Abstract
G-protein coupled receptors (GPCRs) are an important class of membrane protein that transmit extracellular signals invoked by sensing molecules such as hormones and neurotransmitters. GPCR dysfunction is implicated in many diseases and hence these proteins are of great interest to academia and the pharmaceutical industry. Leucine-rich repeat-containing GPCRs contain a characteristic extracellular domain that is an important modulator of intracellular signaling. One member of this class is the leucine-rich repeat-containing G-protein-coupled receptor 5 (LGR5), a stem cell marker in intestinal crypts, and mammary glands. LGR5 modulates Wnt signaling in the presence of the ligand R-spondin (RSPO). The mechanism of activation of LGR5 by RSPO is not understood, nor is the intracellular signaling mechanism known. Recently reported structures of the extracellular domain of LGR5 bound to RSPO reveal a horseshoe-shaped architecture made up of consecutive leucine-rich repeats, with RSPO bound on the concave surface. This review discusses the discovery of LGR5 and the impact it is having on our understanding of stem cell and cancer biology of the colon. In addition, it covers functional relationships suggested by sequence homology and structural analyses, as well as some intriguing conundrums with respect to the involvement of LGR5 in Wnt signaling.
Keywords: GPCR, LGR5, RSPO, Wnt signaling, colon cancer, stem cells
G-Protein Coupled Receptors (GPCRs)
G-protein coupled receptors belong to one of the largest and most diverse families of membrane proteins. In humans GPCRs are encoded by more than 800 genes.1 GPCRs are important signal transducers that control key physiological functions including immune responses, hormone, and enzyme release from endocrine and exocrine glands, neurotransmission, cardiac, smooth muscle contraction, and blood pressure regulation. GPCRs respond to a wide gamut of stimuli ranging from photons of light, to ions (H+ and Ca2+), small organic molecules, peptides, and proteins.2 Once ligand binding has occurred, the receptor undergoes a change that causes the activation of cytosolic signaling molecules, resulting in a cellular response.
Present day drugs for allergies, hypertension, reflux, depression, asthma, and cancer all act by modulating the activity of GPCRs. In reality, 50–60% of all current therapeutic agents directly or indirectly target GPCRs.3 Because of their number, diversity and critical role(s) in signaling, GPCRs offer extraordinary opportunities for development of novel drugs. Defining the molecular changes that accompany function in different classes of GPCRs is not only of fundamental scientific interest, but holds enormous prospects for improving our knowledge of stem cell biology and enhancing human health.
After a short introduction to the description and status of GPCR structural biology, this review focuses on a particular GPCR family, the leucine-rich repeat-containing G-protein coupled receptors (LGRs).
Structure of classical GPCR family members
Structure determination of GPCRs is challenging at all stages, including protein expression, purification, and crystallization. The field is now, however, taking advantage of the high-throughput revolution in structural biology, utilizing an array of methods developed to stabilize and engineer GPCR proteins for crystallization and analysis. These methods include the introduction of T4 lysozyme and apocytochrome into linker regions of GPCRs,4–6 cocrystallization with simplified monoclonal antibody fragments derived from camels and llamas,7 thermostabilization of GPCRs by multiple systematic point scanning mutagenesis8 and protein engineering for example, introduction of non-native disulfide bridges. More standard approaches include removal of flexible portions of the receptor and use of high affinity ligands. All such approaches either reinforce crystal contacts or stabilize one conformational state over another. The use of lipid cubic phase and other bilayer mimetic methods and the availability of new types of solubilizing detergents have further increased the crystallization potential of GPCRs. At the time of writing, 22 unique GPCR structures have been deposited in the protein database.9
The molecular structure of a GPCR comprises three “zones” with respect to the membrane: (1) an extracellular region consisting of the N-terminus and three extracellular loops (ECL1–ECL3), (2) a transmembrane (TM) region consisting of seven α-helical segments (TM1–TM7) and (3) an intracellular region consisting of three intracellular loops (ICL1–ICL3), an intracellular amphipathic helix, and the C-terminus [Fig. 1(A)]. A detailed analysis of the different GPCR structural domains is provided in Venkatakrishnan et al.9
Figure 1.
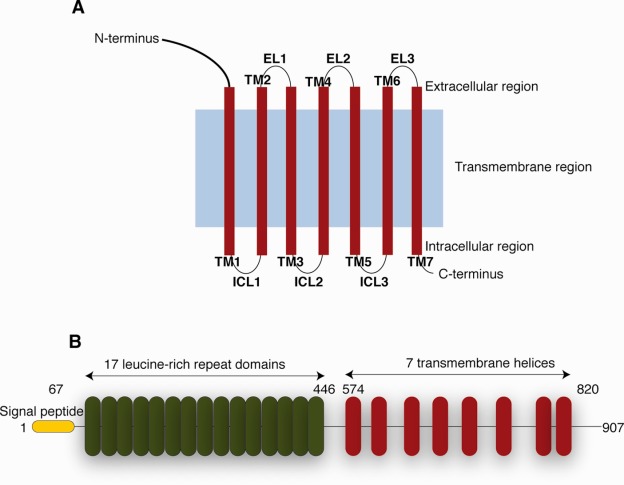
Schematic presentation of the general structure of GPCRs and LGR5. (A) General architecture of GPCRs. (B) LGR5 contains a signal peptide (yellow) followed by 17 leucine-rich repeat (LRR) domains (red). It contains a linker region between the last LRR and the first TM domain, followed by a seven helical TM domain homologs to rhodopsin-like GPCR.
Active, intermediate-active, and inactive states of GPCRs have been observed and have provided important insights into the general mechanism of GPCR activation.10–12 The binding of ligands to the extracellular region appears to result in changes to interactions between the extracellular domain and the transmembrane region. This results in subtle conformational changes in the TM core. It is thought to precede larger structural rearrangements in the membrane cytoplasm that facilitate the binding of intracellular effectors (e.g., heterotrimeric G-proteins and β-arrestins).13
Classification of GPCRs
Nonsensory GPCRs (i.e., those excluding light-, odor-, and taste-receptors) have been classified according to their pharmacological properties: Class A are rhodopsin-like, Class B are secretin-like, Class C are metabotropic glutamate/pheromone, and the fourth Class comprises the frizzled/smoothened receptor families. Class A is the largest and has been further subdivided into four groups α, β, γ, and δ (Table1).14 The δ group contains olfactory receptors as well as purine, MAS-related and the leucine-rich repeat-containing receptors (LGRs).
Table 1.
Classification of Class A GPCRs {Stevens, 2013 #221}
| Class A GPCRs | |||
|---|---|---|---|
| α-group | β-group | γ group | δ group |
| Prostaglandin | Orexin | Somatostatin | Olfactory receptors Purine |
| Amine | Neuropeptide | Opioids | MAS-related |
| Opsin | Neurokinin | Galanin | Leucine-rich repeat-containing receptors |
| Melatonin | Bombesin | Melanin concentrating hormone | |
| Melanocortin | Neurotensin | Chemokine peptides | |
| Cannabinoid | Ghrelin | ||
| Adenosine | Neuromedin | ||
| Arginine | |||
| Vasopressin | |||
| Gonadotropin-releasing hormone | |||
| Oxytocin | |||
Leucine-rich repeat-containing GPCRs (LGRs)
The LGR proteins are a distinct subset of evolutionarily conserved Class A GPCRs, which harbor a rhodopsin-like GPCR and a large extracellular domain with multiple leucine-rich repeats (LRR).15 LRRs are structural motifs that consist of a conserved 11-residue sequence rich in hydrophobic amino acids; often leucines are at defined positions (LxxLxLxxNxL, where x is any amino acid). The tertiary fold of a string of LRR repeats is known as an α/β horseshoe.15 The extracellular domain links ligand binding to modulation of downstream LGR intracellular signaling pathways.16 LGR family proteins have been categorized into three main groups (A, B, and C), according to the relative abundance of LRRs in the ectodomain, the presence of a low-density lipoprotein receptor class A domain (LDLa) and the length of a hinge region connecting the GPCR region to the extracellular domain.17,18
Type A LGR receptors are characterized both by a long hinge region and by having seven to nine LRRs in their ectodomain. The glycoprotein hormone receptors, like follicle stimulating hormone receptor (FSHR), luteinizing hormone receptor (LHR), and thyroid-stimulating hormone receptor (TSHR), belong to the Type A receptor subfamily. Type C receptors have similar number of LRRs to Type A, but are distinguishable by a shorter hinge region than Type A and the presence of an LDLa motif. This subgroup includes the relaxin hormone receptors LGR7 and LGR8.15,19 Signal transduction via Type A and C receptors is thought to occur when hormone binding to the ectodomain triggers conformational changes within the transmembrane domain, which in turn activates heterotrimeric G-proteins bound to the intracellular loop. This sequence of events results in activation of downstream signaling pathways.20 The Type B receptor family LGR4, LGR5, and LGR6 are characterized by the presence of 13–18 LRRs within the extracellular domain [Fig. 1(B)]. There are only three closely related proteins in this family.
