Abstract
The high yields required for the economical production of chemicals and fuels using microbes can be difficult to achieve due to the complexities of cellular metabolism. An alternative to performing biochemical transformations in microbes is to build biochemical pathways in vitro, an approach we call synthetic biochemistry. Here we test whether the full mevalonate pathway can be reconstituted in vitro and used to produce the commodity chemical isoprene. We construct an in vitro synthetic biochemical pathway that uses the carbon and ATP produced from the glycolysis intermediate phosphoenolpyruvate to run the mevalonate pathway. The system involves 12 enzymes to perform the complex transformation, while providing and balancing the ATP, NADPH, and acetyl-CoA cofactors. The optimized system produces isoprene from phosphoenolpyruvate in ∼100% molar yield. Thus, by inserting the isoprene pathway into previously developed glycolysis modules it may be possible to produce isoprene and other acetyl-CoA derived isoprenoids from glucose in vitro.
Keywords: isoprenoids, biofuel, metabolic engineering, green chemistry, commodity chemicals, in vitro synthesis
Introduction
There is currently considerable interest in the metabolic engineering of microbes to produce chemicals and fuels that can replace petroleum based chemicals.1,2 So far, metabolic engineering has had difficulty producing low value commodity chemicals, such as fuels, at low enough costs to compete with petroleum-based chemicals.3,4 Multiple factors plague cellular systems including: myriad off-pathway reactions that reduce yields, product toxicity, slow fermentation, and expensive product separations.5 Recently, cell-free production strategies, which we refer to as synthetic biochemistry, have gained interest as a solution to the problems associated with cellular production.6–8
Synthetic biochemistry offers a great simplification over the complexities of cellular metabolism and can allow greater control over reaction parameters (i.e., pH, temperature, enzyme load), faster reaction rates, and simpler and cleaner product isolation.8 By moving away from cells, competing metabolic pathways are eliminated so that theoretical yields can realistically approach 100%. Traditionally, in vitro bio-transformations have mostly been limited to production of high-value compounds (e.g., cell-free protein synthesis) or the reconstitution of short (less than 3) multistep pathways for the production of high value chemicals or use in assay systems.6 Recently, there has been increasing interest in reconstituting more complex pathways for the production of inexpensive commodity chemicals and fuels.9
Multiple groups have been able to reconstitute glycolysis in vitro to convert glucose to lactate,10 malate,11 ethanol,12–14 isobutanol,13 or butanol15 at high yields. In all these cases, the only cofactor employed for biosynthesis was NAD(P)H. Net ATP production was eliminated by adding arsenate or ATPases12 to rapidly turnover any ATP generated, or by creative pathway redesigns.10,11,13 In one redesign, the ATP-producing glycolytic enzyme glyceraldehyde-phosphate dehydrogenase was replaced with a nonphosphorylating glyceraldehyde–phosphate dehydrogenase, GapN.10 In a more dramatic redesign, a completely non-natural, ATP independent pathway for glucose conversion was developed to produce butanol.13 These efforts highlight the flexibility of synthetic biochemistry, yet the potential diversity of chemicals that could be produced using synthetic biochemistry approaches would be advanced if it were possible to employ other cofactors such as ATP and coenzyme A (CoA).
The biosynthesis of isoprenoids via the mevalonate pathway is an example of an industrially relevant biochemical pathway that utilizes ATP, NAD(P)H, and other cofactors (CoA).16 Isoprenoids are one of the largest families of bioactive compounds that display anti-inflammatory, anti-cancer, and anti-infectious properties and have potential for use as vitamins and biofuels.17 In nature, isoprenoids are made from the building blocks isopentenyl-pyrophosphate (IPP) and dimethylallyl-pyrophosphate (DMAPP) produced from either the 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate pathway (MEP/DOXP pathway) present primarily in bacteria, or the mevalonate pathway present in eukaryotes, archaea, and a few bacteria.16 In the mevalonate pathway, 3 acetyl-CoA molecules along with 2 NADPH and 3 ATP are employed to synthesize either the 5-carbon IPP or DMAPP.
The mevalonate pathway has been reconstituted in vivo in heterologous hosts such as E. coli18 for the biosynthesis of isoprene,19 amorpha-4,11-diene,18 taxadiene,20 limonene,21 carotenoids,22 and long-chain isoprenoid pyrophosphates.23 However, like other in vivo systems, the recombinant mevalonate pathway is limited by the ability of the host to express recombinant proteins, the toxicity of the products and intermediates in vivo, and the presence of competing pathways.3,18 It is possible that if the isoprenoid pathway could be reconstituted in vitro using purified proteins with an appropriate module (glycolysis) supplying ATP, NADPH, and acetyl-CoA, high titers of isoprenoid products could be achieved in a relatively short amount of time at high yields. As a test of this possibility, we reconstituted the mevalonate pathway in vitro and find that high yields of isoprene can be achieved in vitro from the glycolysis products phosphenolpyruvate (PEP) and ATP while recycling NAD(P)+ and CoA.
Results and Discussion
Overview of the in vitro isoprene pathway
As we ultimately want to employ glycolysis to supply carbon, reducing equivalents, and ATP, we designed a system to produce isoprene from the glycolysis intermediate phosphoenolpyruvate (PEP). The biosynthesis of isoprene via the mevalonate pathway requires 2 NADPH and 3 ATP in addition to the 3 acetyl-CoA building blocks (Fig. 1). The canonical glycolysis pathway plus pyruvate dehydrogenase produces a net output of 4 NADH, 2 ATP, and 2 acetyl-CoA. Thus, the mevalonate pathway (1 ATP per acetyl-CoA) perfectly balances the ATP output from glycolysis (1 ATP per acetyl-CoA). On the other hand, an excess of reducing equivalents are produced from glycolysis plus pyruvate dehydrogenase (PDH) than are used in the mevalonate pathway (0.6 per acetyl-CoA) and they are of the wrong kind (NADH vs. NADPH), incompatibilities that we must accommodate.
