88–94). The MSE that we use is closely related to maximizing the Pearson correlation and is often a highly ranked method, particularly when trying to assign a motif to a specific class of transcription factors. In other approaches more emphasis is put on matching high information content positions in the binding sites and low information content positions are scored similar to mismatches. For example, the recently published zinc finger predictor from the Princeton group (82) specifically maximizes the number of correctly predicted positions with high information content, which has advantages for some purposes (see later in the text).
Construction of ZFP recognition motif predictions
We established a Web site that will predict the binding motif for an input ZFP containing any number of fingers (http://stormo.wustl.edu/ZFModels/). ZFP sequences can be submitted in two forms as follows: a concatenation of the four critical recognition residues of each finger (−1, +2, +3 and +6) or the entire protein sequence. In the latter case, the Web site will determine the locations of the recognition residues in each finger based on a HMMER analysis (95) of zinc finger motifs present within the sequence. Three different ZFP motif generation methods are available based on the trained RF regression models: one-finger model, multi-finger model and the average of these models. In the one-finger model, the predictions are based on training of single fingers, and the complete motif is predicted by concatenating the individual predictions. In the multi-finger model, the predictions are based on the two-finger training data, and the complete motif is stitched together from the overlapping two-finger predictions, where the positions of overlap between the motifs are averaged (Supplementary Figure S1). The third method averages together the prediction from the one-finger and two-finger models to generate the final prediction. Generally, the different predictions are in close agreement but sometimes there is a divergence and the most accurate may depend on the specific zinc finger protein; therefore, we advocate testing with each model to examine the inherent variation.
Evaluation of Bcl6 predictive motif for predicting ChIP-seq peaks
The predicted DNA-binding specificity of Bcl6 was estimated using the multi-finger model through the ZFModels interface. The top 100 ChIP-seq peaks for Bcl6 (96) were extracted using Galaxy (97), and a motif for Bcl6 was extracted from these peaks using MEME (zoops mode) (98). MSE was calculated from this PFM against different motifs as described above. FIMO (99) was used to determine the number of the top 100 ChIP peaks containing favorable Bcl6 binding sites (P < 10−4) based on each motif.
RESULTS
Selection and characterization of two-finger modules recognizing GNNGNG target sites
We used OPEN selections (44,45) to identify two-finger modules recognizing 64 different 6-bp target sites of the form GNNGNG (Figure 1B). This set of target sites was chosen to include a focused set of sequences that were available in the OPEN system to explore the quality of the B2H-generated fingers. In addition, for the defined target positions (constant guanines), there are strong expectations about the complementary recognition determinants that would be selected. Deviations from the expected residues in the recovered sequences would be indicative of context-dependent effects. These two-finger modules were selected via the B2H system in the context of a three-finger array harboring a fixed N-terminal anchor finger that recognizes a GCG subsite. Fifty-eight of these selections yielded zinc finger arrays that bound their target site as evidenced by their ability to activate transcription in a B2H lacZ reporter assay (Supplementary Table S1).
We determined the DNA-binding specificity of a representative set of the B2H-selected two-finger modules using the B1H system (49,71). Each two-finger module was characterized using a reporter system containing a 6-bp randomized binding site library adjoining the finger 1 recognition element—GCG (46,71) (Figure 1B). After selection, surviving colonies carrying the functional DNA-sequences for each two-finger module recovered from this library were pooled and characterized by Illumina sequencing from which a preferred recognition motif was determined (46). This analysis yielded motifs for 95 OPEN-selected two-finger modules (Supplementary Figure S2). For 64 of these two-finger modules, the preferred recognition sequence matched the expected target site. The remaining modules are complementary to their target sequence, but actually prefer a related binding site. These modules expand the population of characterized two-finger modules for the construction of artificial zinc finger arrays, and the coupled specificity data provide additional information on the recognition potential of specific determinant combinations for the construction of improved predictive models.