The LGR gene family was originally identified via in silico screens for cDNAs encoding proteins with homology to the Type A glycoprotein hormone receptor.21,22,15 The recent explosion of interest in the LGR group of GPCRs is chiefly due to the their presence on the epithelial stem cells of hair, skin, intestine, and breast tissues.23–27
Discovery and Validation of LGR5 as Adult Stem Cell Marker
LGR5 is a Wnt target gene28 and was discovered by researchers trying to find an interstitial stem cell marker.29 It has been known for many decades that the intestinal epithelium regenerates constantly23 and a small population of stem cells residing at the base of the intestinal crypts drives this regeneration process.30 However, the identity of the crypt stem cells remained elusive because of a lack of specific markers. Epithelial homeostasis in the adult intestine is orchestrated by several signaling pathways including EGFR,31 EpH,32 Notch,33 Hedgehog,34 and Wnt.35 Wnt signaling plays a critical role in maintaining intestinal epithelial cell proliferation.35 Hyperactivation of the Wnt pathway is associated with adenomatous transformation of the intestinal epithelium36 [similar to adenomatous transformation caused by loss of the tumor suppressor gene, adenomatous polyposis coli (APC)36] and is the principal cause of colon cancer in humans.37,38 The role that Wnt signaling plays in the physiology of the intestine suggested that one or more Wnt target genes could be stem cell markers.
Clevers and coworkers identified a Wnt driven genetic programme that is activated in APC-mutant human colon cancer cells.29 The expression programme consists of core set of ∼80 genes. Although the majority of these genes are expressed throughout the proliferative crypt compartment29,28 and in mature Paneth cells,39 the expression of several Wnt target genes appeared to be restricted to the base of the crypts, that is, the stem cell compartment. Of the basally expressed genes, LGR5 is specifically expressed in small wedged-shaped cells present in-between the Paneth cells at the base of the small intestinal crypts. These wedged-shaped cells are known as “crypt base columnar” (CBC) cells and had been identified in 1974 by Cheng and Leblond using electron microscopy.40 CBC cells are morphologically immature cells that gained prominence as a candidate stem cell population following the publication of the “stem cell zone” model by Bjerknes and Cheng.41 LGR5 has now emerged as a candidate stem cell marker in the intestinal crypts. Further examination of LGR5 expression patterns in the mouse found discrete populations of LGR5 expressing cells (LGR5+) in other organs, including skin, large intestine, stomach, mammary gland, tongue, kidney, and endometrium,23–25,42–46 suggesting that LGR5 is a potential “universal epithelial stem cell marker.”44,47
To validate the LGR5+ population as adult epithelial stem cells, in vivo lineage-tracing experiments were conducted on LGR5-expressing CBC cells in mouse small intestine.23 In vivo lineage tracing is a genetic fate-mapping technique in which heritable genetic marks are introduced into candidate stem cell populations in situ in living tissues.48 The descendants of these marked stem cell candidates can be probed in situ for the introduced genetic markers.48 A marked stem cell candidate is said to be multipotent if the entire set of differentiated cell lineages can be traced back to a single stem cell and long-term production of marked cell lineages in a given tissue exhibits the self-renewal capacity of the stem cell candidate.48 Thus a candidate cell demonstrating both multipotency and self-renewal capacity in this system fulfills the requirements to be called an adult stem cell (possessing “stemness”).48
To evaluate the “stemness” of LGR5+ populations in vivo using lineage tracing, a heritable-inducible lacZ reporter gene was introduced into LGR5-expressing cells. Initially resulting in the appearance of lacZ+ cells in the CBC compartment within the crypt base,23 over the course of the week the progressively expanding lacZ+ progeny were observed extending from the crypt base towards the tips of interstitial villi. Similar observations were also made in colon.23 Thus, individual lacZ+ tracing units were present in all epithelial cell lineages and persisted throughout the life of the organism, identifying LGR5+ cells as a truly multipotent, self-renewing population of adult intestinal stem cells. In vitro, small numbers of LGR+ cells are able to generate self-organizing, self-renewing epithelial organoids with an architecture and cell composition that are remarkably similar to in vivo crypts/villus units.49
In vivo and in vitro data identify the LGR5+ cells in the mouse intestine as the proliferating stem cells responsible for the daily self-renewal capacity of the mucous lining. In vivo lineage tracing has also been used to demonstrate “stemness” of LGR5-expressing populations in the adult hair follicle, adult distal stomach, taste buds, and embryonic kidney.24,25,42,43,46 Recently it was shown that mammary glands can be reconstructed efficiently from LGR5+ cells.45 These reconstructed mammary glands exhibit regenerative capacity in serial transplantations.45 Adult tissues that display lower turnover rates, such as the liver,50 respond to acute damage by activating Wnt signaling and consequentially generate LGR5+ stem cells that result in tissue regeneration.51
Mechanism of maintaining epithelial cell homeostasis by LGR5+ stem cells
Validation of LGR5 as a stem cell marker of intestinal epithelial cells allowed the role of stem cells in homeostasis to be studied in greater depth. The stem cell-driven process that maintains the homeostasis of continually renewing intestinal epithelia requires a delicate balance between daily production of committed progeny and new stem cells throughout the lifetime of an organism. Understanding this process in the adult stem cell compartment in vivo is crucial for deciphering how disturbance to this equilibrium contributes to disorders such as cancer.
It has been proposed that adult stem cells within tissues undergo obligate asymmetric division to maintain the balance between production of committed progeny and new stem cells.52 However, recent studies have found compelling evidence of prevalently stochastic, symmetric cell division within the LGR5+ stem cell compartment. In particular, multicolor lineage tracing experiments show that cell division in LGR5+ stem cells is symmetric (Supporting Information Fig. 1). In the short-term, LGR5+ stem cells rarely generate daughter cells that adopt divergent fates. In the long-term, however, the multicolor stem cell pool is converted to a single-color population, indicating a gradual shift towards clonality.53 Thus it appears likely that LGR5+ stem cells double daily and that adoption of stem cell or progenitor fate is determined stochastically. It has been independently demonstrated that the segregation of chromosomes during mitosis of LGR5+ intestinal stem cells is random. At present the molecular mechanisms that stimulate LGR5+ intestinal stem cell division and their subsequent fate are not known.