Figure 1.
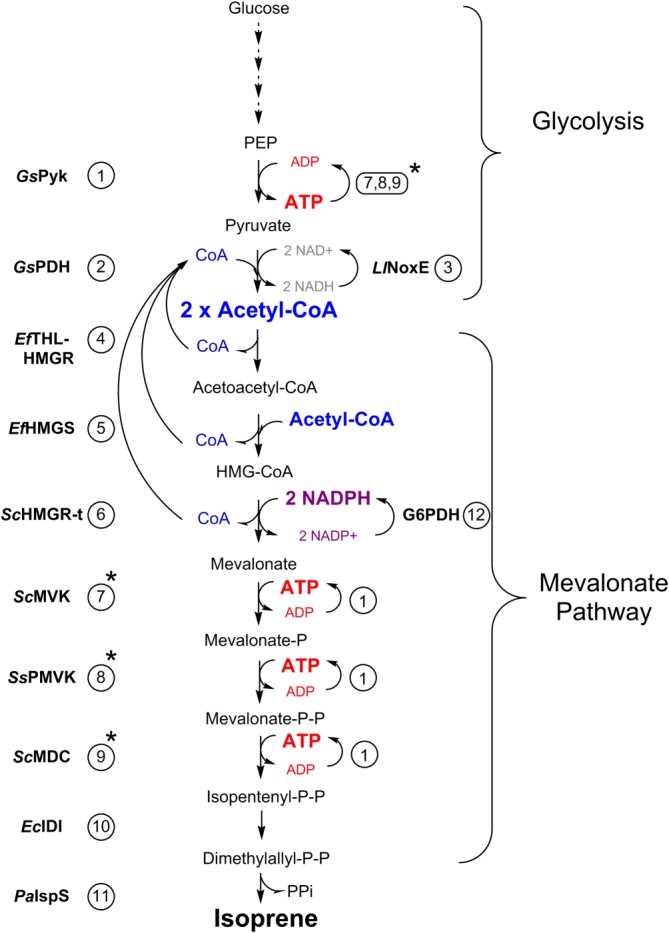
Schematic illustration of the in vitro production of isoprene from glycolysis intermediates. Steps that recycle CoA, ADP, and NADPH are shown in blue, red, and purple, respectively. Enzymes involved in the recycle of ADP and ATP are highlighted with a star. The enzymes used in this study are numbered as in Table1.
In our system, shown in Figure 1, we start from PEP so that we generate the same net 2 ATP produced in glycolysis, but avoid the complication of needing ATP to initiate glycolysis. To produce acetyl-CoA, we use the PDH complex from the thermophile G. stearothermophilus (GsPDH) that is specific for NAD+ [Fig. 2(A)].24 However, a potential limitation of using the PDH complex to produce acetyl-CoA in vitro is feedback inhibition of PDH by NADH.25 Without a pathway to consume NADH, PDH would quickly come to a stop, thereby halting acetyl-CoA biosynthesis. To use the PDH from G. stearothermophilus to generate acetyl-CoA continuously, we employed a water-forming NADH oxidase from Lactobacillus lactis, LlNoxE.26 LlNoxE does not oxidize NADPH at an appreciable rate [Fig. 2(B)], preserving the exogenously added NADPH for isoprene synthesis. The specific enzymes employed in this work are listed in Table1 and shown in Figure 1.
Figure 2.
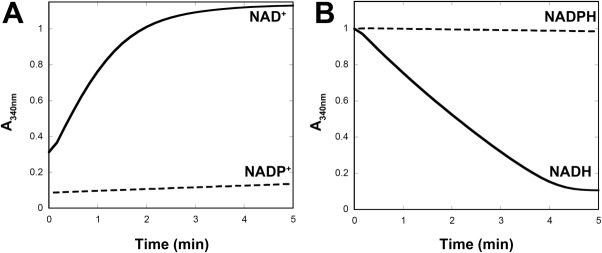
Cofactor specifity of GsPDH and LlNoxE. A: Reduction of NAD+ or NADP+ by GsPDH in the presence of pyruvate and coenzyme A. B: Oxidation of NADH or NADPH by LlNoxE. Reactions for A and B were performed in 50 mM Tris-Cl pH 8.0 containing 2 mM MgCl2 at 37°C and monitored at 340 nm.
Table 1.
List of enzymes employed in this work
| Enzyme | Name | Gene | Accession Number | Plasmid | Tag | Organism | Reference | |
|---|---|---|---|---|---|---|---|---|
| 1 | GsPyk | Pyruvate Kinase | Pyk | CAA40994 | pET28 | N-His | G. stearothermophilus | 27 |
| 2 | GsPDH | Pyruvate Dehydrogenase Complex | G. stearothermophilus | 24 | ||||
| 2a | E1α | Pyruvate Deyhdrogenase Subunit α | E1α | P21873 | pET28 | none | G. stearothermophilus | |
| 2b | E1β | Pyruvate Deyhdrogenase Subunit β | E1β | P21874 | pET28 | none | G. stearothermophilus | |
| 2c | E2 | Dihydrolipoamide Acetyltransferase | E2 | CAA37630 | pET22 | none | G. stearothermophilus | |
| 2d | E3 | Dihydrolipoamide Dehydrogenase | E3 | P11959 | pET28 | none | G. stearothermophilus | |
| 2e | EcLplA | Lipoate Protein Ligase | LplA | NP_418803 | pBAD/ p15A | N-His | E. coli | 28 |
| 3 | LlNoxE | NADH oxidase (H2O forming) | NoxE | YP_007507681 | pET22 | C-His | L. lactis | 26 |
| 4 | EfTHL-HMGR | Thiolase-HMG-CoA Reductase fusion | MvaE | WP_002357755 | pET28 | N-His | E. faecalis | 29 |
| 5 | EfHMGS | HMG-CoA Synthase | MvaS | WP_010785222 | pET28 | N-His | E. faecalis | 30 |
| 6 | ScHMGR-t | truncated HMG-CoA Reductase | HMG1p | EGA77439 | pET28 | N-His | S. cerevisiae | 31 |
| 7 | ScMVK | Mevalonate Kinase | ERG12 | BAA24409 | pET28 | N-His | S. cerevisiae | 32 |
| 8 | SsPMVK | Phosphomevalonate Kinase | ERG8 | NP_344303 | pET28 | N-His | S. sofataricus | 33 |
| 9 | ScMDC | Mevalonate-PP Decarboxylase | MVD1 | NP_014441 | pET28 | N-His | S. cerevisiae | 34 |
| 10 | EcIDI | Isopentenyl-PP Isomerase | IDI | NP_417365 | pET22 | C-His | E. coli | 35 |
| 11 | PaIspS | Isoprene Synthase | IspS | Q50L36 | pET28 | N-His | P. alba | 36 |
| 12 | G6PDH | Glucose-6-Phosphate Dehydrogenase | G6PD | P11412 | S. cerevisiae | Sigma | ||
Testing the mevalonate pathway in vitro
To test whether we could reconstitute the mevalonate pathway in vitro we first divided it into two parts that could each be conveniently monitored separately: (1) the formation of mevalonate from three acetyl-CoA and two NADPH; and (2) the phosphorylation and decarboxylation of mevalonate to IPP using three ATP. It has been shown that the flux through the mevalonate pathway is primarily determined by the first and third steps,29,37 namely the condensation of two acetyl-CoA by thiolase in the first step and reduction and CoA release by HMG-CoA reductase in the third.