Assessing context dependence in our selected two-finger modules
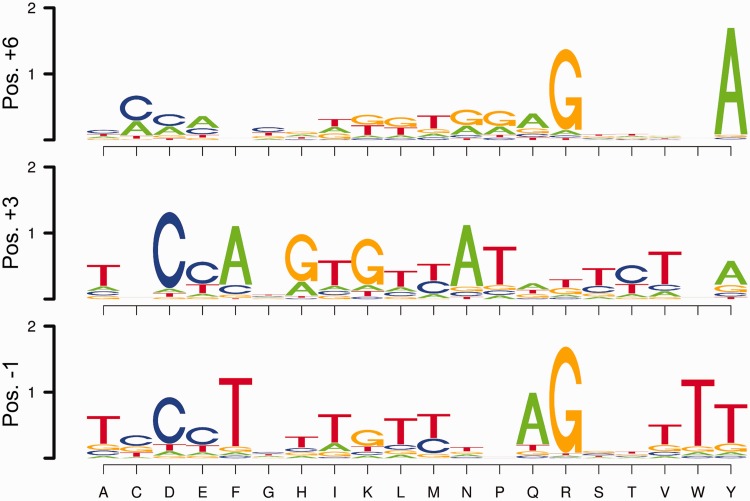
As a basis set for constructing predictive recognition models for ZFPs, we have used quantitative B1H specificity data on a large group of naturally occurring (49,50) and artificial (41,62,71) zinc finger arrays. To facilitate the evaluation of DNA-recognition by these zinc fingers, we have parsed this data set into 1209 different one-finger modules or 678 different two-finger modules. For example, a characterized three-finger array is broken down into three one-finger modules or two overlapping two-finger modules with their associated subsite motifs (Supplementary Figure S1). Figure 2 shows the base preferences at base pair positions 1, 2 and 3 within the core subsite (contacted by specificity determinants at positions +6, +3 and −1, respectively; see Figure 1) for this data set of one-finger modules. In general, the observed amino acid to base correlations at each position are consistent with previous studies of recognition preferences for zinc finger proteins (42,43,50,76–78). The strongest correlations are observed at the central base; amino acid changes at position +3 in the recognition helix primarily influenced recognition at the middle base position of the altered finger subsite in our two-finger modules when examined over the data set (Supplementary Figure S3). The independence of recognition at this position was previously harnessed to expand the recognition diversity of our two-finger modules in a directed manner in many instances (71).
Figure 2.
Base preferences observed across the data set for specificity determinants at each of the canonical recognition positions (+6, +3 and −1). For each amino acid (X-axis) at the finger positions +6 (top), +3 (middle) and −1 (bottom), the corresponding base preferences, averaged over all examples, are garnered from the B1H-determined recognition motifs. Base preferences at binding site position 1 are indicated for position +6 specificity determinants; base preferences at binding site position 2 are indicated for position +3 specificity determinants; base preferences at binding site position 3 are indicated for position −1 specificity determinants.
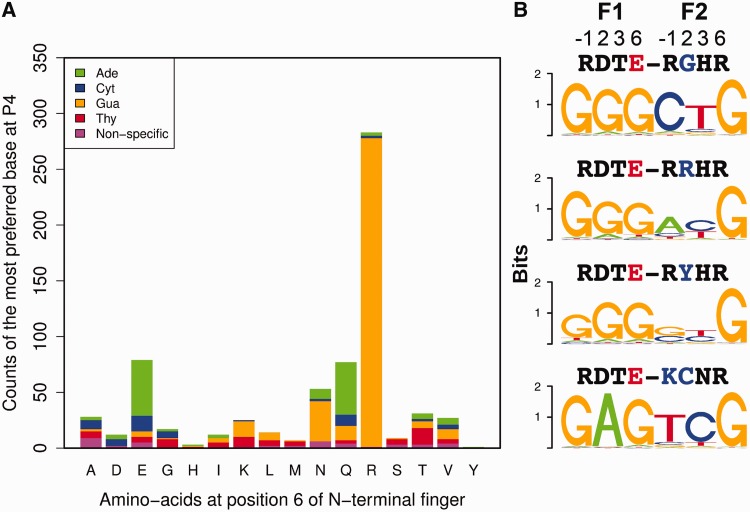
Weaker correlations at other positions highlight the role of context on specificity. The influence of context dependence on the DNA-binding specificity of individual fingers is apparent from a qualitative analysis of finger sets within our data set, particularly at the finger–finger interface for a subset of two-finger modules where residues on both sides of the interface were randomized to more effectively capture these effects (Figure 1A) (62,71). For many individual two-finger modules, the base at position 4 is highly specified. However, when the preferred specificity at this position is binned across the data set based on the type of residue at position +6 of the N-terminal finger (Figure 3A), some amino acids are associated with each of the four bases in different C-terminal finger contexts. Glutamate at position +6 provides a notable example, where two-finger modules containing this residue display distinct preferences for each of the four bases at position 4 (Figure 3B). The potential influence of residues within the C-terminal finger, in particular the residue at position +2, on recognition at base position 4 are well documented (29,31,38,100). Consistent with the potential influence of position +2 on recognition, changes in the residue at position +2 in the recognition helix in many instances appear to influence neighboring base preference, particularly at position 4 (Supplementary Figure S4). These data highlight the need for a predictive model that can capture the influence of each determinant position on multiple base positions within the zinc finger recognition sequence.
Figure 3.
