Abstract
In pathogens, the accurate programming of virulence gene expression is essential for infection. It is achieved by sophisticated arrays of regulatory proteins and ribonucleic acids (sRNAs), but in many cases their contributions and connections are not yet known. Based on genetic, biochemical and structural evidence, we report that the expression pattern of a Staphylococcus aureus host immune evasion protein is enabled by the collaborative actions of RNAIII and small pathogenicity island RNA D (SprD). Their combined expression profiles during bacterial growth permit early and transient synthesis of Sbi to avoid host immune responses. Together, these two sRNAs use antisense mechanisms to monitor Sbi expression at the translational level. Deletion analysis combined with structural analysis of RNAIII in complex with its novel messenger RNA (mRNA) target indicate that three distant RNAIII domains interact with distinct sites of the sbi mRNA and that two locations are deep in the sbi coding region. Through distinct domains, RNAIII lowers production of two proteins required for avoiding innate host immunity, staphylococcal protein A and Sbi. Toeprints and in vivo mutational analysis reveal a novel regulatory module within RNAIII essential for attenuation of Sbi translation. The sophisticated translational control of mRNA by two differentially expressed sRNAs ensures supervision of host immune escape by a major pathogen.
INTRODUCTION
Staphylococcus aureus is a commensal flora, but it can also be an opportunistic pathogen and the cause of nosocomial and community-acquired infections (1). Staphylococcus aureus produces a large number of virulence determinants to survive and to establish an infection. These factors include surface proteins for adherence to host cells and tissues, haemolytic toxins that contribute to tissue damage and dissemination and exoproteins for host immune avoidance (2). For instance, staphylococcal protein A (SpA) recognizes the immunoglobulin Fc domain, which results in inverted tagging and in turn blocks C1q and Fcγ receptor binding sites (3). Another immunoglobulin-binding protein (Sbi) is also expressed by S. aureus (4), and it impairs the host immune response. Sbi acts as a complement inhibitor and forms a tripartite complex with host complement factors H and C3b (5). Recent data indicate that Sbi and extracellular fibrinogen-binding protein recruit human plasmin to degrade complement C3 and C3b (6).
A coordinated and timely expression of multiple virulence factors is essential for S. aureus infection. The process involves two-component sensory transduction systems and global regulatory proteins (7). In addition to protein-mediated gene controls, ribonucleic acids (RNAs) have regulatory functions in many bacterial pathogens (8,9), including S. aureus (10). In S. aureus, ∼250 regulatory RNAs are expressed (11–17), but until now few have demonstrated physiological functions. One is the multifunctional RNAIII, which combines quorum sensing with virulence regulation (18). RNAIII acts as a messenger RNA (mRNA), encoding the 26-aa delta-haemolysin peptide. As a trans-acting RNA, it has multiple regulatory functions (7), including the regulation of the expression of numerous mRNAs at the translational and/or transcriptional levels (19,20) (Figure 1A). The RNAIII 5′-domain activates translation of α-haemolysin by disrupting the inhibitory translation initiation site (TIS) hairpin of hla mRNA (21). The RNAIII 3′-domain represses the translation of several virulence factors and of the transcriptional repressor of toxins (Rot), together reducing early expressed virulence factors (19,20,22), thus facilitating S. aureus infection (19). One RNAIII target is the SpA immune evasion molecule, whose expression is downregulated at both transcriptional (18) and translational (23) levels.
Figure 1.
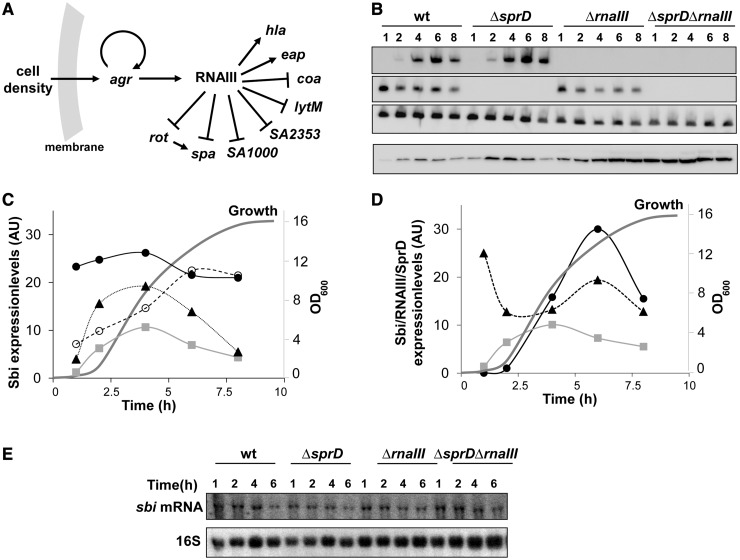
Translational control of Sbi during S. aureus growth is shared by RNAIII and SprD. (A) Simplified overview of the RNAIII regulon. RNAIII is the effector of the accessory gene regulator (agr) quorum sensing system in S. aureus. RNAIII positively regulates hla and eap expression levels (21,22), whereas RNAIII represses the expression of spa, rot, coa, SA2353, SA1000 and lytM by translation initiation inhibition (19,23,24). (B) RNAIII and SprD expression profiles were monitored in wt, ΔsprD, ΔrnaIII and sprD-ΔrnaIII HG001 isogenic strains by northern blots using labelled DNA probes specific for each RNA (upper panels). Immunoblot analysis with anti-Sbi antibodies of the intracellular and membrane proteins in the four strains during S. aureus growth (lower panel). The 5S ribosomal RNAs (rRNAs) were used as loading controls. (C) Quantifications of the Sbi protein levels, during S. aureus growth, in strains expressing, or not, SprD and/or RNAIII. The amounts of Sbi proteins are provided in arbitrary units (AU) and were calculated relative to the quantity of total protein amounts per tracks, evidenced by Coomassie staining in Supplementary Figure S5A. The graphs indicate the Sbi protein levels in wt (grey squares), ΔsprD (triangles), ΔrnaIII (empty circles) and ΔsprD-ΔrnaIII (filled circles) S. aureus strains. The data are representative of at least three independent experiments. The growth curve of strain wt is presented. (D) Expression levels of the Sbi protein (grey squares), RNAIII (circles) and SprD (triangles) in S. aureus strain wt during growth. Quantifications of the Sbi protein was performed as for panel C. The amounts of RNAIII (circles) and SprD (triangles) are shown in arbitrary units (AU) and were calculated relative to the 5S rRNA. (E) Northern blot analysis of the steady-state sbi mRNA levels in wt strain HG001, ΔsprD, ΔrnaIII and ΔsprD-ΔrnaIII mutants during bacterial growth. The 16S rRNAs were used as internal loading controls.
Small pathogenicity island RNA D (SprD) is among the few S. aureus RNAs with identified functions. SprD is expressed from the genome of a converting phage (11), a horizontally acquired pathogenicity island that is a repository for many toxins, adherence and invasion factors, superantigens and secretion systems (25). Using an antisense mechanism, SprD downregulates the translational expression of the Sbi immune evasion molecule (26). Interestingly, SprD significantly contributes to disease incidence in a mouse model of staphylococcal infection, although this effect is not linked solely to deregulation of Sbi production.