To analyze the flux through the first three enzymes (THL, HMGS, HMGR), we monitored the consumption of NADPH at 340 nm, which becomes oxidized in the final step. Figure 3(A) shows the results of employing a mixture of EfTHL-HMGR, EfHMGS, and ScHMGR-t (EfTHL-HMGR is a fusion protein that has both acetyl-CoA acetyltransferase and HMG-CoA reductase activities, but we found that supplementing the reaction with ScHMGR-t enhanced flux). The decrease in A340 reflects a reduction in NADPH levels and indicates that acetyl-CoA can be converted to mevalonate. However, despite using an excess of acetyl-CoA the extent of the reaction only reaches ∼75% conversion based on comparison with the final A340 reached when assayed from pyruvate (below). A similar yield of ∼50% was also obtained when mevalonate was synthesized in vitro from acetyl-CoA using immobilized enzymes in a small bioreactor.38 Most likely a build-up of free CoA or NADP+ (or both) inhibits the forward direction of the mevalonate pathway in vitro.29,39 Indeed, addition of free CoA above 0.5 mM was found to inhibit the reaction (not shown).
Figure 3.
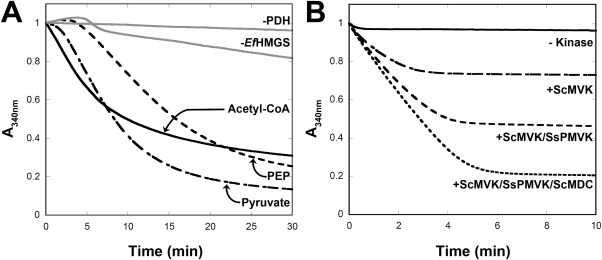
Analysis of the mevalonate pathway in vitro. A: The top portion of the mevalonate pathway (EfTHL-HMGR, EfHMGS, ScHMGR-t) was assayed alone (acetyl-CoA) or in combination with GsPDH/LlNoxE (pyruvate) or GsPyk/GsPDH/LlNoxE (PEP) by monitoring the reduction of HMG-CoA by HMGR. Controls lacking PDH or EfHMGS are shown as gray lines. B: The bottom portion of the mevalonate pathway (ScMVK, SsPMVK, ScMDC) was assayed by limiting mevalonate (0.3 mM). The extent of reaction was monitored by the disappearance of NADH using an ATP regeneration assay consisting of 0.1 mM ADP, 1.5 mM PEP, 5 mM MgCl2, 10 mM KCl, and PK/LDH (Sigma). Addition of each kinase in succession allows the reaction to consume more ATP.
As a solution to the build-up of free CoA, and to test whether it would be possible to couple the mevalonate pathway to glycolysis, we employed the GsPDH/LlNoxE system described above to recycle free CoA and continuously produce mevalonate from pyruvate. Since three CoAs are released, two from the condensation steps and one from the reduction step, CoA (and NAD+) can be recycled through GsPDH. Because NADH (but not NADPH) is quickly oxidized by LlNoxE, NADH never builds up, and the flux through HMG-CoA reductase can be monitored by the disappearance of added NADPH. Figure 3(A) shows that pyruvate can be converted to mevalonate in the presence of NADPH using the GsPDH/LlNoxE system along with EfTHL-HMGR, EfHMGS, and ScHMGR-t. In the conversion of pyruvate to mevalonate shown in Figure 3(A), we started with 1 mM pyruvate, but free CoA and NAD+ were limited at 0.2 mM, meaning that these cofactors were recycled numerous times. When the reaction was started from PEP using GsPyk and ADP, a similar extent of reaction was observed but the reaction was slower than from either pyruvate or acetyl-CoA. It is possible that the build-up of ATP (which we eliminate in the full system below) affects the kinetics of GsPDH.
The second part of the mevalonate pathway was analyzed by starting with a limiting amount of mevalonate and sequentially adding each kinase in an effort to generate DMAPP. As the final step involves an essentially irreversible decarboxylation, we expected to be able to drive the reaction to completion. We employed an ATP regeneration system40 by adding PEP/ADP and GsPyk, which also allowed the reaction to be conveniently monitored by coupling the pyruvate produced to NADH oxidation by lactate dehydrogenase. Figure 3(B) shows the results of adding different combinations of the three kinases. When only the first kinase is added (ScMVK), ATP consumption proceeds to ∼1/3 of the full extent seen for the mixture of all three kinases. When only the first two enzymes (ScMVK and SsPMVK) are added, ATP consumption proceeds to ∼2/3 of the total for the mixture of all three kinases. This result is consistent with the mixture of kinases passing the limiting mevalonate feedstock through the three steps in the pathway.