Context-dependent preferences observed for the base at position 4 (P4) recognized by the two-finger modules across the entire data set. (A) Stacked bar plot showing the distribution of base preferences dictated by each amino acid at position +6 of N-terminal finger in a two-finger module. The height of each bar corresponds to the number of zinc finger modules with the amino acid labeled on the X-axis. The height of each colored bar segment corresponds to number of modules preferring a particular base. Preference was defined as nonspecific if the information content at a position is <0.3. (B) Examples of context-dependent preference at position P4. Logos representing the specificity of four different two-finger modules with Glu at position +6 (red) of N-terminal finger with different base preferences at P4. Above each observed motif are the amino acids at the four canonical recognition positions (−1,+2,+3 and +6) for the N-terminal and C-terminal fingers.
RF recognition models for ZFPs
Zinc fingers have been the focus of several studies on qualitative recognition codes [reviewed in (42,43)]. More recently, several groups have developed models that predict quantitative motifs for zinc finger proteins based on the residues present at canonical recognition positions within each finger (76–79). Although superior to purely qualitative recognition codes, their accuracies leave considerable room for improvement. These models were limited because they were trained primarily on qualitative data: collections of proteins and their binding sites with high binding affinity, but where the preference of each ZFP for its target site relative to other sequences was unknown. Our B1H-characterized zinc finger data provide a much larger training set with quantitative information about the preferences of different proteins for different DNA binding sites, which allows us to train new recognition models to obtain higher accuracy predictions. In pilot studies, we tested the feasibility of creating recognition models using several different machine learning algorithms, including neural networks (78), support vector machines (83), k-nearest neighbors (101), partial linear regression (102) and RF (84). We found that RF-based models performed as well or better than those of other methods and its implementation was computationally less demanding, so we used an RF regression algorithm to create a predictive model for ZFPs. The results of these preliminary studies were similar to those we previously reported for predicting the specificity of homeodomain proteins (85).
We trained RF predictive models on either one-finger or two-finger module specificity data, where the latter model is designed to capture context-dependent effects between neighboring fingers. Training the two-finger model takes as input the amino acids at the eight canonical recognition positions (−1, +2, +3 and +6 of each finger) and builds regression trees to predict recognition preference over the entire 6-bp binding site. (The one-finger model was similarly trained on individual fingers and each 3-bp binding site.) Importantly, these models are not restricted to the canonical interactions between particular finger recognition positions and bases within the binding site, unlike many previous recognition models (76,77). Because we have a much larger training set than was available for previous models, a wider range of potential interactions between these recognition positions and the binding site are allowed within the model to capture context-dependent effects observed within the data. Consequently, each recognition position within the two-finger module contributes to the overall predicted PFM, although the strongest contributions within the model will be between the most highly correlated amino acids and base pairs.
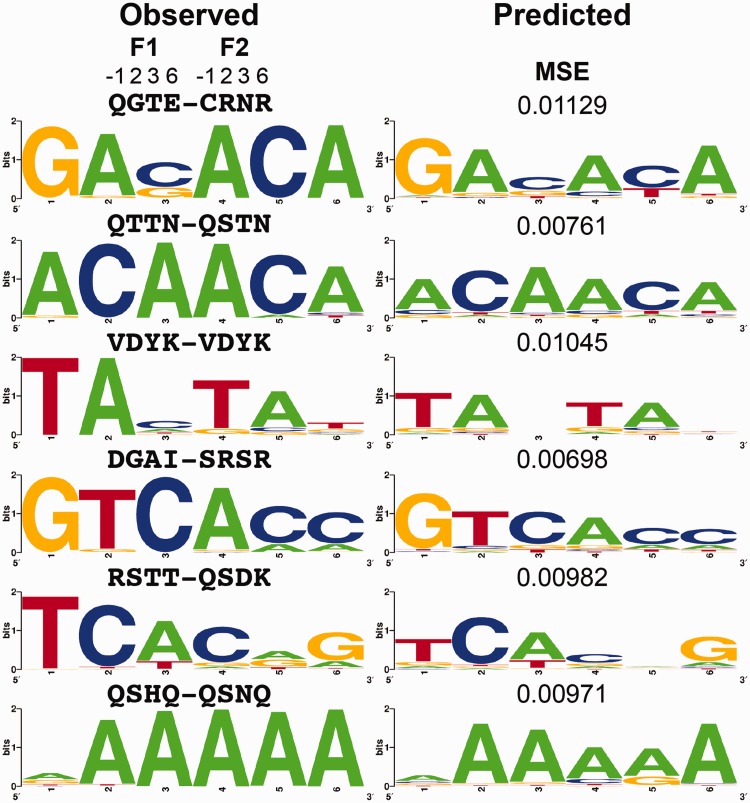
The objective during model training is to minimize the MSE between the observed and predicted PFM values for each two-finger module. Table 1 shows the average value (both the mean and median with standard deviations) obtained in a 10-fold cross-validation of our two-finger model. This was compared with predictions by each of four other published models that were readily available for testing (76–79). The MSE is greatly reduced with the new ZFModels predictions to less than half for means and less than one-third for medians when compared with other prior models. The prediction error is fairly evenly distributed across the positions of the binding sites (Table 2). Figure 4 displays several examples that are near the median value of MSE to show the degree of similarity between observed and predicted PFMs. Many of the highest accuracy examples contain guanine at positions 1 and 6 because the training set was biased with fingers recognizing guanine at these positions. Figure 4 highlights examples deviating from this pattern, demonstrating that our ZFModels can generate accurate predictions for a wide variety of different types of motifs. As expected, the two-finger predictive model can capture the context dependence at the finger–finger junction observed in our data set, such as the motifs in Figure 3B, whereas the one-finger predictive model fails to capture this subtlety (Supplementary Figure S5).