In this article, in vitro and in vivo genetic, biochemical and structural approaches were used to newly identify the Sbi immune evasion protein as a target for RNAIII. RNAIII operates as an antisense RNA annealing sbi mRNA at multiple sites, including at the TIS, thus downregulating the expression of Sbi. We have also identified a new unconventional single-stranded zone in RNAIII that is essential for attenuating Sbi translation. Experimental evidence shows that RNAIII cooperates with a second RNA, SprD, to achieve accurate monitoring of sbi expression during S. aureus growth. Such a sophisticated translational control of virulence factor-encoding mRNA by two differentially expressed RNAs ensures that immune evasion molecules are fabricated early, temporarily and at the appropriate levels to defeat host immune defences.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
The strains used are listed in Supplementary Table S1. The sprD gene was deleted in strain HG001 using pBT2ΔsprD vector as described for strain N315 (26). Inactivation of the rnaIII gene using pBT2ΔrnaIII was performed as described previously by the authors (26), yielding the strain HG001ΔrnaIIIa. Isogenic strains HG001ΔrnaIII and HG001ΔsprDΔrnaIII were constructed by transducting the cat-tagged rnaIII mutation from WA400 (27) into strains HG001 and HG001ΔsprD, respectively, and by selecting for chloramphenicol resistance. The S. aureus strains were cultured at 37°C in brain–heart infusion broth (Oxoid). When necessary, erythromycin was added at a concentration of 10 µg/ml. In pCN38-rnaIII, RNAIII was expressed from its endogenous promoter. The rnaIII sequence was polymerase chain reaction (PCR)-amplified from HG001 genomic DNA as a 1063-bp fragment with flanking PstI and EcoRI restriction sites, then cloned into the vector pCN38 (28). We used mutagenized oligonucleotides to produce RNAIII mutants (Supplementary Table S2).
Protein isolations, immunoblots, DNA and RNA manipulations
Protein extract preparations and detection of the protein Sbi were performed as previously described (26). Culture pellets were washed with Tris-EDTA (TE) buffer (50 mM Tris, pH 7.5, 50 mM EDTA) and suspended in 0.2 ml of this solution supplemented with 0.1 mg/ml lysostaphin. Following 37°C incubation for 10 min, samples were boiled 5 min and separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis. For the immunoblots, proteins were transferred to PolyVinyliDene Fluoride (PVDF) membrane (Immobilon-P, Millipore). Sbi was visualized by anti-Sbi antibodies (Dr van den Elsen, University of Bath, UK), anti-rabbit IgG, peroxidase-conjugated, secondary antibodies (Jackson) and the ‘Amersham ECL’ system (Amersham Pharmacia Biotech). Total S. aureus RNA was prepared as previously described (25). Northern blots targeting SprD and RNAIII were performed with 5 µg of total RNAs (26). RNAs used for probing, gel-shift and toeprint assays were transcribed from PCR fragments amplified from purified genomic DNA using the primers listed in Supplementary Table S2. To produce the template-encoding RNAIII mutants, mutagenized oligonucleotides were used (Supplementary Table S2). RNAs were produced by in vitro transcription using MEGAscript (Ambion). The 5′-labelling of RNAs was performed as described previously (29). RNAs were purified by 8% polyacrylamide gel electrophoresis, eluted, ethanol-precipitated and then stored at −80°C until needed.
RNA probing, gel-shift and toeprint assays
Gel retardation assays were performed (26), and 0.4 pmol of labelled sbi mRNA or RNAIII were incubated with various concentrations (from 1.6 to 50 pmols) of unlabelled RNAs. The toeprint assays were performed as previously described (26). The annealing mixtures used contained 0.2 pmol of sbi mRNA and 1 pmol of labelled SBIrevTR primer in a reaction buffer of 10 mM Tris-acetate, pH 7.5, 60 mM NH4Cl and 1 mM dithiothreitol (DTT). For the competition assays, various concentrations of SprD, RNAIII or RNAIII mutants were added before the purified Escherichia coli 70S ribosomes. Ribosomes were activated for 15 min at 37°C and diluted in the same reaction buffer in the presence of 1 mM MgCl2. Next, 4 pmols of ribosomes were added and incubated for 5 min, then the MgCl2 was adjusted to 10 mM and the reactions were incubated for 5 min. In all, 10 pmols of uncharged transfer RNAfMet (tRNAfMet) were then added for 15 min. Complementary DNAs were synthesized with 2 UI of AMV RT (Biolabs) for 15 min at 37°C. Reactions were ended by the addition of 10 µl of loading buffer II (Ambion). The complementary DNAs were separated in 8% denaturating polyacrylamide gel electrophoresis. Gels were dried and visualized using a STORM 840 Phosphorimager (Molecular Dynamics).
RESULTS
The immune evasion protein Sbi is a newly discovered RNAIII molecular target
Sbi is an immune evasion protein expressed by S. aureus, an expression that must be tightly controlled during staphylococcal infection. The inactivation of the accessory gene regulator (agr) acting as a global virulence regulator increases Sbi abundance in vivo (30), indicating that agr is a negative regulator of sbi expression. Because RNAIII is the effector of the agr regulon, we tested to see whether RNAIII by itself could influence sbi expression in S. aureus cells. We analysed the Sbi protein levels during growth by comparing western blots in wild-type (wt) strain HG001 and in an isogenic ΔrnaIII mutant (Figure 1B). The absence of RNAIII increased Sbi protein levels throughout S. aureus growth, demonstrating that endogenous RNAIII reduces the expression of Sbi in vivo. RNAIII has a specific expression profile during S. aureus growth: low levels are detected early but its expression significantly increases up to the pre-stationary growth phase (Figure 1, panels B and D). In the absence of RNAIII (strain ΔrnaIII), the Sbi levels significantly increase during the post-exponential phase of growth (Figure 1, panels B and C), when RNAIII expression are the highest in strain wt (Figure 1, panels B and D). Therefore, the absence of RNAIII alters the overall expression pattern of Sbi, indicating RNAIII expression is necessary to ensure the temporal control of Sbi expression. Therefore, RNAIII is required for adjustment of Sbi expression during S. aureus growth.
Joint action of two RNAs to adjust Sbi levels during S. aureus growth
SprD controls Sbi expression by antisense pairings, preventing translation initiation in the agr-negative strain N315 (26). The deletion of SprD in the agr-positive HG001 strain (ΔsprD) results in elevated Sbi protein levels, although its overall expression profile during growth remains similar (Figure 1, panels B and C) to what has been reported for strain N315 (26). Because Sbi expression is monitored by at least two sRNAs in S. aureus, we constructed a ΔsprDΔrnaIII double mutant in strain HG001. In this HG001ΔsprDΔrnaIII strain, we detected a considerable increase in Sbi levels throughout growth (Figure 1, panels B and C). This demonstrates that RNAIII and SprD are necessary for lowering and adjusting Sbi protein expression in vivo.
These sRNAs possess specific distinct expression profiles during growth (Figure 1, panels B and D). SprD expression is high at beginning of S. aureus growth and decreases later (Figure 1, panels B and D). RNAIII expression, however, begins later, then gradually increases during growth. SprD and RNAIII act in concert to shape the expression profile of Sbi, which is highest at early- to mid-exponential (E) phase (Figure 1, panels B and D). There is thus a tight functional relationship between Sbi protein levels and the expression profiles of these two RNA regulators during S. aureus growth.