In vitro production of isoprene from pyruvate and phosphoenolpyruvate (PEP)
Having established that the enzymes from the two halves of the mevalonate pathway could work in vitro, we next attempted to reconstitute the entire pathway to generate isoprene. We first tested isoprene production from acetyl-CoA at 30°C, by simply adding ATP exogenously and directly monitoring isoprene production by GC. Starting from 1.5 mM acetyl-CoA, we added an excess of ATP (3 mM) and NADPH (2 mM) to provide energy and reducing equivalents to the mevalonate pathway. The amount of each enzyme added is listed in Table2. A total of ∼0.8 mg/mL final enzyme loading was used. As shown in Figure 4, acetyl-CoA is converted to isoprene at 89.2 ± 1.1% yield. To increase complexity and determine whether NADP(H) could also be recycled, Glucose-6-phosphate dehydrogenase (G6P DH), and Glucose-6-phosphate were added to generate NADPH in situ.41 Using this NADPH recycling method a slightly higher yield of 92.5 ± 6.5% was obtained from acetyl-CoA.
Table 2.
Enzyme amounts used
| Enzyme | Substrate | Coupling Enzyme | OD | Units/mg | mg added | Units added | |
|---|---|---|---|---|---|---|---|
| 1 | GsPyk | PEP/ADP | LDHa | 340 | 55.2 ± 0.08 | 0.012 | 0.66 |
| 2 | GsPDH | Pyruvate/CoA/NAD+ | none | 340 | 153.4 ± 0.01b | – | 0.038 |
| 3 | LlNoxE | NADH | none | 340 | 9.0 ± 0.002 | 0.021 | 0.19 |
| 4 | EfTHL-HMGR | Acetyl-CoA/NADPH | EfHMGS | 340 | 0.06 ± 0.002c | 0.021 | 0.0013 |
| 5 | EfHMGSd | Acetyl-CoA/Acetoacetyl-CoA | none | 303 | 0.6 ± 0.01 | 0.014 | 0.0084 |
| 6 | ScHMGR-t | HMG-CoA | none | 340 | 0.5 ± 0.08 | 0.012 | 0.0060 |
| 7 | ScMVK | Mevalonate/ATP | PK/LDHa | 340 | 47.0 ± 0.9 | 0.003 | 0.14 |
| 8 | SsPMVK | Mevalonate-P/ATP | PK/LDHa | 340 | 0.8 ± 0.02 | 0.013 | 0.011 |
| 9 | ScMDC | Mevalonate-PP/ATP | PK/LDHa | 340 | 4.0 ± .07 | 0.012 | 0.048 |
| 10 | EcIDI | ND | 0.035e | 0.028 | 0.00098 | ||
| 11 | PaIspS | ND | 0.156f | 0.016 | 0.0025 | ||
| 12 | G6PDHa | ND | 200-400c | 0.005 | 1.0 |
From Sigma.
Units/mL.
Assayed in forward direction (synthesis) by coupling to MvaS. NADPH consumption monitored at 340 nm.
Mutant A110G (From Ref. 30).
From Ref. 35.
From Ref. 36.
Figure 4.
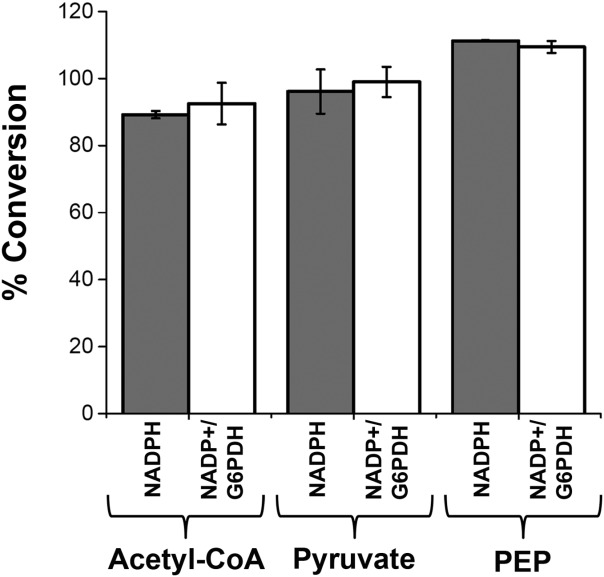
Isoprene production via the in vitro mevalonate pathway. The mevalonate pathway was reconstituted in vitro alone or in combination with GsPDH/LlNoxE or GsPyk/GsPDH/LlNoxE as described in the text and in Table2. NADPH was added to 2 mM from the beginning (gray bars) or regenerated from 0.2 mM NADP+ (white bars) using 6 mM Glucose-6-phosphate and 1 unit of G6PDH. Reactions were incubated for 16 h at 32°C. The % conversion was based on the number of nmol isoprene that should be produced from 1.5 mM Acetyl-CoA, Pyruvate, or PEP in 200 μL reactions. Values represent the average of two independent experiments.
To test whether the mevalonate pathway and isoprene production could be coupled to glycolysis, we used pyruvate as the feedstock, employing the GsPDH/LlNoxE system that recycles CoA and NAD+. As shown in Figure 4 96.1 ± 6.6% of the added pyruvate (1.5 mM) was converted to isoprene when NADPH was added to the reaction, and 99 ± 4.5% was converted to isoprene using the in situ NADPH generating system (NADP+/G6P/G6PDH).