Table 1.
MSE for several prediction programs
Table 2.
MSE for each position, for one-finger and two-finger models (mean/median)
| Nucleotide position | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 finger | 0.016/0.004 | 0.015/0.005 | 0.008/0.001 | |||
| 2 fingers | 0.006/0.001 | 0.007/0.003 | 0.006/0.001 | 0.012/0.004 | 0.010/0.004 | 0.004/0.000 |
Note: The reported median values represent the bin the median value falls in, where the bins are 0.001 wide and labeled with the lower value. So if the median value is reported as 0.000 that means the median is in the bin between 0.000 and 0.001. These values come from training and testing on the complete data rather than from cross-validation, resulting in lower values than in Table 1.
Figure 4.
Examples of observed motifs for two-finger modules that are within our data set, and predicted motifs for these fingers using our final predictive model. Above each observed motif are the amino acids at the four canonical recognition positions (−1, +2, +3 and +6) for the N-terminal and C-terminal fingers. The MSE value between the observed and predicted PFMs is displayed above the predicted motif.
Evaluating the utility of the RF-based zinc finger recognition model
Several published studies have determined specificity of ZFPs using SELEX (26,103–105). None of these examples were included in the training data and so they constitute an independent test set. Supplementary Figure S6 contains the logos from the published PFMs for a subset of these ZFPs and the logos predicted by ZFModels. In every case, the predictions match preferred binding sites from the experiments when we take into account the variable spacing between neighboring fingers due to noncanonical linkers in some instances. However, the quantitative models are less consistent than the average fits to zinc fingers within our data set via cross-validation analysis (Supplementary Table S3). This may be due to the SELEX data being evaluated after multiple rounds of selection where the resulting PFM is heavily weighted toward a subset of the highest affinity sites, leading to an over-specified motif. We also compared the ZFModels predictions on some of the same data sets with the predictions made by a recently published method (zf.princeton.edu) based on support vector machine training (83). ZFModels makes more accurate predictions as measured by MSE (Supplementary Table S4) on these independent test sets than the Princeton model, although the Princeton model often contains more matching positions with high information content (see Discussion).
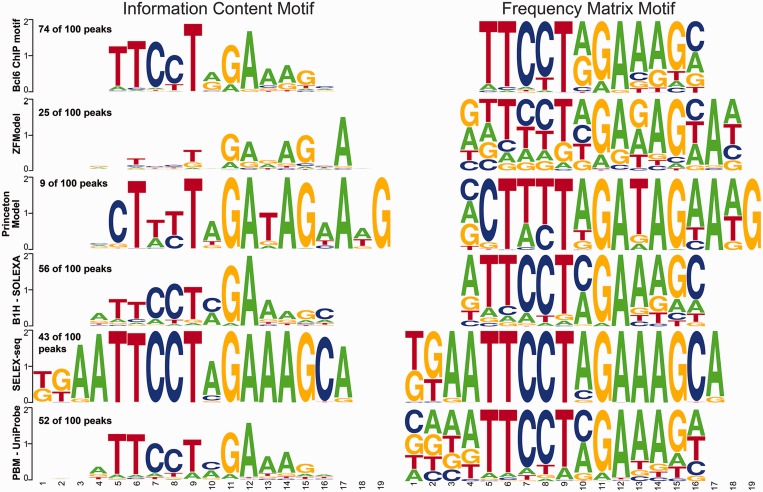
Ideally, our recognition model would also allow prediction of ZFPs with uncharacterized DNA-binding specificity throughout the genome. We chose to evaluate its predictive utility for Bcl6, as this ZFP has been characterized by B1H (50), PBM (47) and SELEX-seq (26), which allows a comparison of our predictive motif against DNA-binding specificities determined via multiple methods, and against ChIP-seq data for this factor (96). The Bcl6 recognition motifs produced by B1H, PBM and SELEX-seq are all similar, although the SELEX-seq motif appears over-specified (Figure 5). We also generated a predicted recognition motif for Bcl6 using the Princeton SVM model for comparison with our model. The Princeton motif has greater information content than our ZFmodel motif, but at many positions, the Princeton motif predicts a particular base with absolute certainty, which much like the SELEX-seq motif suggests that it is over-specified. When judged against an independent source, a MEME (98) motif from the top 100 Bcl6 ChIP-seq peaks (96), the B1H and PBM motifs appear most similar. The ZFModels multi-finger predictive model also shows good similarity to the determined motifs (MSE values 0.04 from the MEME-ChIP motif, 0.05 from either the PBM- or B1H-based motifs, 0.05 from the Princeton motif and 0.08 from the SELEX-seq motif), but it is a bit worse than the average value of <0.01 in our cross validation studies. FIMO analysis (99) of these ChIP peaks using each motif confirms this assessment: the MEME-derived motif from the Bcl6 ChIP data discovers a good Bcl6 binding site (P < 10−4) in 74 of 100 peaks, the B1H motif in 56 of 100 peaks, the PBM motif in 52 of 100, the SELEX-seq motif in 43 of 100, the ZFModels predicted motif in 25 of 100 and the Princeton motif in 9 of 100, where only four would be expected by chance. Thus, our predictive motif has value for the discrimination of binding sites within the genome, and in this example is superior to the Princeton motif, but it