To determine whether sbi regulation by RNAIII occurs at the transcriptional level, we examined the effect of RNAIII on the steady-state levels of sbi mRNA. In wt strain, sbi levels were detected early during growth and decreased during the stationary (S) phase (Figure 1E). Northern blots (Figure 1E) and quantitative PCR (Supplementary Figure S1A) showed that in strains deleted for rnaIII, sprD or for both, the sbi mRNA levels were similar. The implications of RNAIII and SprD on the stability of the sbi mRNA were investigated. In the presence of rifampicin, sbi mRNA half-life was measured in strain HG001 and in an isogenic strain deleted for both sRNAs (HG001ΔsprDΔrnaIII, Supplementary Figure S1B). In both strains, the sbi mRNA half-life was estimated ∼4 min, indicating that neither SprD nor RNAIII affect sbi mRNA stability. It implies that, as previously reported in strain N315 for SprD (26), RNAIII and SprD do not influence sbi expression at the RNA level. Together, these data show that RNAIII negatively controls sbi expression and that the regulation occurs at the post-transcriptional level.
RNAIII 5′- and 3′-domains interact at distinct sites on sbi mRNA
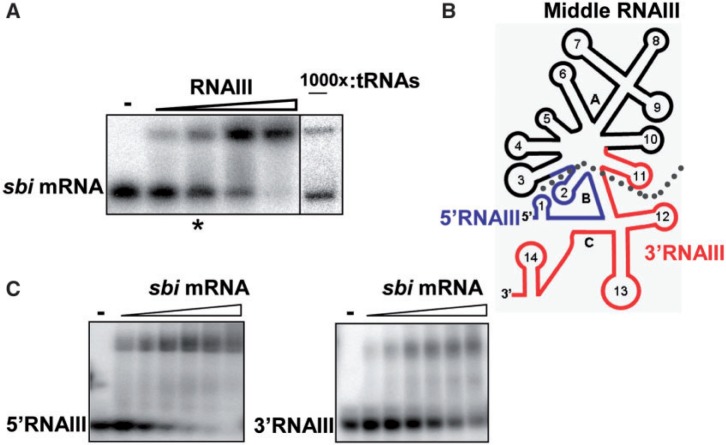
The in vivo data prompted us to test for the existence of a direct interaction between RNAIII and sbi mRNA. We did gel retardation assays to analyse the duplex formation between RNAIII and a 179 nt-long sbi mRNA fragment (sbi1-179) containing its 5′ UTR sequence followed by the first 46 codons. RNAIII binds sbi1-179mRNA, and this interaction was specific because a 1000-fold molar excess of total tRNAs displaced neither the sbi mRNA nor the RNAIII from a preformed RNAIII-sbi mRNA complex (Figure 2A and Supplementary Figure S2A). Therefore, in vitro and without the help of additional molecules, RNAIII forms a specific complex with sbi mRNA, thus demonstrating that sbi mRNA is a newly discovered direct target of RNAIII.
Figure 2.
RNAIII directly interacts with sbi mRNA through its 5′- and 3′-domains. (A) Native gel retardation assays of purified labelled sbi1–179 with increasing amounts of unlabelled RNAIII (0.1, 0.5, 1 and 2 µM). The asterisk indicates the sbi mRNA/RNAIII molar ratio used to perform gel-shift competition assays with a 1000-fold molar excess of total yeast tRNAs. (B) Schematic presentation of RNAIII deletion mutants constructed and produced in this study: Middle RNAIII is indicated by a dotted line and corresponds to stem-loops 3–11, 5′RNAIII is blue and contains stem-loops 1–2 with half B and half C and 3′RNAIII is red and encloses stem-loops 11–14. (C) RNAIII has 5′ and 3′ binding sites that interact with sbi mRNA. Complex formation between increasing concentrations of sbi mRNA (0.1, 0.2, 0.5, 1, 2 and 4 µM) with labelled 5′- or 3′-RNAIII are shown as detected by native gel retardation assays.
RNAIII has an intricate structure made of 14 stem loops (31). To analyse complex formation between RNAIII and the sbi mRNA and to identify the RNAIII domain or domains involved in the interaction, we made three RNAIII deletion mutants (Figure 2B and Supplementary Figure S2B). Each retained either 73 nt at the RNAIII 5′-end (5′RNAIII), 165 nt at the 3′-end (3′RNAIII) or the stem-loops 3–11 (middle RNAIII). We found that middle RNAIII did not interact with sbi mRNA (Supplementary Figure S2C). On the other hand, complex formation between the labelled sbi mRNA and the other two constructs was detected (Figure 2C and Supplementary Figure S2C).
To analyse the sbi mRNA domains involved in the interaction with RNAIII, two shorter fragments were produced: a fragment (sbi1–91) containing nucleotides 1–91 at its 5′-end and including the TISs and another (sbi84–179) including the 86 nt between 84 and 179 (Supplementary Figure S2D). The 5′RNAIII fragment did not interact with sbi1–91 and the 3′RNAIII construct did not recognize sbi84–179 (Supplementary Figure S2E). The 5′RNAIII, however, interacted with sbi84–179 and 3′RNAIII bound to sbi1–91 (Supplementary Figure S2E). Together, these data demonstrate that RNAIII forms a stable complex with the sbi mRNA in vitro and that this interaction may involve several binding sites.
The interaction between sbi mRNA and RNAIII includes the mRNA ribosomal binding site
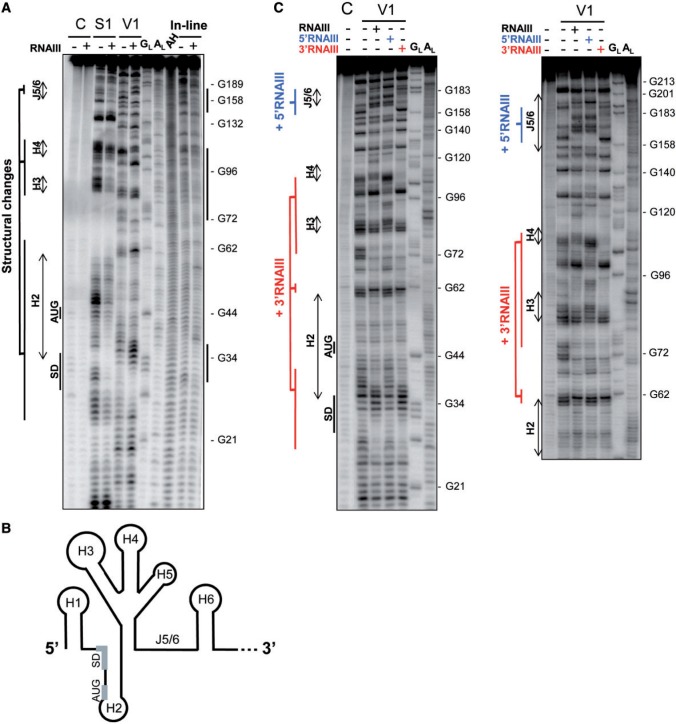
The interaction between RNAIII and the sbi mRNA appears elaborate, as it involves several structural domains from both RNAs. To analyze the molecular basis of complex formation between RNAIII and sbi mRNA, we did a structural analysis of such a complex in solution using S1 and V1 probes (Figure 3). The proposed model was based primarily on probing data and represents 238 nt from the 5′-end of sbi mRNA in the absence of RNAIII (Supplementary Figure S3 and Figure 3B). The data support the existence of six hairpins (H1–H6) and an accessible J5/6 domain is flanked by an internal branching stem and by a 3′ hairpin (H6). S1 and V1 cleavages were detected in identical positions at several locations within the sbi mRNA structure, indicating that this structure is unstable in solution, and probably folds into several ‘fast-exchange’ conformations. S1 and V1 probes and in-line probing of the sbi mRNA in the absence or presence of RNAIII were used to identify sbi mRNA structural changes induced by RNAIII binding. In the presence of RNAIII, sbi mRNA reactivity modifications are concentrated at and around the TIS, but they also appear further downstream from the coding sequence (Figure 3A). The results showed that the presence of wt RNAIII induced cleavage protections within sbi mRNA, protections that are restricted to three domains that correspond to those subjected to reactivity changes with S1 and V1 probes (Figure 3A). The areas involved are A29-U37 (TIS), G72-A118 (H3 and H4) and A157-U191 (J5/6, Supplementary Figure S3).