As a proxy for full recycle of all cofactors using glycolysis, the conversion of isoprene from PEP was accomplished by adding GsPyk to the system to generate ATP. Because ATP and acetyl-CoA generated from PEP are perfectly balanced with their consumption in the mevalonate pathway, the reaction should be self-balancing as long as there is no spontaneous hydrolysis of ATP. As can be seen in Figure 4, full conversion of 1.5 mM PEP to isoprene was obtained while all cofactors were limited (0.2 mM CoA, NAD+, and NADP+, and 0.5 mM ADP). In the active system, CoA and NAD+, were recycled eight times, NADP+ was recycled 10 times, and ADP was recycled three times. Finally, a time course of the conversion of PEP to isoprene was performed (Fig. 5). After a small lag over the first 30 min, isoprene production remains constant at 214.5 μmol L−1 h−1 (14.6 mg L−1 h−1).
Figure 5.
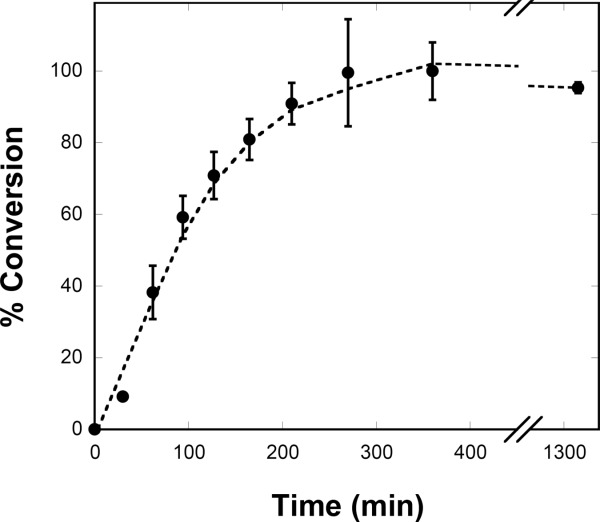
Time course of isoprene production from PEP in vitro. Time course of isoprene production from 1.5 mM PEP. The percent conversion of PEP to isoprene at each time point is shown. The data represent an average of three independent trials. The reaction contained 0.2 mM CoA, 0.2 mM NAD+, 0.2 mM NADP+, 0.5 mM ADP, 5 mM MgCl2, 10 mm KCl, 0.05 mM thiamine pyrophosphate, 0.1 mM AMP, and 6 mM G6P in 100 mM tris-Cl, pH 8.0 and was incubated at 32°C. The amount of each enzyme added is listed in Table2.
Conclusions
The broad application of synthetic biochemistry, where long enzymatic pathways are reconstituted in vitro for the biosynthesis of complex biochemicals from biomass, requires appropriate ATP utilization strategies. Glycolysis produces ATP in addition to reducing equivalents and carbon building blocks in the form of pyruvate, making glycolysis an attractive module to power synthetic biochemistry. The approach described here shows that the ATP produced from glycolysis can be used to produce isoprenoid compounds in vitro at high yields.
The economic viability of bioconversion to isoprene depends on high yields to minimize input materials costs.45 We have shown here that we can obtain essentially quantitative conversion of PEP to isoprene through eight enzymatic steps. Efforts are now underway to fuse the enzyme system developed here to a complete glycolytic system so that the complex conversion of simple sugars to isoprene can be performed. Although the current rate of production (14.6 mg L−1 h−1) is still ∼100-fold below the productivity desired for the industrial production of a commodity chemical (∼2 g L−1 h−1),46 there are many avenues to improve throughput. It should be possible to increase the rates substantially by raising the concentrations of the feedstock, increasing enzyme concentrations, optimizing temperature, pH, and other solution conditions. We expect this approach could also prove useful for generating many other isoprenoid-derived chemicals.
Materials and Methods
Materials
Miller LB media or Miller LB-agar (BD Difco) was used for growth of bacterial strains in liquid or solid media. E. coli BL21Gold(DE3) [B, F-, ompT, hsdSB, (rB–,mB–), dcm+,Tetr,galλ,(DE3) endA Hte] (Agilent) was used as host for both cloning and expression of recombinant proteins using pET vectors. E. coli TOP10(DE3) [F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ−] (Invitrogen) was used for expression of recombinant proteins from the pBAD/p15A vector. Plasmids pET28a(+) and pET22b(+) were purchased from Novagen. Plasmid pBADp15A was described previously.42 HotStart Taq Mastermix (Denville) was used for gene amplification from genomic or plasmid DNA. Phusion DNA polymerase (Finnizymes), Taq DNA ligase (MCLab), and T5 Exonuclease (Epicenter) were purchased separately and used to make the assembly master mix (AMM) used for cloning. ATP, ADP, (±)-α-lipoic acid, phosphoenolpyruvate, pyruvate, coenzyme A, NADPH, and NAD+ were from Sigma. All other chemicals were analytical grade or higher.
Expression plasmids for glycolysis and mevalonate pathway enzymes
The genes encoding glycolysis and mevalonate pathway enzymes (see Table1) were amplified from genomic DNA by the polymerase chain reaction (PCR) using HotStart Taq Mastermix (Denville) except for Isoprene Synthase from P. alba which was synthesized by DNA2.0 without codon optimization. In addition to 15–25 base-pairs complementary to the start and end of the gene necessary for amplification, 15–20 base-pair overhangs were included at the 5′ and 3′ ends that were complementary to the vector to be cloned into. For constructs with an N-terminal 6xHis tag, vector pET28 was first digested with NdeI and XhoI. L. lactis NoxE contained a C-terminal 6xHis-tag and was cloned into pET22 digested NdeI and XhoI. For constructs without affinity tags (GsPDH subunits), pET28 digested with NcoI and XhoI or pET22 digested with NdeI and XhoI was used.