Functions and mechanism of action of LGR5
Much of our understanding of LGR5 function has come from the analysis of null or loss-of-function mutants. A knock-in mouse strain harboring a lacZ reporter gene 5′ to the region that encodes the first transmembrane domain creates a null allele.54 In homozygotes, disruption of LGR5 results in 100% neonatal lethality, characterized by gastrointestinal tract dilation and absence of milk in the stomach. Histological examination of the homozygote mice revealed fusion of the tongue to the floor of the oral cavity (condition called ankyloglossia), while immunostaining showed expression of LGR5 in the epithelia of the tongue and mandibles of wild-type embryos. Thus, neonatal lethality of the LGR5 null mice provided the first firm indication that LGR5 is essential in development. The same LGR5-null strain also demonstrated accelerated maturation of Paneth cells in the developing intestine, indicating that LGR5 may negatively regulate Wnt signaling during neonatal intestinal development.55
Further evidence that LGR5 negatively regulates Wnt signaling has also been indicated in colorectal cancer cell lines by overexpression of LGR5 or reduction of LGR5 expression by RNAi.56 Walker et al. illustrated that overexpressing LGR5 in a colon cancer cell line suppresses the response to Wnt signaling, augments cell–cell adhesion, reduces clonogenicity and attenuates tumorigenicity.56 Conversely, knockdown of LGR5 resulted in enhancement of Wnt signaling attributes such as increased invasion, anchorage-independent growth, and enhanced tumorigenicity.56
R-spondins are ligands of LGR5
In 2011, it was discovered that R-spondin (RSPO) family proteins were ligands of LGR5.57–61 R-spondins are required for the production of crypts in vivo and in vitro49 and have a strong mitogenic effect on LGR5+ cells.62,63 The interaction of RSPOs and LGR5 have been assessed by cell surface binding assays, surface plasmon resonance, cell-free coimmunoprecipitation, and a tandem affinity purification mass spectrometry.57–59 The Kds of binding between different RSPOs and LGR5 are in the nanometer range, (e.g., the Kd of hRSPO1-LGR5 interaction was measured at ∼3.1 nM57,58 and that Kd of RSPO3 and LGR5 ∼3.0 nM).59
R-spondins are secreted proteins of ∼35 kDa and RSPO1-RSPO4 share pair-wise amino-acid similarity of 40–60%. The human RSPO1–4 proteins range from 234 to 272 amino acids in length and feature: (i) a hydrophobic, secretion signal sequence at the N-terminus, (ii) adjacent cysteine-rich furin-like (FU) repeats, (iii) a thrombospondin Type I repeat (TSR) domain that can bind matrix glycosaminoglycans and/or proteoglycans, and (iv) a C-terminus basic amino acid-rich (BR) domain of varying length (Fig. 2). Although RSPOs do not initiate Wnt signaling, they bind LGR5, and presumably release its negative regulation of Wnt signaling, thus potentiating Wnt signaling.58,59,64–66
Figure 2.
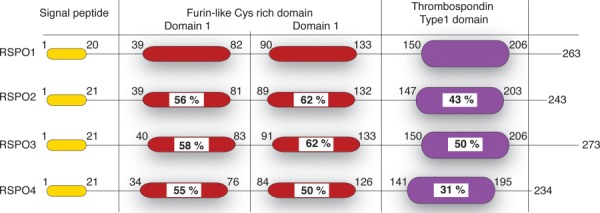
Schematic representation of the domain architecture of RSPO. RSPOs contain a signal peptide followed by two furin-like Cys-rich repeats (red). It contains a thrombospondin type1 domain (violet) and a C-terminal tail of varying lengths. Numbers represent the amino-acid numbers for RSPO. Sequence identity compared to RSPO1 is written as % within the domains.
LGR5, RSPO, and Wnt signaling
Wnt signaling is reviewed in detail elsewhere.67–70 To provide context for the role RSPO and LGR5 in Wnt signaling; however, the canonical Wnt pathway is described briefly here (Fig. 3). The pathway was first identified from genetic screens in Drosophila. The basic molecular signaling framework was further characterized from studies on flies, worms, frogs, fish, and mice.71 In the canonical signaling model, in the absence of Wnt signaling, β-catenin is degraded by a “destruction complex” that comprises of axin, APC, glycogen synthase kinase 3 (GSK3), and casein kinase-1α (CK-1α).72,73 Within this destruction complex β-catenin is multiply phosphorylated, leading to ubiquitination and subsequent proteolytic destruction of β-catenin by the proteasome [Fig. 3(A)].72 Axin has been implicated as the critical component mediating β-catenin degradation.74 However, recent data show that not all phosphorylated β-catenin is degraded and that distinct complexes of phospho-β-catenin are present at different subcellular locations and are likely to have specific functions at these locations,74 for example, phosphorylated β-catenin has been implicated in microtubule regrowth at centrosomes,75 and cell adhesion.76 In addition, it has been suggested that a recently identified Wnt3a-induced phospho-β-catenin-APC-α-catenin complex is involved in Wnt3a-mediated changes in cell–cell adhesion in HEK293 cells.77
Figure 3.
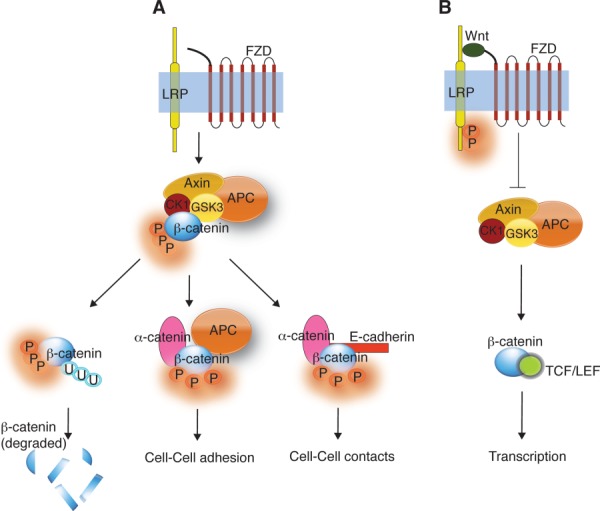
Wnt signaling pathways. (A) In the absence of Wnt, the “destruction complex” (formed by Axin, GSK3, CK1, and APC) phosphorylates β-catenin targeting for ubiquitination and subsequent degradation. In addition, phospho-β-catenin is involved in cell-cell adhesion (with α-catenin and APC) and in cell–cell contacts (with α-catenin and E-cadherin). (B) When Wnt is present, it binds to FZD and LRP forming a ternary complex. This complex inhibits the phosphorylation of β-catenin by the “destruction complex” resulting in translocation of β-catenin into the nucleus. In the nucleus β-catenin binds TCF/LEF resulting in gene transcription.
Wnt initiates signaling by binding to a receptor complex composed of Frizzled (FZD) and lipoprotein receptor-related protein 5/6 (LRP5/6). The Wnt-FZD-LRP5/6 complex inhibits the degradation of β-catenin [Fig. 3(B)].72 In both humans and mice, the FZD receptor family has 10 members belonging to the GPCR superfamily.78 The LRP5/6 receptors are single-pass transmembrane proteins with an extracellular domain containing four EGF (epidermal growth factor)-repeats.72 Formation of a ternary complex of Wnt, FZD, and LRP5/6 switches on β-catenin-TCF-induced transcription72 and changes in cell–cell and cell matrix adhesion.79
Overexpression of LGR5 antagonizes Wnt signaling,56 possibly by reducing access of the Wnt/FZD complex to LRP5/6, but there may also be more indirect effects triggered by signaling from the RSPO-LGR5 complex. The likely outcome of LGR5 antagonism via sequestration of LRP5/6 would be to cause β-catenin phosphorylation and targeting for degradation [Fig. 4(A)]. Over-expression of LGR5 in HEK293 or colon cancer cells stimulates cell–cell adhesion and decreases cell motility.56 Such effects may be associated with the changes in phosphorylation state of β-catenin and subsequent changes in its subcellular distribution. LGR5 also interacts with the tumor suppressor TROY (a member of the tumor necrosis factor receptor superfamily).80 If TROY is recruited to the Wnt/FZD signaling complex via its interaction with LGR580 it could destabilize the cell surface Wnt/FZD/LRP5/6 complex, thereby causing a reduction in Wnt signaling [Fig. 4(B)].80
Figure 4.