Figure 3.
Structural analysis of sbi mRNA conformational changes induced by complex formation with RNAIII, 5′RNAIII and 3′RNAIII. (A) Conformational changes of sbi mRNA when in complex with RNAIII, as detected by structural probes. Autoradiograms of cleavage products of 5′-end-labelled sbi mRNA by RNases S1 and V1 in the presence (+) or absence (−) of RNAIII or obtained by ‘in-line’ probing. Track C, incubation controls; track GL, RNase T1 hydrolysis ladders; track AL, RNase U2 hydrolysis ladders; track AH, alkaline hydrolysis ladders. The sbi mRNA sequence is indexed on the right sides of the panels. On RNAIII binding, the location of the structural changes within the sbi mRNA is indicated on the left side of the panel. (B) Schematic presentation of the structural domains from sbi mRNA 5′-end (1–238 nt) from S. aureus based on the solution probing data. (C) Conformational changes of the sbi238 mRNA induced by either wt RNAIII (blue and red) or by its 5′- (blue) or 3′-domains (red). Short (left) and long (right) runs are presented; the other indications are as in (A).
To individually assess where RNAIII binds to sbi mRNA, we analysed the complexes between sbi mRNA and the RNAIII 5′- and 3′-domains by examining enzymatic cleavages (Figure 3C). Binding to 3′RNAIII induced structural changes in H2, H3 and H4 but not further downstream (Figure 3C and Supplementary Figure S3). The 5′RNAIII fragment, however, induced reactivity changes exclusively at J5/6 (nucleotides 157 to 191), deep in the sbi mRNA internal coding sequence (Figure 3C and Supplementary Figure S3). These results are consistent with binding assays (Figure 2C and Supplementary Figure S2B), indicating that 5′RNAIII interacts with sbi84–179 but not with sbi1–91, and that 3′RNAIII binds to sbi1–91 but not to sbi84–179. An in-depth analysis was done of the structures of the RNAIII domains involved in the sbi mRNA interaction. Binding of sbi mRNA onto the RNAIII 5′-domain induced reactivity changes at stems B, C and H2. Binding onto the RNAIII 3′-domain induced reactivity changes at stem B, hairpins H11 and H12, as well as at position C468 located in a single-stranded region between hairpins 13 and 14 (Supplementary Figure S4). Interestingly, the two RNAIII-sbi mRNA binding sites are self-complementary domains via the B and C stems (Supplementary Figure S4).
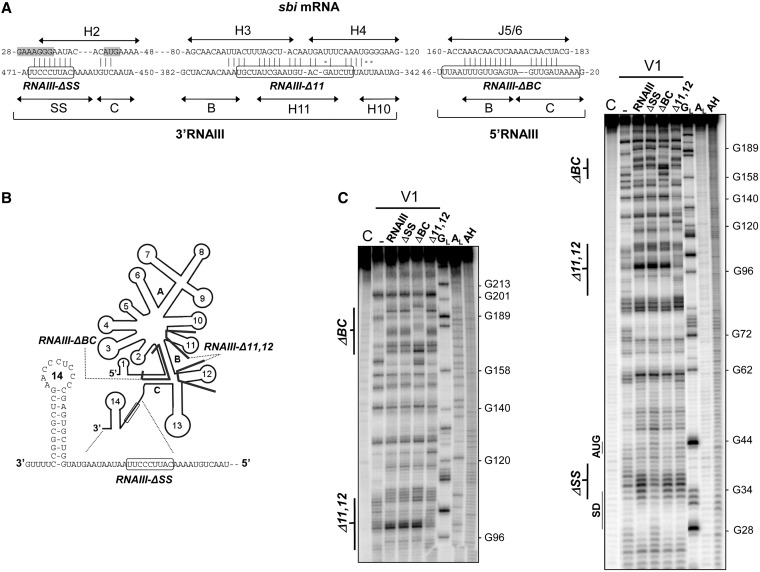
The probing data were then used to propose a model of the pairings between RNAIII and sbi mRNA (Figure 4A), according to which the interaction uses three sites from RNAIII and three from the mRNA target. In our proposed model, nucleotides U25–U41 from the 5′-end of RNAIII (stems B and C) base pair with A163–A181 from sbi mRNA J5/6. Nucleotides U347–A373 from RNAIII, stem H10 and hairpin 11 all pair with U88–G115 from sbi mRNA stem-loops H3 and H4. Finally, nucleotides C454–U469 from the 3′-end of RNAIII (domains C and SS, C462-U470; Supplementary Figure S4B) and situated in a single-strand region between hairpins 13 and 14 were paired with nucleotides A31–U43 from the sbi mRNA (hairpin H2) where both the Shine–Dalgarno (SD) sequence and initiation codon are located.
Figure 4.
The interaction between sbi mRNA and RNAIII involves three structural domains from the two RNAs and covers the mRNA ribosome binding site. (A) Proposed pairings between RNAIII and the sbi mRNA inducing sbi mRNA ribosome binding site sequestration, highlighted in grey. The RNAIII–sbi mRNA interactions are based on native gel retardation assays, deletion analysis and structural mapping of sbi mRNA–RNAIII complexes that included the RNAIII deletion mutants. RNAIII deletions (RNAIII-ΔSS, RNAIII-Δ11 and RNAIII-ΔBC) are circled. (B) Location of three RNAIII mutants on a schematic representation of the RNAIII structure. Inset: the RNAIII-ΔSS mutant is made of nine nucleotides (circled) within the single-strand connecting hairpins 13–14. RNAIII-ΔBC is deleted of one strand in stem B and one in stem C, and RNAIII-Δ11,12 is deleted from hairpins 11 and 12. (C) Structural probing of sbi mRNA in complex with each of the three RNAIII deletion mutants and compared with full-length RNAIII. These additional structural data support the proposed interaction between sbi mRNA and RNAIII as shown in (A). Track C, incubation controls; track GL, RNase T1 hydrolysis ladders; track AL, RNase U2 hydrolysis ladders; track AH, alkaline hydrolysis ladders. The sbi mRNA sequence is indexed on the right sides of the panels.