can still benefit from the incorporation of additional experimental data to improve its quality. Figure 5 displays logos in two formats, the original information-based method (106) and a PFM-based method where the height of each base is proportional to its frequency in the model (107). The frequency representation demonstrates that even in cases where our model does not make a confident (high probability and high information content) prediction, it generally gets the preferred base correct. Combining all of the experimental models with the MEME model from the ChIP-seq data, one finds a consensus sequence of TTCCTnGAAAG (positions 5–15 in the alignment). Our model agrees at every position except 13, where it prefers G slightly to A, but many of those predictions are low confidence. In contrast, the Princeton model has more high information content positions that match the consensus, but it also contains several positions where the preferred base is assigned a very low probability. Our model has an overall better fit to the other models, as evaluated by MSE and similarities to the rank distributions of all possible binding sites, but there are some purposes for which maximizing the number of high confidence, correct predictions is useful (see ‘Discussion’ section).
Figure 5.
Comparison of the MEME motif from the top 100 Bcl6 ChIP peaks (96) with the motif predicted for the five canonically linked fingers by ZFModels and the Princeton SVM method (82) and the recognition motifs determined directly for Bcl6 by B1H (50), SELEX-seq (26) and PBM (47). The left column displays the motifs as information content, whereas the right column displays the motifs as position frequency plots. The frequency of a strong motif match (P < 10−4) for each motif in the top 100 ChIP peaks as determined by FIMO is indicated above each motif.
DISCUSSION
The development of platforms for rapidly characterizing the specificity of transcription factors has dramatically increased the amount of data that is available for all of the major TF families (108), but there are still barriers to generating data for all naturally occurring ZFPs. The average number of fingers in a human ZFP is 8.5 (27), and these polydactyl (i.e. many fingered) ZFPs may have complex binding modes due to the presence of independent DNA-recognition modules. For example, genome-wide ChIP analysis of NRSF (109,110), a 9-finger ZFP, recovered two different types of binding sites: a prominent motif that contains a juxtaposition of two subsites and a set of additional motifs with variable spacing between these subsites. Taipale and colleagues noted the difficulty in characterizing ZFPs by either SELEX-seq or PBM (26): they successfully characterized only 8% of ZFPs and only 3% with more than eight fingers (26). Similarly, our B1H motif set includes only seven naturally occurring ZFPs with ≥8 fingers with a success rate of ∼38% of the attempted Drosophila ZFP genes (50). With the possibility that polydactyl ZFPs use different finger sets to bind multiple distinct motifs, describing their recognition properties is critical to understanding their regulatory mechanisms. The growing body of quantitative specificity data for naturally occurring and artificial ZFPs provides a foundation for the development of improved predictive models for this family to help facilitate a broader understanding of their function as regulators within the genome, where other direct analysis methods may be challenging to use.
Our efforts to construct an improved predictive model have focused on two aspects of the problem as follows: expanding the population of quantitatively characterized finger modules and using new methods for training improved recognition models. We have used OPEN-based ZFP selection methods (44,45) to expand our existing set of B1H-characterized artificial and naturally occurring fingers to 1209 one-finger modules and 678 two-finger modules. The latter group captures context-dependent effects that can occur at the finger–finger interface, allowing the construction of recognition models that span more than a single finger, thereby providing additional information on the recognition potential of specific determinant combinations for the construction of improved predictive models. These finger archives and the underlying data also have value in the design of artificial ZFPs to recognize specific sequences. Thus, the assembly of these modules can be data driven by applying ‘rules’ for recognition of particular sequences to estimate which assembled finger models are likely to provide the desired composite specificities.