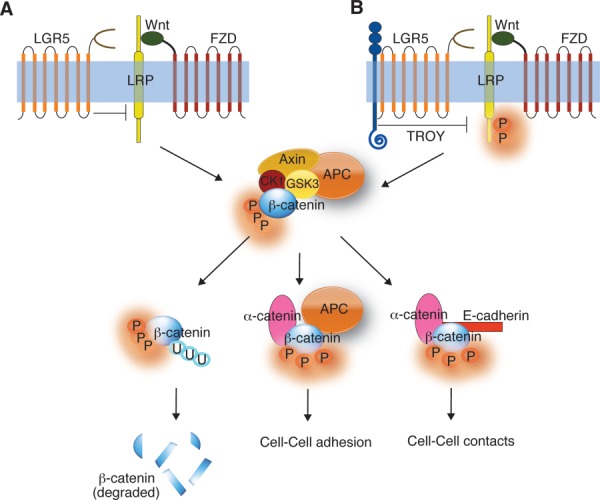
Effect of LGR5 overexpression on Wnt signaling. (A) Overexpression of LGR5 might antagonize Wnt signaling by sequestering LRP5/6, resulting in β-catenin degradation. (B) LGR5 might downregulate Wnt signaling by recruiting TROY that might, in turn, inhibit LRP5/6 leading to the degradation of β-catenin. Scenarios (A) and (B) results in an increase in cell-cell adhesion and cell-cell contacts.
In the presence of RSPO, the inhibitory effect of LGR5 on Wnt signaling appears to be abolished. The formation of the LGR5:RSPO complex potentiates Wnt signaling in HEK293T cells57–59 but the mechanism is unclear; in particular, there is no evidence that binding of RSPO to LGR5 leads to G-protein-mediated activation of typical intracellular messengers such Ca2+ or cAMP.57,58 One model for potentiation of Wnt signaling involves a direct interaction between RSPO:LGR5 and the Wnt/FZD/LRP5/6 complex. When LGR5 receptor is used as bait, a physical interaction between LGR5 and FZD/LRP6 can be detected by mass spectrometric analysis.58 On this basis, it has been suggested that a “Wnt potentiating complex” (RSPO/LGR5/LRP5/6/WNT/FZD) may form at the membrane [Fig. 5(A)].58
Figure 5.
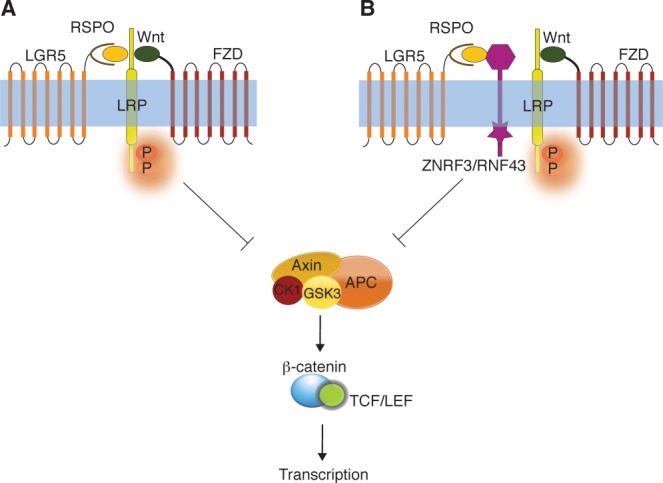
Effect of RSPO:LGR5 complex on Wnt signaling. (A) LGR5:RSPO interacts with FZD, LRP, and Wnt to form a “potentiating complex” that inhibits the phosphorylation of β-catenin by the “destruction complex.” This results in gene transcription (enhance Wnt signaling). (B) The LGR5:RSPO complex might interact with the negative Wnt regulator, ZNRF3/RNF43 to enhance Wnt signaling.
Phosphorylation of a serine residue in LRP6 can be detected within 30 min of RSPO stimulation.57,81 Interestingly, this observation concurs with previous findings that phosphorylation of a serine in LRP is the earliest molecular event occurring during activation of Wnt signaling pathway and that it potentiates the endocytosis of the receptors (LGR5/LRP/FZD) and the ligands (RSPO/WNT).60 In contrast to caveolin-dependent LRP6 endocytosis after WNT stimulation,82 the endocytosis of LGR5, LRP6, and FZD induced by WNT and RSPO cotreatment appears to be mediated by clathrin.59,60
There are conflicting reports as to whether endocytosis of LGR5 and LRP6 are critical for Wnt signal activation. In brief, while one study59 indicates that endocytosis of the receptor complex is critical for WNT signaling, another study60 reports that blocking endocytosis has no effect on the activation of Wnt signaling. The understanding of the role of endocytic pathway during LGR5 signaling is further complicated by a recent study that shows constitutive internalization of LGR5, in the apparent absence of RSPOs, through a dynamin GTPase.83 The internalized LGR5 was then shown to transit through a retromer complex (important in recycling transmembrane receptors from endosomes84) that regulates retrograde trafficking to the trans-golgi network.83 Further investigation is needed to map out the role of endocytosis in both Wnt and LGR5 signaling.
It is also possible that the LGR5:RSPO complex enhances Wnt signaling by interacting with the cell-surface transmembrane E3 ubiquitin ligases, zinc, and ring finger 3 (ZNRF3) and/or its homologs ring finger 43 (RNF43).85 Recent studies have implicated ZNRF3 and RNF43 in fine-tuning Wnt signaling in the intestinal stem cell compartment.85,86 ZNRF3 and RNF43 are negative feedback regulators of Wnt signaling that appear to promote the ubiquitinylation of the FZD and LRP6 receptors on the cell surface.85,86 As for the LRP5/6 interaction, association of LGR5:RSPO with ZNRF3/RNF43 may promote removal of ZNRF3/RNF43 from the plasma membrane and, consequentially, increase the levels of FZD and LRP5/6 enhancing the Wnt signaling response [Fig. 5(B)].85
At present it appears that LGR5 acts as an intrinsic negative regulator of Wnt signaling. In the presence of RSPO, LGR5 inhibition of Wnt signaling is removed, leading to an amplified cellular response to the presence of Wnt. Understanding the critical molecular mechanisms associated with the RSPO:LGR5 regulation of Wnt signaling is a key goal in stem cell biology. It is also important to determine whether the RSPO-LGR5 complex activates intracellular signaling pathways independently of the Wnt-FZD complex.
Structural comparison of LGR5 to other LGRs and other glycoprotein hormone receptors
LGR5 is closely related to LGR4 and LGR6 with ∼50% sequence identity. In comparison, it has 33% identity to glycoprotein hormone receptors. LGR5 and LGR4 have 17 LRR in contrast to 13 in LGR6 and nine in glycoprotein hormone receptors. The leucine-rich repeat region of mammalian LGRs is flanked by cysteine-rich segments. The C-terminal flanking segment of LGR4 and LGR5 contains a cysteine-rich, chemokine-like domain, similar to the consensus CF3 subtype domain found in 45 glycoprotein hormone receptors.17 The core sequences of this consensus CF3 domain (CCAF and FK/NPCE sequences) are completely conserved but the number of residues separating the conserved cysteines in LGR4 and LGR5 (CC-4X-C-4/54X-C) differs from that in the three known human glycoprotein hormone receptors (CC-15/23X-C-31/88X-C).21
Crystal structures of complexes incorporating the FU1-FU2 fragment of RSPO1 were determined in the presence (2 Å) [Fig 6(A)] or absence (to 3.2 Å) of the ectodomain of LGR5.87 In RSPO1, each FU domain has an essentially β-fold of hairpin-like elements interconnected by disulfide bonds, in the manner of cysteine-knot proteins. The hydrogen-bonding pattern is atypical. The two FU domains are orthonormal. When bound to the LGR5 ectodomain, RSPO1 undergoes a conformational change, approximately aligning the FU domains and resulting in a flatter morphology [Fig. 6(B)]. In the same study the LGR5:RSPO complex was crystallized in four independent crystal forms. In all four structures, the LGR5:RSPO complex exists as a dimer-of-heterodimers (i.e., 2:2), even though size-exclusion chromatography had indicated a 1:1 LGR5:RSPO complex. This is consistent with oligomerization of the ectodomain being a concentration-dependent process. Alternatively, the 2:2 interfaces may be held together by low affinity interactions that do not survive gel filtration. The LGR5:RSPO structures from the four different crystal forms superimpose closely, with an RMSD of 1.0 Å over the entire Cα of LGR5 [Fig. 6(C)]. However, the structures diverge at or near the C-termini. This might be due to an absence of structural constraints provided by the transmembrane domain of LGR5 or by the lipid bilayer itself.