To analyse these suggested interactions in details, each of the three domains of RNAIII proposed to interact with the sbi mRNA were individually deleted. Three RNAIII mutants were constructed (Figure 4A and B): a mutant lacking 9 nt within the single strand located between hairpins 13 and 14 (RNAIII-ΔSS), another missing hairpins 11 and 12 (RNAIII-Δ11,12) and the final one lacking 25 nt in stems B and C of the RNAII 5′-domain (RNAIII-ΔBC). Labelled sbi mRNA was added in complex with each of these mutants, and we then performed statistical V1 cleavages (Figure 4C). In agreement with the proposed model, the RNAIII structural deletions within each mutant led to its inability to induce the sbi mRNA structural domain reactivity changes that full-length RNAIII induced. Mutants RNAIII-ΔSS, RNAIII-Δ11 and RNAIII-ΔBC did not induce reactivity changes at H2, H3, H4 and J5/6, respectively (Figure 4C), showing that each RNAIII mutant lacks one of the recognition domains for sbi mRNA (Figure 4A). Together, these data indicate that the RNAIII 5′- and 3′-domains, linked by intramolecular pairings within the RNAIII structure, interact with the sbi mRNA at three distinct sites: one that includes the SD-sequence and initiation codon and two others located further along the sbi mRNA internal coding sequence.
The RNAIII 3′-domain is necessary and sufficient for inhibition of sbi mRNA translation initiation
The RNAIII 3′-binding site interacts with nucleotides that contain the SD sequence and sbi mRNA initiation codon (Figure 4A). It coincides with the mRNA portion covered by the ribosomes for translation initiation (32). This suggests that the 3′-terminal domain of RNAIII prevents initiation of sbi mRNA translation. We used toeprint assays to determine whether full-length RNAIII or its independent 5′- or 3′-domains could prevent ribosomes from loading onto sbi mRNA. Ternary initiation complexes made of purified ribosomes, initiator tRNAfMet and sbi1–179 mRNA were assembled. A ribosome toeprint was detected on the sbi mRNA 15–17 nt downstream from the initiation codon, as previously reported (26). The toeprint was reduced by full-length RNAIII in a concentration-dependent manner (Figure 5A). Therefore, our results demonstrate that RNAIII prevents sbi mRNA translation initiation.
Figure 5.
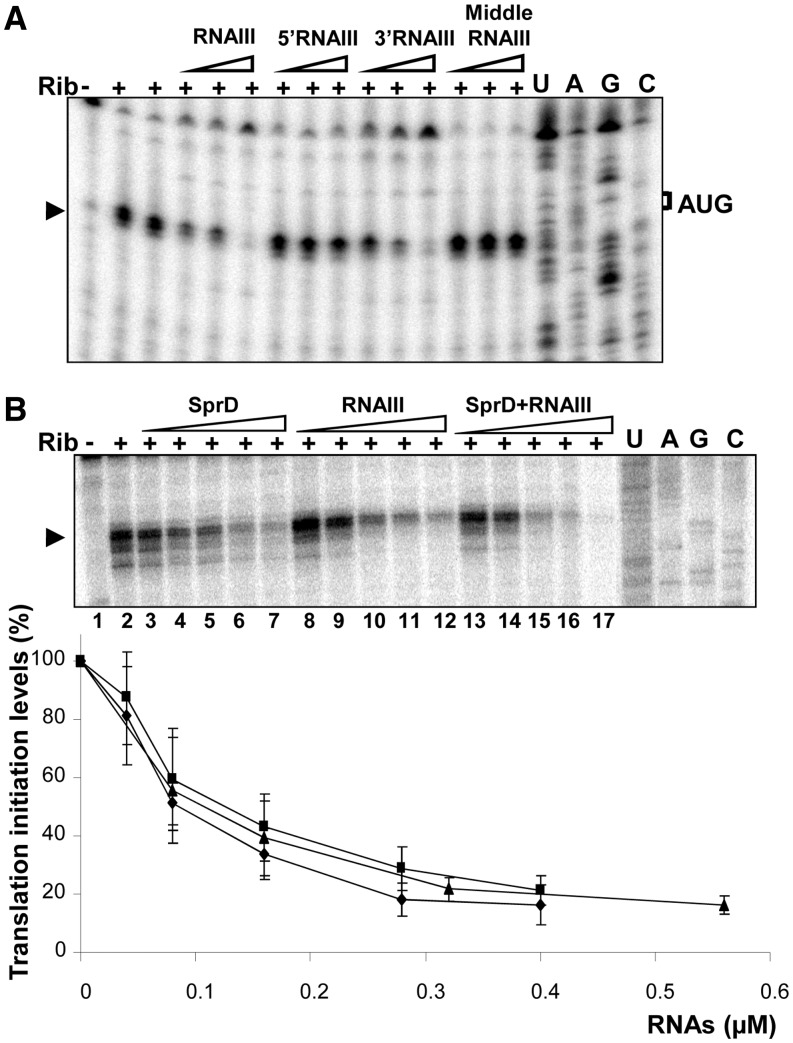
RNAIII regulates sbi translation in vitro (A) Toeprint assays showing that the RNAIII 3′-domain prevents ribosome loading and translation initiation onto the sbi mRNA, whereas the other domains from RNAIII do not. ‘Plus/minus’ indicates the ‘presence/absence’ of purified ribosomes added to RNAIII, 5′RNAIII, 3′RNAIII or middle RNAIII (at concentrations of 0.4, 2 and 10 µM, respectively). The black arrow indicates the toeprints; ‘U’, ‘A’, ‘G’ and ‘C’ refer to the sbi mRNA sequence. (B) Similar inhibitory effects of RNAIII and SprD on initiation of sbi mRNA translation. (Upper panel) Ribosome toeprint assays of the sbi mRNA with increasing amounts of SprD, RNAIII or both RNAs added simultaneously. The presence/absence of purified ribosomes with SprD (tracks 2–7), RNAIII (tracks 8–12) or SprD and RNAIII at identical concentrations (equimolar mixture, tracks 13–17) is indicated with plus/minus. Concentrations of SprD, RNAIII or of an equal mix of SprD/RNAIII were 0.4 µM (tracks 3 and 8), 0.8 µM (tracks 4 and 9), 1.6 µM (tracks 5 and 10), 2.8 µM (tracks 6 and 11) and 4 µM (tracks 7 and 12). Concentrations of an equal SprD/RNAIII mix were 0.8, 1.6, 3.2, 5.6 and 8 µM (tracks 13–17). The arrow points to the toeprints. U, A, G and C refer to the sbi mRNA sequence. (Lower panel) Quantification of the inhibition of sbi mRNA translation by increasing concentrations of SprD (diamonds), RNAIII (squares) and by an equimolar amount of the two RNAs (triangles).
Because sbi translation is controlled by two regulatory RNAs, we tested the ribosome loading onto the sbi mRNA by adding SprD and RNAIII simultaneously, and we compared this with the contribution of each RNA. When SprD and RNAIII were added individually, the toeprint was also reduced in a concentration-dependent manner (Figure 5B). The effect of either RNAIII or SprD on initiation of sbi translation was similar for each concentration tested. In fact, the decrease in sbi translation caused by an equimolar SprD-RNAIII mix was equivalent to the decrease caused by the same concentration of each RNA on its own (Figure 5B). Therefore, RNAIII reduces sbi translation initiation independently of SprD and both RNAs possess a similar ability to lower sbi translation in vitro.