Our assessment of ZFModels shows that the motif predictions obtained are superior to previously published predictors. This is likely due to our larger and better (i.e. quantitative) training sets that allow us to consider more interactions, not just the canonical ones that have been primarily used in the past. We have also leveraged our two-finger module data to extend the model construction beyond a one-finger to two-finger units, where the two-finger model constructs motifs by assembling interfaces via a stitching assembly (62) to try to minimize edge effects of the two-finger module data on the resulting motif. This model is accessible to the community though our Web site (http://stormo.wustl.edu/ZFModels/). Users can input a protein sequence and an HMM-based algorithm will extract the determinants in each finger for construction of a recognition motif. Users can use either the one-finger or multi-finger model, or a hybrid (average) of these two models for generating a motif for their factor. On an independent test set, the hybrid model performed slightly better (Supplementary Table S3), although the results from each method are similar.
There is still room for improvement in our predictive model, especially for some classes of C2H2 ZFs with noncanonical linkers that may lead to alternate finger sequences or binding modes, but in nearly every case tested the predictions are at least partially correct and allow for the alignment of the individual fingers with the segments of the binding motifs that they interact with. A recently reported large compendium of zinc finger proteins selected for binding to specific DNA sequences (74), and then with their specificities determined by B1H, may provide additional, more diverse information to improve the predictive models further, but this has not been tested yet. Currently, predictions from our models are not accurate enough on their own to make reliable regulatory networks, but may be useful in conjunction with accessibility data and DNaseI footprinting data (12) to identify their regulatory sites. They can also aid in assigning ZF-TFs to particular motifs that are discovered through computational analysis of other genomic features, although for that particular problem, the alternative SVM approach of the Princeton group (82) will sometimes work better. Their approach trains their model to maximize the number of high information content positions that are correctly predicted. By then applying string matching methods, one can sometimes identify a ZF-TF that is likely to bind to a known motif [e.g. PRDM9 (58)] in cases where our model may yield a less definitive consensus because it may predict many low information content positions. In some cases, these approaches may also allow us to determine whether only a subset of ZFs are used to recognize DNA, or if different subsets are used to recognize different classes of binding sites, as when ZFPs use alternative modes of binding for interacting with different sequences. Given the rapid diversification of ZFPs during evolution and the technical challenges associated with experimental determination of their specificities, the continued refinement of predictive models will likely play an important role in understanding the roles of these proteins in transcriptional regulatory networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. National Institutes of Health (NIH) [GM068110 to S.A.W., HG000249 to G.D.S., HG004744 to M.H.B. and S.A.W., GM078369 to J.K.J., S.A.W., G.D.S.]. Funding for open access charge: U.S. National Institutes of Health (NIH).
Conflict of interest statement. J.K.J. has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Brodsky, Joung, Stormo and Wolfe laboratories for their assistance with these studies.
REFERENCES
- 1.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kundaje A, Kyriazopoulou-Panagiotopoulou S, Libbrecht M, Smith CL, Raha D, Winters EE, Johnson SM, Snyder M, Batzoglou S, Sidow A. Ubiquitous heterogeneity and asymmetry of the chromatin environment at regulatory elements. Genome Res. 2012;22:1735–1747. doi: 10.1101/gr.136366.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Natarajan A, Yardimci GG, Sheffield NC, Crawford GE, Ohler U. Predicting cell-type-specific gene expression from regions of open chromatin. Genome Res. 2012;22:1711–1722. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arvey A, Agius P, Noble WS, Leslie C. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 2012;22:1723–1734. doi: 10.1101/gr.127712.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 11.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yip KY, Cheng C, Bhardwaj N, Brown JB, Leng J, Kundaje A, Rozowsky J, Birney E, Bickel P, Snyder M, et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc. Natl Acad. Sci. USA. 2011;108:18318–18323. doi: 10.1073/pnas.1110731108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaeger SA, Chan ET, Berger MF, Stottmann R, Hughes TR, Bulyk ML. Conservation and regulatory associations of a wide affinity range of mouse transcription factor binding sites. Genomics. 2010;95:185–195. doi: 10.1016/j.ygeno.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marbach D, Roy S, Ay F, Meyer PE, Candeias R, Kahveci T, Bristow CA, Kellis M. Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. Genome Res. 2012;22:1334–1349. doi: 10.1101/gr.127191.