Abstract
Maize ChitA chitinase is composed of a small, hevein-like domain attached to a carboxy-terminal chitinase domain. During fungal ear rot, the hevein-like domain is cleaved by secreted fungal proteases to produce truncated forms of ChitA. Here, we report a structural and biochemical characterization of truncated ChitA (ChitA ΔN), which lacks the hevein-like domain. ChitA ΔN and a mutant form (ChitA ΔN-EQ) were expressed and purified; enzyme assays showed that ChitA ΔN activity was comparable to the full-length enzyme. Mutation of Glu62 to Gln (ChitA ΔN-EQ) abolished chitinase activity without disrupting substrate binding, demonstrating that Glu62 is directly involved in catalysis. A crystal structure of ChitA ΔN-EQ provided strong support for key roles for Glu62, Arg177, and Glu165 in hydrolysis, and for Ser103 and Tyr106 in substrate binding. These findings demonstrate that the hevein-like domain is not needed for enzyme activity. Moreover, comparison of the crystal structure of this plant class IV chitinase with structures from larger class I and II enzymes suggest that class IV chitinases have evolved to accommodate shorter substrates.
Keywords: chitinase, chitin, crystal structure, plant defense, mass spectrometry, enzyme catalysis
Introduction
More than 800 million tons of maize (Zea mays L.) are produced worldwide each year (http://www.fas.usda.gov/psdonline/). Harvested maize kernels are used for food, as feed for livestock and other animals, and for fuel production. Plants protect developing kernels as they mature within the ears by having an effective system of defense. This system is composed of physical barriers, chemical messengers, and proteins that detect and respond to pathogen attack. Despite these defenses, some specialized fungal pathogens can parasitize developing kernels and cause ear rot.1 Maize ear rot not only reduces yield but also results in contamination of harvested kernels with fungal mycotoxins. Mycotoxin contamination reduces the value of harvested kernels and threatens human and animal health.2,3 An increased understanding of maize defense mechanisms and how they overcome by fungal ear rot pathogens is essential for development of commercial maize hybrids with improved disease resistance and reduced mycotoxin contamination.
ChitA chitinase has been identified as a protein involved in maize defense against ear rot. Encoded by the chiA gene GRMZM2G051943,4 ChitA is a basic 28 kDa chitinase that cleaves chitin, a β-1,4-linked homopolymer of N-acetylglucosamine (GlcNAc) (EC 3.2.1.14). It is composed of a small, amino-terminal domain that resembles the chitin binding peptide hevein and a larger, carboxy-terminal catalytic domain that cleaves chitin.5 ChitA was identified initially as an abundant protein that has in vitro antifungal activity in healthy maize seeds.6 It was further implicated in defense against fungal pathogens when it was identified as the target of secreted fungal proteases termed chitinase modifying proteins.7 These proteases, secreted by fungal pathogens during ear rot, truncate ChitA by cleaving peptide bonds in either the amino-terminal domain or the linker region between domains to produce truncated chitinases.8,9 Structural and biochemical knowledge of ChitA and its individual domains is needed to understand the biological role of ChitA and its fungal-protease-truncated forms.