Next, we tested the involvement of different parts of RNAIII in the inhibition of sbi translation. As anticipated, increasing amounts of middle RNAIII, which was unable to interact with sbi mRNA (Figure 2C), did not prevent ribosome loading (Figure 5A). Increasing amounts of 5′RNAIII had no influence on ribosome loading (Figure 5A), indicating that this site is, by itself, insufficient for control of sbi translation. On the other hand, increasing amounts of 3′RNAIII, which interacts with sbi mRNA domains containing the TIS (Figure 4A), prevented ribosome loading in a concentration-dependent manner (Figure 5A). Taken together, our results demonstrate that RNAIII prevents initiation of sbi mRNA translation through antisense pairings at the TIS, and that the RNAIII 3′-domain is required and sufficient for this regulation in vitro.
A novel regulatory element within RNAIII prevents sbi mRNA translation
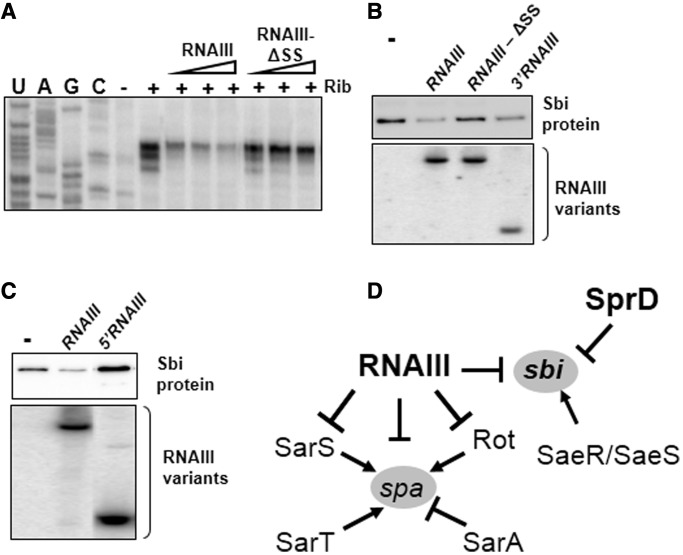
According to our proposed RNAIII-sbi mRNA interaction model (Figure 4A), the region of RNAIII that interacts with the sbi mRNA TIS is situated in the single strand between hairpins 13 and 14. The involvement of this newly-discovered RNAIII element in the control of sbi translation was investigated. We first tested to see whether a 9-nt C462-U470 deletion mutant found between hairpins 13 and 14 (RNAIII-ΔSS, Figure 4) and which is unable to interact with the sbi mRNA TIS region might reduce translation initiation. Toeprint assays showed that unlike wt RNAIII, RNAIII-ΔSS cannot control sbi mRNA translation initiation (Figure 6A). This indicates that the 9 nt at the 3′-end of RNAIII are actually necessary for regulation of sbi translation. To demonstrate the necessity of the RNAIII SS domain in the lowering of Sbi protein levels in vivo, full-length RNAIII, 3′RNAIII, 5′RNAIII and the RNAIII-ΔSS mutants were expressed in a HG001ΔrnaIIIa strain. As evidenced in vitro, western blots showed that both RNAIII and 3′RNAIII repressed sbi translation (Figure 6B), indicating that the expression of the 3′-domain of RNAIII is sufficient to decrease the Sbi protein levels. In accordance with these results, an RNAIII mutant lacking 25 nt in stems B and C from its 5′-domain (RNAIII-ΔBC) was able to decrease the Sbi expression levels in vivo (Supplementary Figure S5). Unexpectedly, expressing the 5′-domain of RNAIII on its own increased the levels of Sbi protein in vivo (Figure 6C). The mechanism of such effect is unknown and will require further investigations. Nevertheless, in vitro data indicate that RNAIII 5′ does not affect Sbi translation initiation suggests that this positive effect probably occurs at the mRNA level.
Figure 6.
RNAIII adjusts sbi translation through a novel regulatory element located at its 3′-end. (A) Ribosome toeprint assays of the sbi mRNA in the presence of increasing amounts of full-length RNAIII or RNAIII-ΔSS mutant. A 9-nt single-strand at the RNAIII 3′-end is required to control Sbi translation initiation. The concentrations of RNAIII and its variant were 0.4, 1.2 and 2.5 µM. The other indications are as in (A). (B) A 9-nt single strand at the RNAIII 3′-end is indispensable for governance of Sbi protein expression in vivo. Here, we show the in vivo expression of protein Sbi at the exponential phase of growth in strain HG001ΔrnaIII (in complement with pCN38ΩRNAIII (RNAIII), pCN38ΩRNAIII-ΔSS (RNAIII-ΔSS) or pCN38Ω3′RNAIII (3′RNAIII). Total protein amounts loaded per track are shown in Supplementary Figure S5B. The lower panel depicts the northern blot expression levels of RNAIII and its mutants. (C) Stimulatory effect of the 5′-domain from RNAIII onto the expression of the Sbi proteins in vivo. Sbi expression of HG001ΔrnaIII transformed with pCN38ΩRNAIII (RNAIII) or with pCN38Ω5′RNAIII (5′RNAIII). The total protein amounts loaded per track are shown in Supplementary Figure S5C. The lower panel depicts the expression levels of RNAIII and the RNAIII 5′ domain by northern blots. (D) Model depicting the regulatory network controlling the expression of two S. aureus immune evasion molecules, Sbi and SpA. The sbi is positively regulated at the transcriptional level by the two-component system SaeR/SaeS (33) and repressed at the translational level by the two sRNAs, SprD and RNAIII. The spa transcription is negatively regulated by SarA and positively regulated by Rot, SarS and SarT (34–36). The rot and spa translations are inhibited by RNAIII, by a direct interaction at translational level (19,23).
Unlike full-length RNAIII, mutant RNAIII-ΔSS was unable to lower Sbi protein levels in vivo (Figure 6B), in agreement with in vitro data. Northern blots indicated that RNAIII-ΔSS and full-length RNAIII are expressed at similar levels (Figure 6B), ruling out the possibility that the absence of RNAIII-ΔSS regulation might be due to its lower expression or stability when compared with wt RNAIII. Altogether, our in vitro and in vivo results highlight an elaborate and direct interaction between RNAIII and sbi mRNA, bringing to light a novel regulatory function module within the RNAIII structure.