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazemian M, Blatti C, Richards A, McCutchan M, Wakabayashi-Ito N, Hammonds AS, Celniker SE, Kumar S, Wolfe SA, Brodsky MH, et al. Quantitative analysis of the Drosophila segmentation regulatory network using pattern generating potentials. PLoS Biol. 2010;8:e1000456. doi: 10.1371/journal.pbio.1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Q, Kazemian M, Pham H, Blatti C, Celniker SE, Wolfe SA, Brodsky MH, Sinha S. Computational identification of diverse mechanisms underlying transcription factor-DNA occupancy. PLoS Genet. 2013;9:e1003571. doi: 10.1371/journal.pgen.1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 26.Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laity JH, Dyson HJ, Wright PE. DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J. Mol. Biol. 2000;295:719–727. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- 29.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 30.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 31.Fairall L, Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 32.Houbaviy HB, Usheva A, Shenk T, Burley SK. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl Acad. Sci. USA. 1996;93:13577–13582. doi: 10.1073/pnas.93.24.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim CA, Berg JM. A 2.2 A resolution crystal structure of a designed zinc finger protein bound to DNA. Nat. Struct. Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- 34.Wolfe SA, Grant RA, Elrod-Erickson M, Pabo CO. Beyond the “recognition code”: structures of two Cys2His2 zinc finger/TATA box complexes. Structure. 2001;9:717–723. doi: 10.1016/s0969-2126(01)00632-3. [DOI] [PubMed] [Google Scholar]
- 35.Segal DJ, Crotty JW, Bhakta MS, Barbas CF, 3rd, Horton NC. Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. J. Mol. Biol. 2006;363:405–421. doi: 10.1016/j.jmb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc. Natl Acad. Sci. USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 38.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 39.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. An affinity-based scoring scheme for predicting DNA-binding activities of modularly assembled zinc-finger proteins. Nucleic Acids Res. 2009;37:506–515. doi: 10.1093/nar/gkn962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choo Y. End effects in DNA recognition by zinc finger arrays. Nucleic Acids Res. 1998;26:554–557. doi: 10.1093/nar/26.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu C, Smith T, McNulty J, Rayla AL, Lakshmanan A, Siekmann AF, Buffardi M, Meng X, Shin J, Padmanabhan A, et al. Evaluation and application of modularly assembled zinc-finger nucleases in zebrafish. Development. 2011;138:4555–4564. doi: 10.1242/dev.066779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Ann. Rev. Biophys. Biomol. Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- 43.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Ann. Rev. Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 44.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): an ‘open-source' protocol for making customized zinc-finger arrays. Nat. Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christensen RG, Gupta A, Zuo Z, Schriefer LA, Wolfe SA, Stormo GD. A modified bacterial one-hybrid system yields improved quantitative models of transcription factor specificity. Nucleic Acids Res. 2011;39:e83. doi: 10.1093/nar/gkr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpaa MJ, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enuameh MS, Asriyan Y, Richards A, Christensen RG, Hall VL, Kazemian M, Zhu C, Pham H, Cheng Q, Blatti C, et al. Global analysis of Drosophila Cys2-His2 zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 2013;23:928–940. doi: 10.1101/gr.151472.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tadepally HD, Burger G, Aubry M. Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains. BMC Evol. Biol. 2008;8:176. doi: 10.1186/1471-2148-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas JH, Emerson RO. Evolution of C2H2-zinc finger genes revisited. BMC Evol. Biol. 2009;9:51. doi: 10.1186/1471-2148-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat. Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- 62.Zhu C, Gupta A, Hall VL, Rayla AL, Christensen RG, Dake B, Lakshmanan A, Kuperwasser C, Stormo GD, Wolfe SA. Using defined finger-finger interfaces as units of assembly for constructing zinc-finger nucleases. Nucleic Acids Res. 2013;41:2455–2465. doi: 10.1093/nar/gks1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 64.Dreier B, Segal DJ, Barbas CF., 3rd Insights into the molecular recognition of the 5′-GNN-3′ family of DNA sequences by zinc finger domains. J. Mol. Biol. 2000;303:489–502. doi: 10.1006/jmbi.2000.4133. [DOI] [PubMed] [Google Scholar]
- 65.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 67.Isalan M, Klug A, Choo Y. Comprehensive DNA recognition through concerted interactions from adjacent zinc fingers. Biochemistry. 1998;37:12026–12033. doi: 10.1021/bi981358z. [DOI] [PubMed] [Google Scholar]
- 68.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Q, Xia Z, Zhong X, Case CC. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J. Biol. Chem. 2002;277:3850–3856. doi: 10.1074/jbc.M110669200. [DOI] [PubMed] [Google Scholar]