Figure 6.
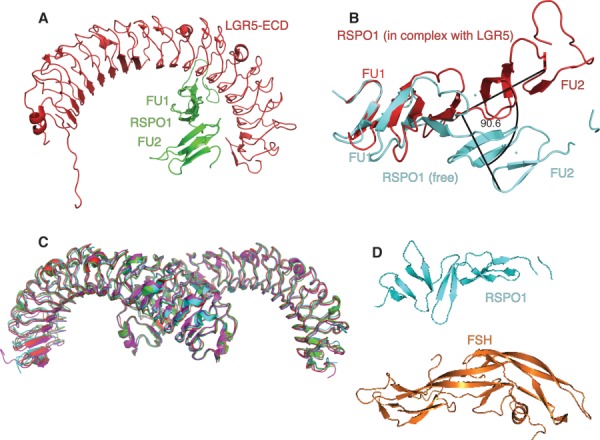
Crystal structures of LGR5-ectodomain:RSPO1 complexes. (A) X-ray crystal structure of the LGR5-ECD (red) in complex with the two furin-like domains (FU1-FU2) of RSPO1 (green) (PDB code: 4BSS). (B) The structures of the FU1-FU2 domains from free RSPO1 (cyan, PDB code: 4BSO) and RSPO1 in complex with LGR5 (red, PDB code: 4BSS) show a 90.5° change in orientation relative to each other. (C) Overlay (Cα over 482 residues LGR5:RSPO complex) of the four crystal forms of LGR5:RSPO complex. P61224 (green, PDB code:4BST), C2 (cyan, PDB code: 4BSU), P22121 (magenta, PDB code: 4BSR), P21 (red, PDB code: 4BSS). (D) Structure of RSPO1 (cyan; PDB code: 4BSO) as compared to FSH structure (orange; PDB code: 1FL7).
Similarly to FSHR, the LGR5 ectodomain adopts a horseshoe-shaped architecture with C- and N-terminal caps.88 The linker between LGR5 repeats 10 and 11 has two phenylalanines at positions usually occupied by leucines. The binding site of RSPO1 on LGR5 is reminiscent of the FSH binding site on the N-terminal leucine-rich repeat region of FSHR, despite the ligands being quite distinct [Fig. 6(D)]. A significant difference between the binding sites; however, is that of FSHR is bipartite; in FSHR, an additional C-terminal hinge domain clamps FSH in place,88 whereas in LGR5 the C-terminal region does not contact RSPO1 directly.
The LGR5:RSPO interface
The FU1 and FU2 domains of RSPO1 both contact LGR5 in the region containing LRR 3–9. A string of residues (R165–W168) on leucine-rich repeat 5 make close contacts with residues 106–110 of RSPO1-FU2 [Fig. 7(A)]. The flanking phenylalanines, F106 and F110, protrude into a cleft in the surface of the LGR5 ectodomain [Fig. 7(B)]. Residues forming the binding site are conserved in LGR4, LGR5, and LGR6 [Fig. 7(B)], suggesting that all three receptors bind RSPO1 in a similar way. The recently determined structure of the LGR4 ectodomain in complex with the FU1–FU2 fragment of RSPO1 verifies that the RSPO1 binding mode is similar in LGR4.89 Key RSPO1 residues at the binding interface, R87, F106, and F110, are conserved in all four RSPOs (Supporting Information Fig. 2) and are likely to be important for binding to LGR4 and LGR6. Recent mutational studies have shown that truncating the side chains of R87, F106, and F110 decreases both RSPO1 binding to LGR4 and, consequentially, Wnt signaling.89
Figure 7.
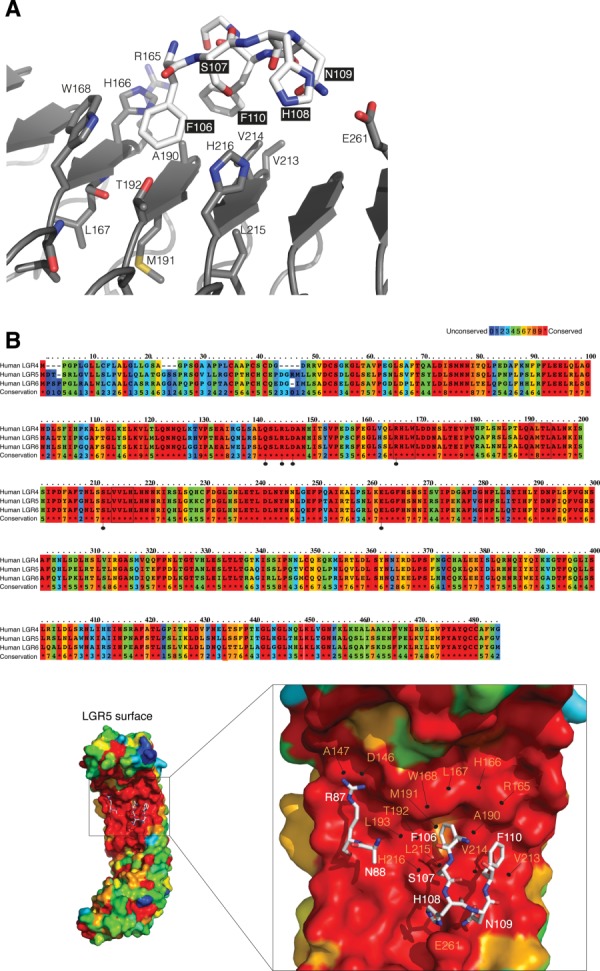
LGR5:RSPO interface. (A) Residues R165 to W168 on LGR5 (gray) make close contacts with residues F106 to F110 on RSPO1 (white). (B) Sequence alignment of human LGR4–6. Residues are colored according to conservation (Highly conserved (Red) to poorly conserved (Blue). Residues that make a H-bond with RSPO1 are marked with a dotted-line (black) (Top). The surface representation of LGR5 colored according to the sequence conservation with RSPO residues in stick representation (white) (bottom). Residues 106–110 in RSPO1 (stick representation; white) are lined by residues in LRR5 (R165, H166, L167, and W168), LRR6 (A190, M191, T192, and L193) and LRR7 (V213, V214, L215, and H216) of LGR5 (surface representation).
In 2013, the structure of a trimeric complex consisting of the ectodomain of LGR5, the FU1:FU2 domains of RSPO1 and the ectodomain of RNF43 [Fig. 8(A)] was reported.90 This structure showed a direct physical interaction between RNF43 and the LGR5:RSPO complex.90 The LGR5 ectodomain from LGR5:RSPO:RNF43 (PDB code: 4BSS) superimposes closely with the LGR5 component of the LGR5:RSPO complex (PDB code: 4KNG) [Fig. 8(B)]. In the trimeric complex, LGR5 does not directly contact RNF43. Instead it binds to the FU1 domain while RNF43 binds the FU2 domain. The affinity of RNF43 for LGR5:RSPO1 has been measured at 10 times higher than its affinity for free RSPO1.90 This suggests that LGR5 reorients RSPO or otherwise potentiates its binding to RNF43, in agreement with previous studies that have shown that the LGR is needed for RSPO1-induced ZNRF3 membrane clearance.85
Figure 8.