Based on primary amino acid sequence, ChitA belongs to glycoside hydrolase family 19 (GH19).10 By an alternative plant chitinase classification system, GH19 chitinases are further divided into three types—plant classes I, II, and IV. ChitA is a plant class IV chitinase (PC4). These chitinases have smaller catalytic domains than those of classes I and II. The larger catalytic domains of class I and II chitinases are indistinguishable; class I chitinases have amino-terminal hevein-like domains while class II do not. It has been suggested that ChitA and other PC4 chitinases evolved from class I chitinases through a series of three deletions in their catalytic domains.11 There are eight plant GH19 chitinase crystal structures, two plant class I (2Z37, 2DKV), four plant class II (1DXJ, 1CNS, 4DWX and 3W3E) and one plant class IV (3HBD). The substrate specificity of the GH19 chitinases has been studied in some detail.12–15 GH19 chitinases act by a single displacement mechanism, which results in an inversion at the anomeric carbon.16–18 The glycosidase activities occur between the internal β-linked N-acetylglucosamine residues within chitin polysaccharides and are generally characterized as endoglycosidases. In vitro studies with short oligosaccharide polymers of GlcNAc have shown that a major difference in catalytic activity between ChitA and a class II chitinase from barley seeds (Hordeum vulgare L.) is that ChitA catalyzes efficient degradation of tetrameric GlcNAc into dimers while the larger barley chitinase prefers longer substrates.15,19 The details of the Zea mays chitinase specificity have been studied on small chitooligosaccharides and indicate that ChitA and its homologues are endoglycosidases that hydrolyze chitobiose units (dp 2) from larger chitin oligosaccharide substrate.19 The cleavage is selective and releases chitobiose units from the reducing end of chito-oligosaccharides larger than chitotetraose (dp 4), suggesting that the smaller catalytic domains evolved to cleave shorter substrates.
In this article, we investigate the catalytic mechanism and binding characteristics of a Family GH19, PC4 chitinase and report the first crystal structure of a chitinase from Zea mays. Structural studies of this catalytic domain provide evidence for the likely catalytic residues. Kinetic investigations indicate that the N-terminal carbohydrate-binding module (CBM) is not necessary for hydrolysis, and that the truncated ChitA enzyme yields similar products to that of the full-length enzyme.
Results
Hydrolytic capabilities of truncated chitinase A
The hydrolytic abilities of ChitA ΔN and ChitA ΔN-EQ were tested by incubation of chitinase with chitohexaose substrate (dp6; Supporting Information Fig. S1). ChitA cleaved dp6 to dp2 and dp3 products, with transient production of dp4. ChitA ΔN demonstrated similar hydrolysis of dp6 with the observed products of dp2 and dp3. ChitA ΔN-EQ, in which Glu62 has been mutated to Gln, did not degrade dp6. Incubation of each chitinase with purified chitin from crab shells followed by colorimetric determination of released soluble carbohydrate yielded the equivalent result. ChitA produced 68.92 nM soluble products (±3.20) from chitin while ChitA ΔN produced 62.32 nM products (±2.19) under conditions specified in materials and methods. Mutation of Glu62 abolished activity in both full-length and truncated proteins.
Full-length and truncated ChitAs with the Glu62 to Gln mutation were also tested for substrate binding (Supporting Information Fig. S2). The ability of ChitA, ChitA-EQ, and ChitA ΔN-EQ to bind crab shell chitin and chitin agarose were analyzed by binding assays. SDS-PAGE analysis of samples showed that ChitA bound both substrates in this assay (left panel). The bound protein samples showed a single band at 28 kDa. Major bands of 28 kDa were evident for both chitin and chitin agarose bound proteins. Notably, ChitA ΔN-EQ also bound both substrates.
To investigate the influence of a modification to the non-reducing end of chitin based substrates and if they are able to act as inhibitors, two modified chitomer substrates were treated with ChitA in the enzyme assay. Tetra-N-acetylchitopentaose (GlcN-GlcNAc4) is a dp 5 chitopentaose oligosaccharide that contains a free, non-acetylated glucosaminyl residue at the non-reducing terminus. When treated with ChitA in the enzyme assay, the molecular ion for GlcN-GlcNAc4 (m/z 1014.2) disappeared over a period of 1 h to give GlcN-GlcNAc2 trisaccharide (dp 3, m/z 608.5), plus a minor ion due to chitobiose (dp 2, m/z 447.0) (Supporting Information Fig. 1A). The same type of cleavage was observed with modified chitotriomycin-pMP (m/z 937.42) as a substrate, which gave NMe3-chitiotriose (dp 3, m/z 627.3) arising from the non-reducing end [Fig. 1(B)]. Although these modified oligosaccharides are known potent inhibitors of β-N-acetyl-hexosaminidases,20 they have not been tested previously against a type 19 chitinase. We observed no inhibition of ChitA, and rather GlcN-GlcNAc4 and chitotriomycin-pMP are both cleaved by ChitA between residues 2 and 3, similar to the cleavage specificity that we observed on chitopentaose (Supporting Information Fig. S3).