A modified Gibson method was used to assemble all expression plasmid constructs.43 In general, 1 μL (10–20 ng) of vector DNA that had been digested with appropriate restriction enzymes was mixed with 1.5 μL (20–50 ng) of PCR product, and 7.5 μL of assembly mix (0.1M Tris-Cl, pH 7.5, 0.2 mM dNTPs, 1 mM NAD+, 5% PEG 8k, 10 mM DTT, 10 mM MgCl2, 0.00375 U/μL T5 Exonuclease, 0.012 μL Phusion DNA polymerase, 4 U/μL TaqDNA ligase) and incubated at 50°C for 2 h. Totally, 5 μL of the assembly mixture was then used to transform either E. coli TOP10(DE3) (EcLplA) or BL21Gold(DE3) (all other enzymes) and plated on selective media (LB-agar containing 50 μg/mL kanamycin, 100 μg/mL ampicillin, or 34 μg/mL chloramphenicol). Mutagenesis of EfHMGS to introduce the A110G mutation30 was done using the Quickchange Site-Directed Mutagenesis Kit (Stratagene).
Overexpression and purification of enzymes and reconstitution of active PDH complex in vitro
All E. coli strains were grown at 37°C in LB-media supplemented with appropriate antibiotic (100 μg/mL ampicillin, 50 μg/mL kanamycin, or 34 μg/mL chloramphenicol). For all constructs, 5 mL of overnight starter culture was used to inoculate 1 L of LB-media. Once the OD600 reached 0.6, 0.3 mM IPTG (pET vectors) or 0.02% arabinose (pBAD/p15A vectors) was added to induce protein expression. After 16 h, cells were harvested, resuspended (4 mL/g wet cells) in 50 mM Tris-Cl pH 7.5, 0.1M NaCl (Buffer A), lysed by sonication, and cell debris removed by centrifugation at 30,000g for 20 min.
The E. coli lysates containing overexpressed 6xHis-tagged proteins were purified by affinity chromatography. Lysate containing 6xHis-tagged protein was loaded onto 3 mL Ni-NTA resin (Qiagen), washed with 25 mL Buffer A containing 10 mM imidazole, and eluted with 5 mL Buffer A containing 250 mM imidazole.
E. coli lysates containing the overexpressed non-tagged individual domains of GsPDH were partially purified by heat prior to modification and reconstitution of the PDH complex. E1αβ−, E2-, or E3-containing cell-free lysates were incubated at 65°C for 35 min to heat denature E. coli proteins followed by centrifugation at 100,000g for 60 min to separate precipitated proteins. Nearly all of the GsPDH domains remain in the supernatant. Next, the E2 domain was lipoated by addition of 1 mM (±)-α-lipoic acid, 2 mM ATP, 3 mM MgCl2, and 50 μg of purified 6xHis-LplA.44 The lipoation reaction was then allowed to proceed with gentle inversion overnight at 25°C yielding lipoated E2 (E2lip). After lipoation, E1αβ, E2lip, and E3 were mixed in a 3 : 1 : 3 molar ratio and incubated for at least 1 h at 25°C to form the active GsPDH complex. The GsPDH complex was then isolated by ultracentrifugation (Beckman) for 4 h at 95,000g. The resulting yellow pellet was resuspended in 20 mM Tris-Cl, pH 7.5 in 1/50 the starting volume and assayed for activity.24 SDS-PAGE analysis confirmed the presence of all four domains. The reconstituted complex was stored at 4°C until use.
Enzyme assays
All measurements of enzymatic activity were performed in 200 μL of 50 mM Tris-Cl pH 8.0 at 37°C on a SpectraMax M5 plate reader. Enzymes that consumed ATP (ScMVK, SsPMVK, ScMDC) were assayed by monitoring the oxidation of NADH at OD340 using an ATP regeneration system with PEP and PK/LDH (Sigma). Activity of EfTHL-HMGR and ScHMGR-t were assayed in the synthetic direction (production of mevalonate) by monitoring the oxidation of NADPH by HMGR at 340 nm.29 To monitor the conversion of acetyl-CoA to mevalonate, the reaction with EfTHL-HMGR was supplemented with EfHMGS30 and incubated with 0.6 mM acetyl-CoA, 0.4 mM NADPH, 1 mM MgCl2, and 2 mM KCl. EfHMGS was assayed at 303 nm with 0.5 mM acetyl-CoA and 0.1 mM acetoacetyl-CoA.30 For monitoring the synthesis of mevalonate from pyruvate, 1 mM pyruvate, 0.2 mM CoA, 0.2 mM NAD+, and 0.6 mM NADPH were mixed with GsPDH complex, LlNoxE, EfTHL-HMGR, EfHMGS, and ScHMGR-t at 37°C and the oxidation of NADPH was monitored at 340 nm as above. The activity of EcIDI and PaIspS were confirmed by analyzing the production of isoprene from 1 mM mevalonate and 3 mM ATP in the presence of ScMVK, SsPMVK, ScMDC, 5 mM MgCl2 and 10 mM KCl at 37°C. The production of isoprene was confirmed by Gas Chromatography (GC).
In vitro production of isoprene from pyruvate or PEP
To monitor in vitro isoprene production from glycolysis intermediates, 200 μL reactions were set up in gas-tight sealed vials. 1.5 mM substrate, 0.2 mM CoA, 0.2 mM NAD+, 0.2 mM NADP+ (or 2 mM NADPH), 0.5 mM ADP, 5 mM MgCl2, 10 mm KCl, 0.05 mM thiamine pyrophosphate, 0.1 mM AMP, 6 mM G6P in 100 mM tris-Cl, pH 8.0 buffer were mixed with the amount of each enzyme listed in Table2 and incubated at 32°C. Isoprene production was monitored by sampling the headspace using solid phase microextraction (SPME)36 followed by GC-FID (HP5890II) equipped with a GS-GasPro column (0.32 mm × 30 m, Agilent). The SPME fiber was directly applied to the inlet and isoprene was identified by comparison to an authentic isoprene standard. The carrier gas was helium with a flow rate of 5 mL/min. The oven temperature was kept at 120°C for 3 min and then raised to 230 at 5°C/min then to 250 at 20°C/min and held at 250°C for 9 min. The inlet and detector temperatures were kept at 270 and 330°C, respectively. The amount of isoprene was determined by comparison to a standard curve of various isoprene concentrations.