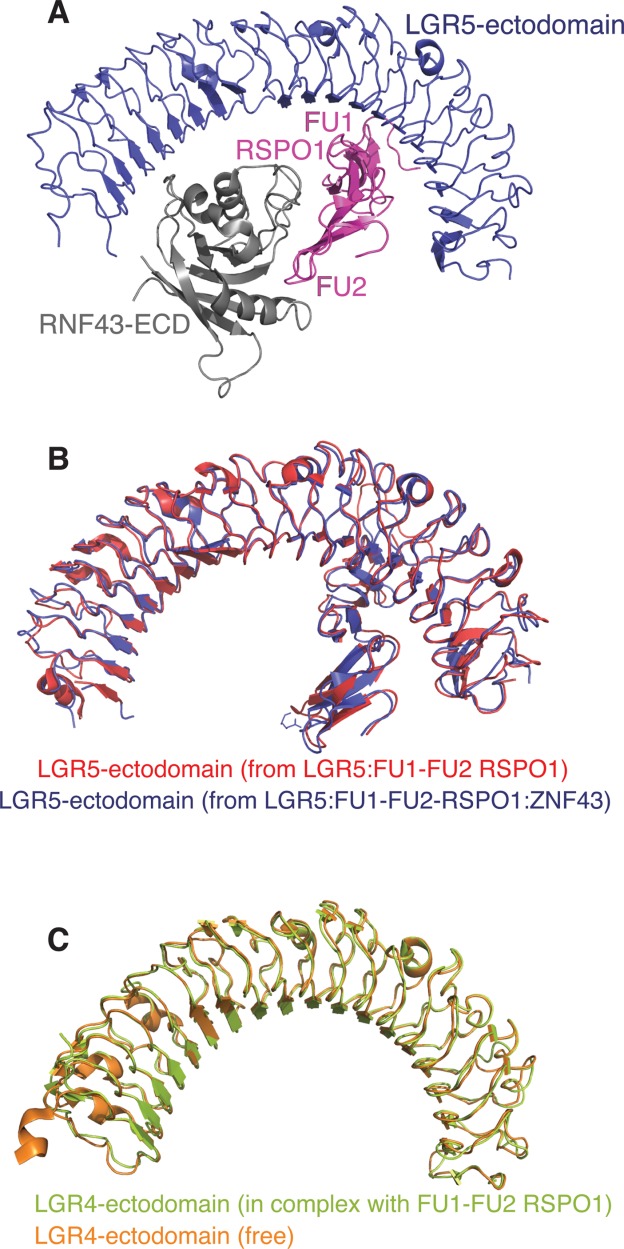
Structures of LGR5/4-ectodomain:RSPO1 complexes. (A) Structure of LGR5-ECD (blue) in a ternary complex with FU1-FU2 domains of RSPO1 (magenta) and RNF43-ECD (gray) (PDB code: 4KNG). (B) Overlay of LGR5-ectodomain:RSPO1 (PDB code: 4BSS) and LGR5-ectodomain:RSPO1:RNF43-ectodomain (PDB code: 4KNG) (Cα 543). (C) The structures of free LGR4 (orange, PDB code: 4LI1) and LGR4 in complex with FU1-FU2 domains of RSPO1 (light green, PDB code: 4LI2) overlay with a RMSD of 0.6 Å (Cα 452).
While RSPO binding does not significantly alter the conformation of LGR4 or LGR5, it disrupts the dimerization of LGR4 [Fig. 8(C)].89 On this basis, it has been hypothesized that RSPO binding alters the receptor oligomerization state of LGR4 and/or its orientation on the cell surface and that this might be important for signal transduction. The role of GPCR oligomerization in signaling is not well characterized, though experimental and theoretical data have proposed roles for GPCR oligomerization in a range of processes from ligand binding and receptor signaling to cell maturation and trafficking.91–93 Further studies are required to investigate LGR4 and LGR5 oligomerization in the light of RSPO effects on Wnt signal transduction.
Intriguingly, a recent study has shown that when the transmembrane domain of LGR5 is replaced by an unrelated single-pass membrane protein, Wnt signaling is reduced to basal levels.87 This shows that binding of RSPO to the LGR5 ectodomain is of itself insufficient to perpetuate Wnt signaling, suggesting that the membrane GPCR domain has a role in signal transduction. The implication, that the α-helical membrane domain plays a role in antagonizing Wnt signaling in its unliganded state, is yet to be tested directly.
Ligand binding to the ectodomain seems likely to facilitate signaling by causing changes within the membrane, similarly to other GPCRs. Agonist-bound structures of the related GPCRs rhodopsin,94 β2-adrenergic receptor (β2-AR),11 and the A2 adenosine receptor12 have helped elucidate the type of structural changes occurring in transmembrane regions of GPCRs during activation. Specifically, these studies have concluded a rearrangement of the TM5-TM6 interface, resulting from movement of a segment of TM6 located in the inner leaflet of the bilayer. The extent of relative TM6 displacement observed between structures varies, but superimposition of two complexes of the β2-adrenergic receptor reveals significant displacement: TM6 of an agonist-bound β2-AR–G-protein complex (PDB code: 3SN6) is 14 Å away from TM6 of an antagonist-bound β2-AR complex (PDB code: 2RH1).10 When agonist is bound, the displacement of TM6 opens up a cleft in the surface where signaling molecules can bind.
To understand whether comparable structural changes in the membrane domain of LGR5 are responsible for triggering downstream signaling events, structure determination of the relevant full-length complexes is vital. No full-length protein structures are yet available for LGR GPCRs. While there are obvious challenges in achieving this, the structures would provide unprecedented insights into its biological role. Additionally, comparing structures of full-length LGR5 with those of other GPCRs would help in elucidating universal principles underlying GPCR signaling.
Until recently there had been no evidence that LGR5 signaling was coupled to G-proteins, In 2013, however, evidence suggesting that LGR5 activates the Gα12/13-Rho GTPase pathway was reported.95 Unexpectedly, the activation of LGR5 was reported to be RSPO-independent, implying that RSPOs are not the ligands relevant to the LGR5:Gα12/13-Rho pathway and opening up the search for other ligands that may couple LGR5 to Gα12/13 pathway. However, it must be noted that in these experiments the possibility of autocrine stimulation by an endogenous RSPO was not considered.
In recent years, so-called biased ligands to other GPCRs selectively activating G-proteins or β-arrestin have been discovered.96 For example, a β-arrestin-biased ligand of the parathyroid hormone receptor results in increased bone density without activating the usual catabolic pathways.97 Another example is a novel angiotensin II Type 1 receptor agonist (TRV120027) that selectively signals via β-arrestins, leading to increased cardiac performance with a reduction in blood pressure98: in the normal circumstance, stimulation with angiotensin causes the angiotensin II Type 1 receptor to signal through the G-protein pathway, resulting in vasoconstriction, increased blood pressure, and decreased cardiac output.98 Biased agonists can and are being used as tools to probe downstream signaling.99 Discovery of biased ligands for directing LGR5 signaling towards the Gα12/13 -Rho pathway would be of great value in illuminating the role of LGR5 in vivo.
Conclusions
LGR5 is a specialized member of the GPCR family that marks stem cells in the epithelia of the colon. It also acts as a negative modulator of Wnt signaling. It was recently discovered that R-spondins are high affinity ligands of LGR4, LGR5, and LGR6. Recent crystal structures of LGR:RSPO complexes define a binding interface where two phenylalanine residues, conserved in RSPOs, project into a cleft on the surface of the ectodomain. The primarily hydrophobic interface is augmented by electrostatic and hydrogen-bonding interactions. In binding, RSPO removes the ability of LGR5 to inhibit FZD based Wnt signals. It is likely that the antagonism results from competing interactions for LGR5 by LRP5/6 and/or RNF43. At present, the antagonism cannot be explained by LGR5-based activation of either G-proteins or β-arrestin. Whilst it is possible that LGR5 ligands other than RSPOs exist, the role of autocrine RSPO stimulation in cell lines needs further investigation. Deducing the links between Wnt signaling, LGR5 signaling and cell-to-cell adhesion will take us significantly further along the path to understanding the role of GPCR signaling in positioning and migration of both normal and cancerous stem cells.