DISCUSSION
RNAIII is a useful paradigm for studying RNA-mediated virulence gene regulations in bacteria. As the effector of the accessory gene regulator (agr) two-component system, RNAIII governs the expression of a large number of virulence factors including cell wall-associated proteins, exoproteins and global regulators. Therefore, RNAIII is a multitasking RNA, coupling quorum sensing with virulence regulation (7,8). In this report, we identified a novel direct RNAIII target: the immune evasion protein Sbi that traps host IgGs and has a region triggering complement C3 activation (37). We recently reported the major implication of another sRNA, SprD, in the control of sbi expression levels (26). Here, we provide experimental evidence to show that SprD collaborates with RNAIII to achieve an optimal and efficient monitoring of sbi expression during S. aureus growth. The regulation of sbi expression profiling needs at least two sRNAs (RNAIII and SprD), as the absence of one of these causes strong alterations in Sbi levels and expression profiles during bacterial growth (Figure 1). We propose that the two sRNAs RNAIII and SprD reduce sbi expression according to their individual expression profiles. During S. aureus growth, SprD is continually expressed with a drop at the beginning of the E phase, whereas RNAIII is undetectable at first and slowly accumulates during growth (Figure 1, panels B and D). In a ΔsprD mutant, the Sbi protein levels are higher during growth while maintaining an overall expression profile similar than isogenic wt (Figure 1, panels B and C). SprD probably sets the basal levels of Sbi expression through growth, whereas RNAIII intervenes at later stages to restrict protein expression when immune evasion is no longer needed. In vivo, the sum of their actions permits the early expression of an immune evasion molecule and its repression at later stages of the infectious process.
Such collaboration between these two sRNAs to control the expression of a virulence gene is a new vision of S. aureus physiology. However, the use of several sRNAs to control the expression of a target gene has been identified in other bacteria. For instance, in enterobacteria, expression-level monitoring of a single mRNA that encodes outer membrane proteins is performed by multiple RNAs (38–40). In E. coli, translation of the RpoS alternative sigma factor is regulated by at least four sRNAs. DsrA, RprA and ArcZ sRNAs all stimulate RpoS translation (4,41–44), whereas a fourth sRNA, OxyS, negatively regulates RpoS expression (45,46). Another remarkable case recently reported is that of at least five sRNAs that work together to control the expression of the CsgD transcription factor, essential in E. coli adhesion and biofilm production (47).
The two regulatory RNAs SprD and RNAIII control the expression of a common target through a shared mechanism. They both prevent translation initiation by antisense pairings: exclusively at the mRNA TIS for SprD (26), and at three distinct sites, including the mRNA TIS, for RNAIII. The SprD-sbi mRNA interaction involves the first 41 nt at the sbi mRNA 5′-end that includes its SD sequence and AUG initiation codon [(26) and Supplementary Figure S7]. The RNAIII-sbi mRNA pairing positioned deep in the sbi mRNA coding sequence also includes the SD sequence and AUG codon. There is a 13 nt overlap between RNAIII and SprD at the mRNA TIS (Supplementary Figure S7), implying that each sRNA acts independently to reduce sbi translation and that their regulations are mutually exclusive. This is in agreement with our in vitro results, which demonstrate that RNAIII reduces sbi translation initiation independently of SprD (Figure 5B). Moreover, it is the first evidence to our knowledge that RNAIII controls the expression of a target exclusively at translational level.
In this report, we propose that RNAIII uses three binding sites to control sbi expression. One of these is an imperfect duplex that sequesters the mRNA TIS, whereas the two others involve sbi mRNA nucleotides from deep in its coding sequence (Figure 4A). The RNAIII-sbi mRNA pairing interactions extend to the 46th codon of the target mRNA, far downstream from the initiation signals, a distance which is unprecedented. The involvement of a novel regulatory module within RNAIII structure that interacts with the sbi mRNA TIS to repress Sbi translation was demonstrated in vivo. The roles of the other two interacting domains from RNAIII in the regulation of Sbi expression are, however, unknown. Remarkably, the 5′domain of RNAIII increases the Sbi expression levels in vivo (Figure 6B). Expressing the RNAIII 5′ domain in vivo increases the sbi mRNA levels (data not shown), and additional investigations will be required to understand the mechanism, enhancing mRNA stability and/or transcription. The use of multiple RNA binding sites is probably essential for such a multifunctional RNAIII to achieve accurate and coordinated regulations of its numerous direct molecular targets and to increase specificity for binding its molecular targets. Several other mRNA targets of RNAIII use two distant domains for the interaction. The first, RNAIII-dependent rot mRNA translational repression, involves two domains remote from the rot mRNA 5′-leader region and recognized by the loop–loop pairings involving the RNAIII H7 and H14 hairpins (19). RNAIII also arrests translation of staphylocoagulase (coa), a virulence factor promoting human plasma clotting, by interacting at two sites on coa mRNA: the TIS and further along at the 13th codon (20). Multiple mRNA target regulations by RNAIII are achieved by specific interactions outside the TIS of each mRNA, which can surprisingly extend >130 nt downstream from the start codon, as we have shown here. An extreme case was also documented in E. coli, where trans-acting DsrA interacts with the hns mRNA initiation and termination codons located 400 nt away (48). Another spectacular example of sRNA regulation was reported in Bacillus subtilis. The SR1 sRNA inhibits translation initiation of the ahrC mRNA and induces structural changes downstream from the TIS of its mRNA target (49).
The molecular mechanisms of RNAIII-mediated downregulation of several early virulence factors (such as coagulase, SpA, the peptidoglycan hydrolase LytM, Rot and Sbi) are strikingly similar to each other, with antisense pairings preventing translation initiations. The structural domains of RNAIII involved in these regulations, however, are variable (50), thus allowing a single RNAIII molecule to regulate them all. For the RNAIII-sbi mRNA interaction, a remarkable array of RNAIII domains is involved, including its 5′- and 3′-domains. This contrasts with previous reports in which only three hairpins from RNAIII (H7, H13 and H14) were reported to be needed to arrest translation of several virulence factors (19–20,23). Moreover, in this work we have uncovered a novel ‘CU-rich’ regulatory module within RNAIII, in a single strand between hairpins 13 and 14 and essential for Sbi translation control. Alignment of RNAIII gene sequences from various staphylococcal species (51) indicates that this 9-nt stretch is strictly conserved but its flanking sequences are not (Figure 7), suggesting positive selection pressure. This novel regulatory module within RNAIII is predicted to control the expression of other target genes.
Figure 7.

Sequences conservations of the novel regulatory element at RNAIII 3′-end. Sequence alignments of the 3′-end of RNAIII from various species of the Staphylococcus genus illustrating nucleotide conservation in its additional regulatory domain (in grey). For internal positioning within the RNAIII sequence, the helix H14 is indicated. The asterisks indicate the conserved nucleotides.
Interestingly, the RNAIII ‘B’ and ‘C’ self-complement domains are subjected to reactivity changes on complex formation with the sbi mRNA. The unfolding of RNAIII 5′ and 3′-ends will provide access to these interacting sequences located next one another in RNAIII tertiary structure, resulting in an efficient interaction between RNAIII and the sbi mRNA. RNAIII unfolding of its 5′- and 3′-ends allow the concomitant recognition and pairing with the interspaced sbi mRNA binding sites, for regulation.
At both transcriptional and translational levels, RNAIII reduces the expression of a second immune evasion molecule (18,23), SpA (Figure 6D). SpA cloaks the bacteria with IgGs, blocking interactions with neutrophil Fc receptors thus hindering phagocytosis (52). The Sbi and SpA immune evasion molecules are expressed early on, when RNAIII is not yet transcribed, and they then switch off when RNAIII becomes expressed during growth. Concurrent monitoring of the expression of two immune evasion molecules by a single regulatory RNA is an elegant way of regulating the expression of functionally linked molecules during S. aureus growth to achieve a united physiological impact. The expression levels of both the immune evasion molecules are also positively and negatively controlled by additional transcription regulators (Figure 6D), some of which (SarS and Rot) are also directly downregulated by RNAIII, a ‘double-check’ mechanism that probably ensures the regulation process. From the results reported here, we anticipate the existence of additional joint regulators of Sbi and SpA expression.