- 70.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. An optimized two-finger archive for ZFN-mediated gene targeting. Nat. Methods. 2012;9:588–590. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lam KN, van Bakel H, Cote AG, van der Ven A, Hughes TR. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Res. 2011;39:4680–4690. doi: 10.1093/nar/gkq1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl Acad. Sci. USA. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Persikov AV, Rowland EF, Oakes BL, Singh M, Noyes MB. Deep sequencing of large library selections allows computational discovery of diverse sets of zinc fingers that bind common targets. Nucleic Acids Res. 2013;42:1497–1508. doi: 10.1093/nar/gkt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benos PV, Lapedes AS, Stormo GD. Probabilistic code for DNA recognition by proteins of the EGR family. J. Mol. Biol. 2002;323:701–727. doi: 10.1016/s0022-2836(02)00917-8. [DOI] [PubMed] [Google Scholar]
- 77.Kaplan T, Friedman N, Margalit H. Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput. Biol. 2005;1:e1. doi: 10.1371/journal.pcbi.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J, Stormo GD. Context-dependent DNA recognition code for C2H2 zinc-finger transcription factors. Bioinformatics. 2008;24:1850–1857. doi: 10.1093/bioinformatics/btn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cho SY, Chung M, Park M, Park S, Lee YS. ZIFIBI: Prediction of DNA binding sites for zinc finger proteins. Biochem. Biophys. Res. Commun. 2008;369:845–848. doi: 10.1016/j.bbrc.2008.02.106. [DOI] [PubMed] [Google Scholar]
- 80.Persikov AV, Osada R, Singh M. Predicting DNA recognition by Cys2His2 zinc finger proteins. Bioinformatics. 2009;25:22–29. doi: 10.1093/bioinformatics/btn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Persikov AV, Singh M. An expanded binding model for Cys2His2 zinc finger protein-DNA interfaces. Phys. Biol. 2011;8:035010. doi: 10.1088/1478-3975/8/3/035010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Persikov AV, Singh M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014;42:97–108. doi: 10.1093/nar/gkt890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vapnik VN. An overview of statistical learning theory. IEEE Trans. Neural Netw. 1999;10:988–999. doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- 84.Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 85.Christensen RG, Enuameh MS, Noyes MB, Brodsky MH, Wolfe SA, Stormo GD. Recognition models to predict DNA-binding specificities of homeodomain proteins. Bioinformatics. 2012;28:i84–i89. doi: 10.1093/bioinformatics/bts202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39:381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- 88.Benson G. A new distance measure for comparing sequence profiles based on path lengths along an entropy surface. Bioinformatics. 2002;18(Suppl. 2):S44–S53. doi: 10.1093/bioinformatics/18.suppl_2.s44. [DOI] [PubMed] [Google Scholar]
- 89.Tanaka E, Bailey T, Grant CE, Noble WS, Keich U. Improved similarity scores for comparing motifs. Bioinformatics. 2011;27:1603–1609. doi: 10.1093/bioinformatics/btr257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang T, Stormo GD. Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics. 2003;19:2369–2380. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- 91.Mahony S, Auron PE, Benos PV. DNA familial binding profiles made easy: comparison of various motif alignment and clustering strategies. PLoS Comput. Biol. 2007;3:e61. doi: 10.1371/journal.pcbi.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Narlikar L, Hartemink AJ. Sequence features of DNA binding sites reveal structural class of associated transcription factor. Bioinformatics. 2006;22:157–163. doi: 10.1093/bioinformatics/bti731. [DOI] [PubMed] [Google Scholar]
- 93.Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 94.Schones DE, Sumazin P, Zhang MQ. Similarity of position frequency matrices for transcription factor binding sites. Bioinformatics. 2005;21:307–313. doi: 10.1093/bioinformatics/bth480. [DOI] [PubMed] [Google Scholar]
- 95.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Isalan M, Choo Y, Klug A. Synergy between adjacent zinc fingers in sequence-specific DNA recognition. Proc. Natl Acad. Sci. USA. 1997;94:5617–5621. doi: 10.1073/pnas.94.11.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alleyne TM, Pena-Castillo L, Badis G, Talukder S, Berger MF, Gehrke AR, Philippakis AA, Bulyk ML, Morris QD, Hughes TR. Predicting the binding preference of transcription factors to individual DNA k-mers. Bioinformatics. 2009;25:1012–1018. doi: 10.1093/bioinformatics/btn645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abdi H. Partial least squares regression and projection on latent structure regression (PLS Regression) Wiley Interdiscip. Rev. Comput. Stat. 2010;2:97–106. [Google Scholar]
- 103.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stormo GD, Zhao Y. Determining the specificity of protein-DNA interactions. Nat. Rev. Genet. 2010;11:751–760. doi: 10.1038/nrg2845. [DOI] [PubMed] [Google Scholar]
- 109.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 110.Otto SJ, McCorkle SR, Hover J, Conaco C, Han JJ, Impey S, Yochum GS, Dunn JJ, Goodman RH, Mandel G. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.