Acknowledgments
JMG is a NHMRC Senior Research fellow, AWB acknowledges funding from the NHMRC Program Grant 487922 and funds from the Operational Infrastructure Support Program provided by the Victorian Government, Australia.
Additional Supporting Information may be found in the online version of this article.
References
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Ji TH, Grossmann M, Ji I. G protein-coupled receptors. I. Diversity of receptor-ligand interactions. J Biol Chem. 1998;273:17299–17302. doi: 10.1074/jbc.273.28.17299. [DOI] [PubMed] [Google Scholar]
- Lundstrom K. Latest development in drug discovery on G protein-coupled receptors. Curr Protein Pept Sci. 2006;7:465–470. doi: 10.2174/138920306778559403. [DOI] [PubMed] [Google Scholar]
- Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, Kunken J, Xu F, Cherezov V, Hanson MA, Stevens RC. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Structure of the agonist-bound neurotensin receptor. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samama P, Cotecchia S, Costa T, Lefkowitz RJ. A mutation-induced activated state of the beta 2-adrenergic receptor. Extending the ternary complex model. J Biol Chem. 1993;268:4625–4636. [PubMed] [Google Scholar]
- Stevens RC, Cherezov V, Katritch V, Abagyan R, Kuhn P, Rosen H, Wuthrich K. The GPCR Network: a large-scale collaboration to determine human GPCR structure and function. Nat Rev Drug Discov. 2013;12:25–34. doi: 10.1038/nrd3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, van der Spek PJ, van Duin M, Hsueh AJ. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol. 2000;14:1257–1271. doi: 10.1210/mend.14.8.0510. [DOI] [PubMed] [Google Scholar]
- Barker N, Tan S, Clevers H. Lgr proteins in epithelial stem cell biology. Development. 2013;140:2484–2494. doi: 10.1242/dev.083113. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Vassart G, Wodak SJ. Modeling of the three-dimensional structure of proteins with the typical leucine-rich repeats. Structure. 1995;3:867–877. doi: 10.1016/S0969-2126(01)00222-2. [DOI] [PubMed] [Google Scholar]
- Van Hiel MB, Vandersmissen HP, Van Loy T, Vanden Broeck J. An evolutionary comparison of leucine-rich repeat containing G protein-coupled receptors reveals a novel LGR subtype. Peptides. 2012;34:193–200. doi: 10.1016/j.peptides.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Kumagai J, Hsu SY, Matsumi H, Roh JS, Fu P, Wade JD, Bathgate RA, Hsueh AJ. INSL3/Leydig insulin-like peptide activates the LGR8 receptor important in testis descent. J Biol Chem. 2002;277:31283–31286. doi: 10.1074/jbc.C200398200. [DOI] [PubMed] [Google Scholar]
- Vassart G, Pardo L, Costagliola S. A molecular dissection of the glycoprotein hormone receptors. Trends Biochem Sci. 2004;29:119–126. doi: 10.1016/j.tibs.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Hsu SY, Liang SG, Hsueh AJ. Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol Endocrinol. 1998;12:1830–1845. doi: 10.1210/mend.12.12.0211. [DOI] [PubMed] [Google Scholar]
- McDonald T, Wang R, Bailey W, Xie G, Chen F, Caskey CT, Liu Q. Identification and cloning of an orphan G protein-coupled receptor of the glycoprotein hormone receptor subfamily. Biochem Biophys Res Commun. 1998;247:266–270. doi: 10.1006/bbrc.1998.8774. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgard R. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Barker N, Clevers H. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology. 2010;138:1681–1696. doi: 10.1053/j.gastro.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, Stange DE, Toftgard R, Clevers H. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- Leushacke M, Barker N. Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. Oncogene. 2012;31:3009–3022. doi: 10.1038/onc.2011.479. [DOI] [PubMed] [Google Scholar]
- Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M, Van Gijn ME, Suijkerbuijk S, Van de Wetering M, Marra G, Clevers H. The Intestinal Wnt/TCF Signature. Gastroenterology. 2007;132:628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Jaks V, Kasper M, Snippert H, Toftgard R, Clevers H. Very long-term self-renewal of small intestine, colon, and hair follicles from cycling Lgr5+ve stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:351–356. doi: 10.1101/sqb.2008.72.003. [DOI] [PubMed] [Google Scholar]
- Yamaoka T, Yan F, Cao H, Hobbs SS, Dise RS, Tong W, Polk DB. Transactivation of EGF receptor and ErbB2 protects intestinal epithelial cells from TNF-induced apoptosis. Proc Natl Acad Sci U S A. 2008;105:11772–11777. doi: 10.1073/pnas.0801463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Wang B. Eph/ephrin signaling in epithelial development and homeostasis. Int J Biochem Cell Biol. 2009;41:762–770. doi: 10.1016/j.biocel.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development. 2005;132:279–289. doi: 10.1242/dev.01576. [DOI] [PubMed] [Google Scholar]
- Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, Clarke AR, Winton DJ. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, Taketo MM, Clevers H. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- Bjerknes M, Cheng H. The stem-cell zone of the small intestinal epithelium. III. Evidence from columnar, enteroendocrine, and mucous cells in the adult mouse. Am J Anat. 1981;160:77–91. doi: 10.1002/aja.1001600107. [DOI] [PubMed] [Google Scholar]
- Barker N, Rookmaaker MB, Kujala P, Ng A, Leushacke M, Snippert H, van de Wetering M, Tan S, Van Es JH, Huch M, Poulsom R, Verhaar MC, Peters PJ, Clevers H. Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep. 2012;2:540–552. doi: 10.1016/j.celrep.2012.08.018. [DOI] [PubMed] [Google Scholar]
- de Visser KE, Ciampricotti M, Michalak EM, Tan DW, Speksnijder EN, Hau CS, Clevers H, Barker N, Jonkers J. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228:300–309. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- Gil-Sanchis C, Cervello I, Mas A, Faus A, Pellicer A, Simon C. Leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5) as a putative human endometrial stem cell marker. Mol Hum Reprod. 2013;19:407–414. doi: 10.1093/molehr/gat014. [DOI] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, Werb Z. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3:70–78. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee KK, Li Y, Redding KM, Iwatsuki K, Margolskee RF, Jiang P. Lgr5-EGFP marks taste bud stem/progenitor cells in posterior tongue. Stem Cells. 2013;31:992–1000. doi: 10.1002/stem.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh M, Smith JL, Huh YH, Sherley JL. A resource for discovering specific and universal biomarkers for distributed stem cells. PLoS One. 2011;6:e22077. doi: 10.1371/journal.pone.0022077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Sanchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, Haft A, Vries RG, Grompe M, Clevers H. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Morita H, Mazerbourg S, Bouley DM, Luo CW, Kawamura K, Kuwabara Y, Baribault H, Tian H, Hsueh AJ. Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol Cell Biol. 2004;24:9736–9743. doi: 10.1128/MCB.24.22.9736-9743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MI, Ghiani M, Lefort A, Libert F, Strollo S, Vassart G. LGR5 deficiency deregulates Wnt signaling and leads to precocious Paneth cell differentiation in the fetal intestine. Dev Biol. 2009;331:58–67. doi: 10.1016/j.ydbio.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Walker F, Zhang HH, Odorizzi A, Burgess AW. LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLoS One. 2011;6:e22733. doi: 10.1371/journal.pone.0022733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, van Es JE, Guardavaccaro D, Schasfoort RB, Mohri Y, Nishimori K, Mohammed S, Heck AJ, Clevers H. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, Niehrs C. LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmon KS, Lin Q, Gong X, Thomas A, Liu Q. LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/beta-catenin signaling. Mol Cell Biol. 2012;32:2054–2064. doi: 10.1128/MCB.00272-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner H, Sprunger J, Charlat O, Leighton-Davies J, Grosshans B, Salathe A, Zietzling S, Beck V, Therier M, Isken A, Xie Y, Zhang Y, Hao H, Shi X, Liu D, Song Q, Clay I, Hintzen G, Tchorz J, Bouchez LC, Michaud G, Finan P, Myer VE, Bouwmeester T, Porter J, Hild M, Bassilana F, Parker CN, Cong F. R-Spondin potentiates Wnt/beta-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One. 2012;7:e40976. doi: 10.1371/journal.pone.0040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KA, Kakitani M, Zhao J, Oshima T, Tang T, Binnerts M, Liu Y, Boyle B, Park E, Emtage P, Funk WD, Tomizuka K. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanskaya O, Glinka A, del Barco Barrantes I, Stannek P, Niehrs C, Wu W. R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev Cell. 2004;7:525–534. doi: 10.1016/j.devcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Kim KA, Wagle M, Tran K, Zhan X, Dixon MA, Liu S, Gros D, Korver W, Yonkovich S, Tomasevic N, Binnerts M, Abo A. R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol Biol Cell. 2008;19:2588–2596. doi: 10.1091/mbc.E08-02-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hutchins BI, Kalil K. Wnt5a induces simultaneous cortical axon outgrowth and repulsive axon guidance through distinct signaling mechanisms. J Neurosci. 2009;29:5873–5883. doi: 10.1523/JNEUROSCI.0183-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Basler K. Wnt trafficking: new insights into Wnt maturation, secretion and spreading. Traffic. 2010;11:1265–1271. doi: 10.1111/j.1600-0854.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus H. Three decades of Wnts: a personal perspective on how a scientific field developed. EMBO J. 2012;31:2670–2684. doi: 10.1038/emboj.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of beta-catenin in mice. Genes Dev. 2008;22:2308–2341. doi: 10.1101/gad.1686208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Nusse R. Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem. 2006;281:22429–22433. doi: 10.1074/jbc.R600015200. [DOI] [PubMed] [Google Scholar]
- Burgess AW, Faux MC, Layton MJ, Ramsay RG. Wnt signaling and colon tumorigenesis—a view from the periphery. Exp Cell Res. 2011;317:2748–2758. doi: 10.1016/j.yexcr.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Huang P, Senga T, Hamaguchi M. A novel role of phospho-beta-catenin in microtubule regrowth at centrosome. Oncogene. 2007;26:4357–4371. doi: 10.1038/sj.onc.1210217. [DOI] [PubMed] [Google Scholar]
- Medrek C, Landberg G, Andersson T, Leandersson K. Wnt-5a-CKI{alpha} signaling promotes {beta}-catenin/E-cadherin complex formation and intercellular adhesion in human breast epithelial cells. J Biol Chem. 2009;284:10968–10979. doi: 10.1074/jbc.M804923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton MJ, Faux MC, Church NL, Catimel B, Kershaw NJ, Kapp EA, Nowell C, Coates JL, Burgess AW, Simpson RJ. Identification of a Wnt-induced protein complex by affinity proteomics using an antibody that recognizes a sub-population of beta-catenin. Biochim Biophys Acta. 2012;1824:925–937. doi: 10.1016/j.bbapap.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Schulte G, Bryja V. The Frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:518–525. doi: 10.1016/j.tips.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fafilek B, Krausova M, Vojtechova M, Pospichalova V, Tumova L, Sloncova E, Huranova M, Stancikova J, Hlavata A, Svec J, Sedlacek R, Luksan O, Oliverius M, Voska L, Jirsa M, Paces J, Kolar M, Krivjanska M, Klimesova K, Tlaskalova-Hogenova H, Korinek V. Troy, a tumor necrosis factor receptor family member, interacts with lgr5 to inhibit wnt signaling in intestinal stem cells. Gastroenterology. 2013;144:381–391. doi: 10.1053/j.gastro.2012.10.048. [DOI] [PubMed] [Google Scholar]
- Wei Q, Yokota C, Semenov MV, Doble B, Woodgett J, He X. R-spondin1 is a high affinity ligand for LRP6 and induces LRP6 phosphorylation and beta-catenin signaling. J Biol Chem. 2007;282:15903–15911. doi: 10.1074/jbc.M701927200. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Komekado H, Kikuchi A. Caveolin is necessary for Wnt-3a-dependent internalization of LRP6 and accumulation of beta-catenin. Dev Cell. 2006;11:213–223. doi: 10.1016/j.devcel.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Snyder JC, Rochelle LK, Lyerly HK, Caron MG, Barak LS. Constitutive internalization of the leucine-rich G protein-coupled receptor-5 (LGR5) to the trans-Golgi network. J Biol Chem. 2013;288:10286–10297. doi: 10.1074/jbc.M112.447540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough IJ, Cullen PJ. Recent advances in retromer biology. Traffic. 2011;12:963–971. doi: 10.1111/j.1600-0854.2011.01201.x. [DOI] [PubMed] [Google Scholar]
- Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, Mao X, Ma Q, Zamponi R, Bouwmeester T, Finan PM, Kirschner MW, Porter JA, Serluca FC, Cong F. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM, Clevers H. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- Peng WC, de Lau W, Forneris F, Granneman JC, Huch M, Clevers H, Gros P. Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 2013;3:1885–1892. doi: 10.1016/j.celrep.2013.06.009. [DOI] [PubMed] [Google Scholar]
- Jiang X, Liu H, Chen X, Chen PH, Fischer D, Sriraman V, Yu HN, Arkinstall S, He X. Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc Natl Acad Sci U S A. 2012;109:12491–12496. doi: 10.1073/pnas.1206643109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu Y, Rajashankar KR, Robev D, Nikolov DB. Crystal structures of Lgr4 and its complex with R-spondin1. Structure. 2013;21:1683–1689. doi: 10.1016/j.str.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PH, Chen X, Lin Z, Fang D, He X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013;27:1345–1350. doi: 10.1101/gad.219915.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin JP, Galvez T, Prezeau L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol Ther. 2003;98:325–354. doi: 10.1016/s0163-7258(03)00038-x. [DOI] [PubMed] [Google Scholar]
- Harikumar KG, Morfis MM, Sexton PM, Miller LJ. Pattern of intra-family hetero-oligomerization involving the G-protein-coupled secretin receptor. J Mol Neurosci. 2008;36:279–285. doi: 10.1007/s12031-008-9060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of molecular switches in GPCRs—theoretical and experimental studies. Curr Med Chem. 2012;19:1090–1109. doi: 10.2174/092986712799320556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss J, Edwards PC, D'Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon MS, Park BO, Kim HM, Kim S. Leucine-rich repeat-containing G-protein coupled receptor 5/GPR49 activates G12/13-Rho GTPase pathway. Mol Cells. 2013;36:267–272. doi: 10.1007/s10059-013-0173-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends Mol Med. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesty-Palmer D, Flannery P, Yuan L, Corsino L, Spurney R, Lefkowitz RJ, Luttrell LM. A beta-arrestin-biased agonist of the parathyroid hormone receptor (PTH1R) promotes bone formation independent of G protein activation. Sci Transl Med. 2009;1:1ra1. doi: 10.1126/scitranslmed.3000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin JD, DeWire SM, Yamashita D, Rominger DH, Nguyen L, Schiller K, Whalen EJ, Gowen M, Lark MW. Selectively engaging beta-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J Pharmacol Exp Ther. 2010;335:572–579. doi: 10.1124/jpet.110.173005. [DOI] [PubMed] [Google Scholar]
- Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