References
- Yadav VG, De Mey M, Giaw Lim C, Kumaran Ajikumar P, Stephanopoulos G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metabol Eng. 2012;14:233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AC. Metabolic engineering is key to a sustainable chemical industry. Nat Prod Rep. 2011;28:1406–1425. doi: 10.1039/c1np00029b. [DOI] [PubMed] [Google Scholar]
- Cardinale S, Arkin AP. Contextualizing context for synthetic biology—identifying causes of failure of synthetic biological systems. Biotechnol J. 2012;7:856–866. doi: 10.1002/biot.201200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C. Big oil turns on biofuels. Nat Biotech. 2013;31:870–873. doi: 10.1038/nbt.2704. [DOI] [PubMed] [Google Scholar]
- Kittleson JT, Wu GC, Anderson JC. Successes and failures in modular genetic engineering. Curr Opin Chem Biol. 2012;16:329–336. doi: 10.1016/j.cbpa.2012.06.009. [DOI] [PubMed] [Google Scholar]
- You C, Zhang Y-HP. Cell-Free Biosystems for Biomanufacturing. In: Zhong J-J, editor. Future trends in biotechnology. Advances in biochemical engineering/biotechnology. Berlin Heidelberg: Springer; 2013. pp. 89–119. Available at: http://link.springer.com/chapter/10.1007/10_2012_159. [DOI] [PubMed] [Google Scholar]
- Hodgman CE, Jewett MC. Cell-free synthetic biology: thinking outside the cell. Metabol Eng. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollin JA, Tam TK, Zhang Y-HP. New biotechnology paradigm: cell-free biosystems for biomanufacturing. Green Chem. 2013;15:1708–1719. [Google Scholar]
- Oroz-Guinea I, García-Junceda E. Enzyme catalysed tandem reactions. Curr Opin Chem Biol. 2013;17:236–249. doi: 10.1016/j.cbpa.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Ye X, Honda K, Sakai T, Okano K, Omasa T, Hirota R, Kuroda A, Ohtake H. Synthetic metabolic engineering-a novel, simple technology for designing a chimeric metabolic pathway. Microbial Cell Factories. 2012;11:120. doi: 10.1186/1475-2859-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Honda K, Morimoto Y, Okano K, Ohtake H. Direct conversion of glucose to malate by synthetic metabolic engineering. J Biotechnol. 2013;164:34–40. doi: 10.1016/j.jbiotec.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Welch P, Scopes RK. Studies on cell-free metabolism: ethanol production by a yeast glycolytic system reconstituted from purified enzymes. J Biotechnol. 1985;2:257–273. [Google Scholar]
- Guterl JK, Garbe D, Carsten J, Steffler F, Sommer B, Reiße S, Philipp A, Haack M, Rühmann B, Koltermann A, Kettling U, Brück T, Sieber V. Cell-free metabolic engineering: production of chemicals by minimized reaction cascades. ChemSusChem. 2012;5:2165–2172. doi: 10.1002/cssc.201200365. [DOI] [PubMed] [Google Scholar]
- Stevenson BJ, Liu J-W, Kuchel PW, Ollis DL. Fermentative glycolysis with purified Escherichia coli enzymes for in vitro ATP production and evaluating an engineered enzyme. J Biotechnol. 2012;157:113–123. doi: 10.1016/j.jbiotec.2011.09.019. [DOI] [PubMed] [Google Scholar]
- Krutsakorn B, Honda K, Ye X, Imagawa T, Bei X, Okano K, Ohtake H. In vitro production of n-butanol from glucose. Metabol Eng. 2013;20:84–91. doi: 10.1016/j.ymben.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Lombard J, Moreira D. Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol Biol Evol. 2011;28:87–99. doi: 10.1093/molbev/msq177. [DOI] [PubMed] [Google Scholar]
- Dewick PM. The biosynthesis of C5–C25 terpenoid compounds. Nat Prod Rep. 1997;14:111–144. doi: 10.1039/np9971400111. [DOI] [PubMed] [Google Scholar]
- Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotech. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhao G, Sun Y, Zheng Y, Jiang X, Liu W, Xian M. Bio-isoprene production using exogenous MVA pathway and isoprene synthase in Escherichia coli. Bioresource Tech. 2012;104:642–647. doi: 10.1016/j.biortech.2011.10.042. [DOI] [PubMed] [Google Scholar]
- Ajikumar PK, Xiao W-H, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Gutierrez J, Chan R, Batth TS, Adams PD, Keasling JD, Petzold CJ, Lee TS. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metabolic Eng. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Yoon S-H, Lee Y-M, Kim J-E, Lee S-H, Lee J-H, Kim J-Y, Jung K-H, Shin Y-C, Keasling JD, Kim S-W. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol Bioeng. 2006;94:1025–1032. doi: 10.1002/bit.20912. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhou J, Jang H-J, Yoon S-H, Kim J-Y, Lee S-G, Choi E-S, Kim S-W. Engineered heterologous FPP synthases-mediated Z,E-FPP synthesis in E. coli. Metabolic Eng. 2013;18:53–59. doi: 10.1016/j.ymben.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Domingo GJ, Chauhan HJ, Lessard IAD, Fuller C, Perham RN. Self-assembly and catalytic activity of the pyruvate dehydrogenase multienzyme complex from Bacillus stearothermophilus. Eur J Biochem. 1999;266:1136–1146. doi: 10.1046/j.1432-1327.1999.00966.x. [DOI] [PubMed] [Google Scholar]
- Visser J, Kester H, Huigen A. Purification and some properties of the pyruvate dehydrogenase complex of Bacillus subtilis. FEMS Microbiol Lett. 1980;9:227–232. [Google Scholar]