CONCLUSION
Our report reveals an unprecedented case in S. aureus: the direct control of a single mRNA by two sRNAs. It further reinforces the essential role of the RNome in regulating virulence gene expression. In Gram-positive bacteria, S. aureus becomes a model organism for identifying the targets and regulatory mechanisms of the nearly 250 sRNAs expressed (53), most of which have unknown biological functions. The next challenge will be to understand how these sRNAs are integrated into the intertwined protein-based regulatory networks that are involved in stress adaptation and virulence. At present, the few known examples of regulatory RNA actions on shared targets probably correspond to the tip of the iceberg. We anticipate that similar sRNA collaborations will turn out to be a widespread phenomenon in living organisms.
SUPPLEMENTARY DATA
Supplementary Data are available Online.
FUNDING
Salary support was provided by the Institut National de la Santé et de la Recherche Medicale and through the regional ‘Fonds de maturation’ of Bretagne (to S.C.). Agence Nationale de la Recherche [ANR-09-MIEN-030-01 to B.F.]; Funding for open access charge: Inserm dotation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr J. van den Elsen (University of Bath, UK) for the anti-Sbi antibodies and to M. Hallier and N. Amiot from our laboratory for the purified ribosomes.
REFERENCES
- 1.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nat. Rev. Microbiol. 2008;6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moks T, Abrahmsen L, Nilsson B, Hellman U, Sjoquist J, Uhlen M. Staphylococcal protein A consists of five IgG-binding domains. Eur. J. Biochem. 1986;156:637–643. doi: 10.1111/j.1432-1033.1986.tb09625.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Jacobsson K, Vasi J, Lindberg M, Frykberg L. A second IgG-binding protein in Staphylococcus aureus. Microbiology. 1998;144(Pt. 4):985–991. doi: 10.1099/00221287-144-4-985. [DOI] [PubMed] [Google Scholar]
- 5.Haupt K, Reuter M, van den Elsen J, Burman J, Halbich S, Richter J, Skerka C, Zipfel PF. The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement Factor H and C3b. PLoS Pathog. 2008;4:e1000250. doi: 10.1371/journal.ppat.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koch TK, Reuter M, Barthel D, Bohm S, van den Elsen J, Kraiczy P, Zipfel PF, Skerka C. Staphylococcus aureus proteins Sbi and Efb recruit human plasmin to degrade complement C3 and C3b. PLoS One. 2012;7:e47638. doi: 10.1371/journal.pone.0047638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu. Rev. Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 8.Romby P, Vandenesch F, Wagner EG. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 2007;10:182–188. doi: 10.1016/j.mib.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Romby P, Charpentier E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol. Life Sci. 2010;67:217–237. doi: 10.1007/s00018-009-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–1092. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–7257. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezee-Durant E, Barbet R, Jacquet E, Jacq A, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–6636. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abu-Qatouseh LF, Chinni SV, Seggewiss J, Proctor RA, Brosius J, Rozhdestvensky TS, Peters G, von Eiff C, Becker K. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J. Mol. Med. 2010;88:565–575. doi: 10.1007/s00109-010-0597-2. [DOI] [PubMed] [Google Scholar]
- 16.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, Francois P. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One. 2010;5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chevalier C, Boisset S, Romilly C, Masquida B, Fechter P, Geissmann T, Vandenesch F, Romby P. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 2010;6:e1000809. doi: 10.1371/journal.ppat.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Mu C, Ying X, Li W, Wu N, Dong J, Gao Y, Shao N, Fan M, Yang G. RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. FEBS Lett. 2011;585:899–905. doi: 10.1016/j.febslet.2011.02.021. [DOI] [PubMed] [Google Scholar]
- 23.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chunhua M, Yu L, Yaping G, Jie D, Qiang L, Xiaorong T, Guang Y. The expression of LytM is down-regulated by RNAIII in Staphylococcus aureus. J. Basic Microbiol. 2012;52:636–641. doi: 10.1002/jobm.201100426. [DOI] [PubMed] [Google Scholar]
- 25.Novick RP, Christie GE, Penades JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chabelskaya S, Gaillot O, Felden B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010;6:e1000927. doi: 10.1371/journal.ppat.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antal M, Bordeau V, Douchin V, Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 30.Jones RC, Deck J, Edmondson RD, Hart ME. Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J. Bacteriol. 2008;190:5265–5278. doi: 10.1128/JB.00383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benito Y, Kolb FA, Romby P, Lina G, Etienne J, Vandenesch F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA. 2000;6:668–679. doi: 10.1017/s1355838200992550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huttenhofer A, Noller HF. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 1994;13:3892–3901. doi: 10.1002/j.1460-2075.1994.tb06700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J. Infect. Dis. 2010;201:241–254. doi: 10.1086/649570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustafsson E, Karlsson S, Oscarsson J, Sogard P, Nilsson P, Arvidson S. Mathematical modelling of the regulation of spa (protein A) transcription in Staphylococcus aureus. Int. J. Med. Microbiol. 2009;299:65–74. doi: 10.1016/j.ijmm.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Gao J, Stewart GC. Regulatory elements of the Staphylococcus aureus protein A (Spa) promoter. J. Bacteriol. 2004;186:3738–3748. doi: 10.1128/JB.186.12.3738-3748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oscarsson J, Harlos C, Arvidson S. Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC 8325-4. Int. J. Med. Microbiol. 2005;295:253–266. doi: 10.1016/j.ijmm.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 37.Burman JD, Leung E, Atkins KL, O'Seaghdha MN, Lango L, Bernado P, Bagby S, Svergun DI, Foster TJ, Isenman DE, et al. Interaction of human complement with Sbi, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus. J. Biol. Chem. 2008;283:17579–17593. doi: 10.1074/jbc.M800265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 40.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 41.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 42.Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 43.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Harb. Perspect. Biol. 2010;3:pii: a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang A, Altuvia S, Storz G. The novel oxyS RNA regulates expression of the sigma s subunit of Escherichia coli RNA polymerase. Nucleic Acids Symp. Ser. 1997:27–28. [PubMed] [Google Scholar]
- 46.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 47.Boehm A, Vogel J. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol. Microbiol. 2012;84:1–5. doi: 10.1111/j.1365-2958.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- 48.Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl Acad. Sci. USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heidrich N, Moll I, Brantl S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007;35:4331–4346. doi: 10.1093/nar/gkm439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Felden B, Vandenesch F, Bouloc P, Romby P. The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 2011;7:e1002006. doi: 10.1371/journal.ppat.1002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tegmark K, Morfeldt E, Arvidson S. Regulation of agr-dependent virulence genes in Staphylococcus aureus by RNAIII from coagulase-negative staphylococci. J. Bacteriol. 1998;180:3181–3186. doi: 10.1128/jb.180.12.3181-3186.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foster TJ. Immune evasion by staphylococci. Nat. Rev. Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 53.Guillet J, Hallier M, Felden B. Emerging functions for the Staphylococcus aureus RNome. PLoS Pathog. 2013;9:e1003767. doi: 10.1371/journal.ppat.1003767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.