Figure 1.
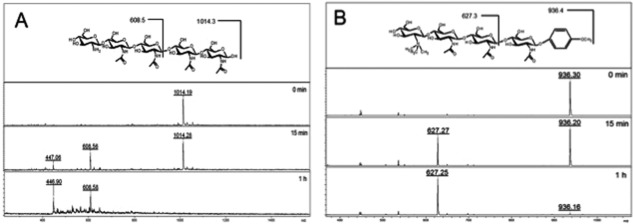
Enzyme activity of ChitA on modified chitin substrates. A: Enzyme activity of ChitA on the modified chitopentaose substrate GlcN-GlcNAc4, which contains a non-acetylated glucosamine residue at the nonreducing end (structure shown at top). The disappearance of the substrate (m/z 1014.4) and appearance of cleavage products (GlcN-GlcNAc2, m/z 608.5; and chitobiose, m/z 447.0) as monitored by MALDI-TOF MS are apparent over the assay time course, with the [M + Na]+ masses revealing the cleavage position. B: Enzyme activity of ChitA on the modified chitotetraose substrate TMG-chitotriomycin-pMP, which contains a N-trimethylamino glucosamine (TMG) residue at the non-reducing end, and a p-methyoxyphenyl protecting group at the anomeric position (structure shown at top). The disappearance of the substrate (m/z 937.4) and appearance of a cleavage product (TMG-GlcNAc2, m/z 627.3) as monitored by MALDI-TOF mass spectrometry is apparent over the assay time course, with the [M + Na]+ masses revealing the cleavage position.
Structure of Zea Mays chitinase
The structure for ChitA ΔN-EQ (4MCK) was successfully solved and is characterized as having two lobes separated by a large cleft. This structure is highly α-helical, comprising eight α-helices, each 6-19 residues in length. Similar to other GH19 chitinases, ChitA ΔN-EQ has five large loops (I-V), stabilized by three-conserved di-sulfide bonds, Cys18-Cys67, Cys79-Cys88, and Cys166-Cys198. The mutation in ChitA ΔN-EQ does not disrupt the overall structure of the catalytic domain of ChitA. Rather, the effects of this mutation are limited to an altered orientation of the side-chain of residue 62 [Fig. 2(A)].
Figure 2.
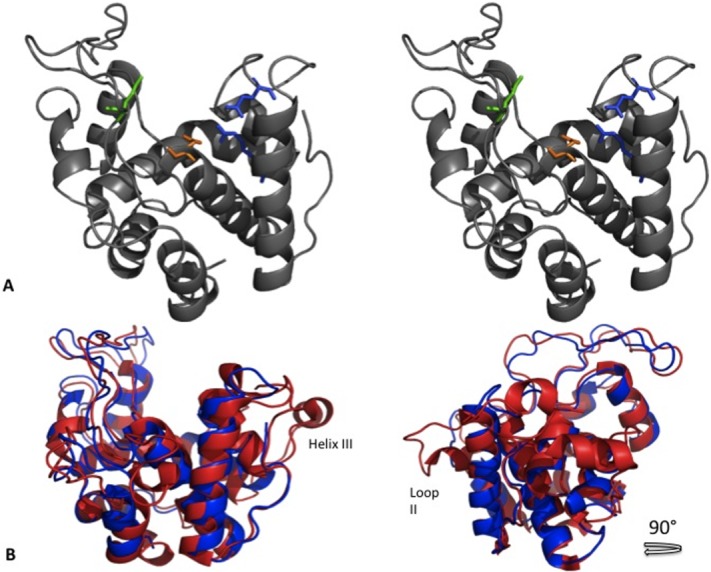
Structure of ChitA ΔN-EQ and comparison of class IV chitinases. A: Stereo image of ribbon representation of ChitA ΔN-EQ. Highlighted catalytic residues for ChitA ΔN-EQ. Glu62 (orange), Glu71 (green), Glu165 (blue), and Arg177. B: Comparison of Secale cereal chitinase (red) with ChitA ΔN-EQ (blue).