- Lopez de Felipe F, Hugenholtz J. Purification and characterisation of the water forming NADH-oxidase from Lactococcus lactis. Intl Dairy J. 2001;11:37–44. [Google Scholar]
- Sakai H, Suzuki K, Imahori K. Purification and properties of pyruvate kinase from Bacillus stearothermophilus. J Biochem. 1986;99:1157–1167. doi: 10.1093/oxfordjournals.jbchem.a135579. [DOI] [PubMed] [Google Scholar]
- Morris TW, Reed KE, Cronan JE. Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J Biol Chem. 1994;269:16091–16100. [PubMed] [Google Scholar]
- Hedl M, Sutherlin A, Wilding EI, Mazzulla M, McDevitt D, Lane P, Burgner JW, Lehnbeuter KR, Stauffacher CV, Gwynn MN, et al. Enterococcus faecalis acetoacetyl-coenzyme A thiolase/3-hydroxy-3-methylglutaryl-coenzyme A reductase, a dual-function protein of isopentenyl diphosphate biosynthesis. J Bacteriol. 2002;184:2116–2122. doi: 10.1128/JB.184.8.2116-2122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steussy CN, Robison AD, Tetrick AM, Knight JT, Rodwell VW, Stauffacher CV, Sutherlin AL. A structural limitation on enzyme activity: the case of HMG-CoA synthase†,‡. Biochemistry. 2006;45:14407–14414. doi: 10.1021/bi061505q. [DOI] [PubMed] [Google Scholar]
- Polakowski T, Stahl U, Lang C. Overexpression of a cytosolic hydroxymethylglutaryl-CoA reductase leads to squalene accumulation in yeast. Appl Microbiol Biotechnol. 1998;49:66–71. doi: 10.1007/s002530051138. [DOI] [PubMed] [Google Scholar]
- Porter JW. In: Methods in Enzymology. Law HCR, editor. Vol. 110. Academic Press; 1985. pp. 71–78. Mevalonate kinase. In. Steroids and Isoprenoids, Part A. Available at: http://www.sciencedirect.com/science/artic le/pii/S0076687985100613. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Azami Y, Miyagawa M, Hashimoto C, Yoshimura T, Hemmi H. Biochemical evidence supporting the presence of the classical mevalonate pathway in the thermoacidophilic archaeon Sulfolobus solfataricus. J Biochem. 2013;153:415–420. doi: 10.1093/jb/mvt006. [DOI] [PubMed] [Google Scholar]
- Gogerty DS, Bobik TA. Formation of isobutene from 3-hydroxy-3-methylbutyrate by diphosphomevalonate decarboxylase. Appl Environ Microbiol. 2010;76:8004–8010. doi: 10.1128/AEM.01917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters J, Oudjama Y, Ghosh S, Stalon V, Droogmans L, Oldfield E. Structure and mechanism of action of isopentenylpyrophosphate-dimethylallylpyrophosphate isomerase. J Am Chem Soc. 2003;125:3198–3199. doi: 10.1021/ja029171p. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Ohara K, Yazaki K. Gene expression and characterization of isoprene synthase from Populus alba. FEBS Lett. 2005;579:2514–2518. doi: 10.1016/j.febslet.2005.03.066. [DOI] [PubMed] [Google Scholar]
- Soto G, Stritzler M, Lisi C, Alleva K, Pagano ME, Ardila F, Mozzicafreddo M, Cuccioloni M, Angeletti M, Ayub ND. Acetoacetyl-CoA thiolase regulates the mevalonate pathway during abiotic stress adaptation. J Exp Bot err. 2011;287 doi: 10.1093/jxb/err287. [DOI] [PubMed] [Google Scholar]
- Sutherlin A, Rodwell VW. Multienzyme mevalonate pathway bioreactor. Biotechnol Bioengin. 2004;87:546–551. doi: 10.1002/bit.20157. [DOI] [PubMed] [Google Scholar]
- Durr IF, Rudney H. The reduction of β-hydroxy-β-methylglutaryl coenzyme A to mevalonic acid. J Biol Chem. 1960;235:2572–2578. [PubMed] [Google Scholar]
- Crans DC, Kazlauskas RJ, Hirschbein BL, Wong C-H, Abril O, Whitesides GM. Enzymatic regeneration of adenosine 5′-triphosphate: acetyl phosphate, phosphoenolpyruvate, methoxycarbonyl phosphate, dihydroxyacetone phosphate, 5-phospho-α-d-ribosyl pyrophosphate, uridine-5′-diphosphoglucose. In: Klaus Mosbach., editor. Methods in enzymology. Vol. 136. Academic Press; 1987. pp. 263–280. Immobilized enzymes and cells, Part C. Available at: http://www.sciencedirect.com/science/article/pii/S0076687987360276. [DOI] [PubMed] [Google Scholar]
- Chenault HK, Whitesides GM. Regeneration of nicotinamide cofactors for use in organic synthesis. Appl Biochem Biotechnol. 1987;14:147–197. doi: 10.1007/BF02798431. [DOI] [PubMed] [Google Scholar]
- Massey-Gendel E, Zhao A, Boulting G, Kim H-Y, Balamotis MA, Seligman LM, Nakamoto RK, Bowie JU. Genetic selection system for improving recombinant membrane protein expression in E. coli. Protein Sci. 2009;18:372–383. doi: 10.1002/pro.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Meth. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Lessard IAD, Domingo GJ, Borges A, Perham RN. Expression of genes encoding the E2 and E3 components of the Bacillus stearothermophilus pyruvate dehydrogenase complex and the stoichiometry of subunit interaction in assembly in vitro. Eur J Biochem. 1998;258:491–501. doi: 10.1046/j.1432-1327.1998.2580491.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y-HP, Myung S, You C, Zhu Z, Rollin JA. Toward low-cost biomanufacturing through in vitro synthetic biology: bottom-up design. J Mater Chem. 2011;21:18877–18886. [Google Scholar]
- Lau MW, Gunawan C, Balan V, Dale BE. Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 for cellulosic ethanol production. Biotechnol Biofuels. 2010;3:11. doi: 10.1186/1754-6834-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