Catalytic mechanism of Zea Mays chitinase
Family GH19 chitinases operate by an inverting glycoside mechanism. The two residues involved in this mechanism have both been identified as glutamate residues, as seen in previously solved GH19 chitinase structures.21–25 The structure reported here suggests the proton donor and general base are Glu62 and Glu71 respectively for ChitA. A catalytic triad highly conserved in GH19 chitinases, Glu62, Arg177, and Glu165, is positioned to participate in catalysis [Fig. 2(A)].22,26,27
Proposed substrate binding for ChitA ΔN-EQ
To visualize the structural interactions between Z. mays chitinases and the chitin substrates discussed in this article, models of ChitA ΔN-EQ were examined in complex with various chitin-derived substrates (Fig. 3), based on the papaya chitinase (3CQL) structure. With the protein structures superimposed in the program, Coot,28 the substrates were aligned in the active site of ChitA, and the docked complex were refined using ligand restraints.25,28 The predicted sugar residues were bound in the sub-sites represented as C, D, E, and F in hen egg white lysozyme and correspond to binding positions-2,-1, +1, and +2 in ChitA ΔN-EQ, respectively.29 This model supports the prediction that residue Glu62 is primarily involved in the hydrolysis of this substrate [Fig. 3(B)]. In addition, the residue Ser103 was also observed to have a strong interaction. Aspects of ChitA ΔN specificity were examined with modified chitin substrates p-methylphenyl TMG-chitotriomycin and GlcNGlcNAc4. In comparison with the modeled dp4, GlcNGlcNAc4 implicates additional resides in substrate binding, noticeably Asn161, His61, Tyr106, and Phe140 [Fig. 3(C)]. TMG-pMP was observed to interact with Tyr106 and Arg177 [Fig. 3(D)].
Figure 3.
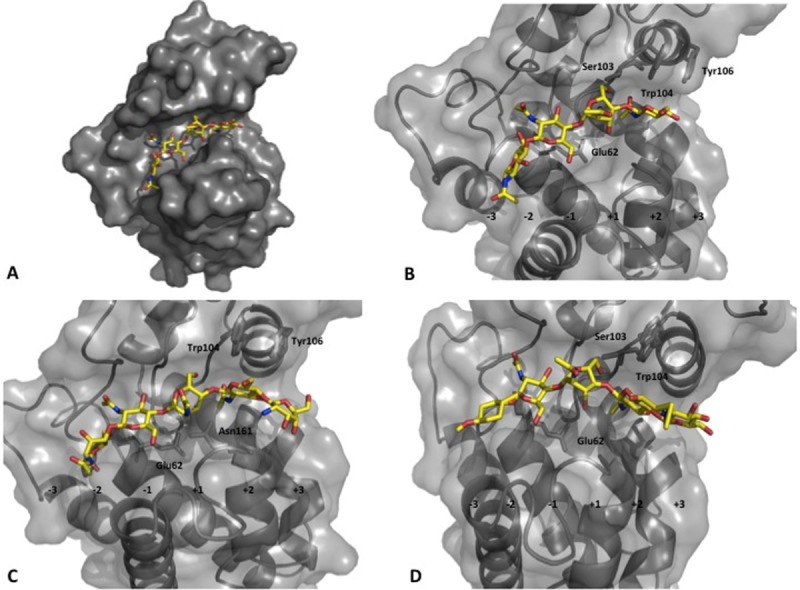
Modeled chitin substrates in ChitA ΔN-EQ structure. A: Surface representation of docked model of ChitA ΔN-EQ with the substrate chitintetraose. B: Active site of the docked model of ChitA ΔN-EQ with the substrate chitintetraose, (C) modified-chitopentaose, and (D) TMG-chitotriomycin-OMP.
Discussion
Analysis of the hydrolytic capabilities of the truncated ChitA revealed that the catalytic domain of Zea mays chitinase was able to hydrolyze chitotetraose into chitobiose products. Truncated ChitA was also observed to release soluble carbohydrate oligomers from crab shell chitin. The N-terminal domain of ChitA is not necessary for catalysis and is proposed to be an evolutionary change in this enzyme due to the selective pressure from the fungal modification. Furthermore, the shorter truncated forms of this enzyme produced by fungal proteases in nature retain chitinase activity.
Mutational studies of the residue Glu62 to Gln in both full length and truncated ChitA support its key role in catalysis in PC4 enzymes. This mutation in Chit-A ΔN resulted in an abolishment of activity, but without a loss of substrate binding. An increase in the observed chitin binding ability of ChitA-EQ compared to catalytically active ChitA is also observed for full-length protein. Therefore, binding of ChitA to chitin appears to result predominantly from binding to the catalytic domain.
Structure of Zea Mays chitinase
Glu62 and Glu71 are well positioned in the x-ray structure of Chit-A ΔN–EQ to be the catalytic residues. Mutational studies in barley and mustard seed chitinase support that Glu62 (Glu67 in barley and Glu212 in mustard seed) is essential for the hydrolysis of chitin substrates, while interestingly Glu71 (Glu89 and Glu234, respectively) was not required.26,30 Arg177 and Glu165 are positioned to have a role in hydrolysis and are predicted to function in a triad mechanism with Glu62.
Comparison to Family 19 chitinases
In comparative analysis of the structures reported here and previously characterized chitinase structures from GH19, it was observed that there are highly conserved structural components for these enzymes. Conserved di-sulfide bonds stabilize five loops and two large lobes on either side of the catalytic cleft, which dominates all structures. The cleft consists of multiple sub-sites for substrate binding and hydrolysis. The class of enzymes within this family demonstrates variations of this catalytic cleft. The highest degree of sequence similarity of the Zea mays chitinase structure is to the PC4 chitinase from Norway spruce (3HBD)21 (RMS: 1.026, Cα #: 181 atoms). The key elements of the catalytic domain are observed in the nine helices and five loops; however, ChitA ΔN-EQ lacks a small three-residue β-strand present in 3HBD.
Comparison of Z. mays chitinase to the recently reported Secale cereal PC2chitinase (4DYG)31 (RMS: 1.398, Cα #: 180 atoms) revealed that 4DYG has an additional α-helix (IV), part of which loops out from the base of the active site cleft, [Fig. 2(C)]. This structural divergence reflects some of the sequence differences that characterize these different classes of GH19 enzymes (Class II verse IV). This specific α-helix and Loop II were reported to stabilize chitin oligomers in the binding site in the catalytic domain of 4DYG. It was also shown with rice chitinase (Class I) that the removal of Loop II enhanced hydrolysis of chitin substrates, revealing this loop's role in the specific activity of these chinases.14 The ability of ChitA ΔN-EQ to bind chitin despite not having this ninth helix and Loop II, suggests an evolutionary advantage within this glycoside hydrolase family at accommodating smaller chitin oligomeric substrates in its binding cleft (Refer to Supporting Information Fig. S4 for sequence alignment).
Proposed substrate binding for ChitA ΔN-EQ
Analysis of chitin assays with modified chitin substrates combined with structural docking, identified specific residues involved in the binding of chitin-based substrates. Earlier work has shown that dp5 and dp5-C-glycoside substrates are preferentially cleaved at a single location to release dp2 from the non-reducing end, irrespective of the termini modifications. This is in contrast with dp6-C-glycoside, which is cleaved at any of 3 different glycosidic bonds. It also suggests that the-2 site has relatively low specificity, and is able to accommodate the p-methoxyphenyl group on the chitotriomycin-pMP. The results here suggest that Ser103 plays an important role in the binding of chitin substrates and Tyr106 contributes to the binding of longer substrates in the +3 binding site. Tyr106 has previously been implicated in the chitinase mechanism by chemical modification.32 This residue appears to be conserved in plant chitinases; however, some tolerance to mutation at this position suggests that this Tyr does not participate directly in catalysis, but probably has a secondary role in substrate binding.33 We speculate that the lack of Loop II in PC4 chitinases has led to the evolution of roles for these residues in the binding and stabilization of the substrates.
This study of the Zea mays chitinase mechanism, supported by activity data and structural studies, provides important information on GH19, PC4 chitinases and contributes to the understanding of the functionality of this enzyme family. Continued research of Zea mays chitinase and its truncated forms can provide additional information regarding its biological role and its contribution to plant defenses. In addition, further understanding of the specifics of hydrolysis and binding of chitin-based substrates can help to understand how this family of enzymes has evolved into three distinct classes. Continued mutational studies and structural studies with substrates can help to clarify these unanswered questions and contribute to the understanding of Family GH19 plant chitinases.
Materials and Methods
Recombinant ChitA-EQ, ChitA DN, and ChitA DN-EQ expression strains
The ChitA cDNA from maize inbred LH82 was excised from ChitA expression plasmid pTAN121 with restriction endonucleases EcoRI and XbaI.8 The excised cDNA was ligated into cloning plasmid pBluescript KS (+) (Stratagene, La Jolla, CA) to create pTAN132. A PCR-based method was used to change a single nucleotide in pTAN132 to create pTAN132(EQ). This method employed overlapping oligonucleotide primers at the site of mutagenesis (OverlapFwd and OverlapRev) and outer primers beyond unique NcoI (OutsideFwd) and BglII (OutsideRev) sites.34 Truncated ChitA cDNAs were made from both pTAN132 and pTAN132(EQ) by PCR cloning (oligonucleotide primers ChitADNFwd and ChitADNRev) to create plasmids pTAN132(DN) and pTAN132(DN-EQ). The cDNAs were excised from pTAN132(EQ) and pTAN132(DN) and pTAN132(DN-EQ) with EcoRI and XbaI, and ligated into expression plasmid pPICZaA (Invitrogen, Carlsbad, CA). These expression plasmids were named pTAN121(EQ) and pTAN121(DN) and pTAN121(DN-EQ); they encode expression of full-length ChitA with a point mutation in the catalytic domain (ChitA-EQ), truncated ChitA with only the catalytic domain (ChitA DN), and truncated ChitA with the catalytic domain containing a point mutation(ChitA DN-EQ). For each, plasmids were isolated from cultures of Escherichia coli DH5a, linearized by digestion with restriction endonuclease SacI, and transformed into Pichia pastoris X-33 by electroporation. P. pastoris isolates with integrated cDNAs were selected by plating on agar with zeocin.
Protein expression and purification
Small (5 mL) cultures of yeast extract peptone dextrose medium with zeocin (1% yeast extract, 2% peptone, 2% dextrose, 100 mg/L zeocin) were inoculated from isolated colonies on an agar plate. Cultures were grown to saturation (30°C; 250 RPM). A 1 mL aliquot was then used to inoculate a 1 L culture of buffered glycerol complex medium [1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 6), 1.34% yeast nitrogen base, 4 × 10−5% biotin, 1% glycerol]. Cells were grown at 25°C to an optical density of 10. Cells were pelleted by centrifugation (22,500g) and resuspended in buffered methanol complex medium to initiate induction [1% yeast extract, 2% peptone, 100 mM potassium phosphate (pH 6), 1.34% yeast nitrogen base, 4 × 10−5% biotin, 0.5% methanol]. Cells were grown in a shake incubator for 48 h at 25°C and 250 RPM to allow expression and secretion of chitinase. Cells were then removed from the media by centrifugation. Chitinases were purified from the cell-free media, essentially as described previously for ChitA.7 Briefly, proteins were precipitated from media by addition of ammonium sulfate (390 g/L) followed by centrifugation (22,500g). Precipitated proteins were resuspended in cation exchange binding buffer [10 mM sodium acetate (pH 5.2), 100 mM NaCl] and dialyzed. After dialysis, chitinases were bound to a cation exchange column [HiTrap SP XL (5 mL), GE Healthcare, Waukesha, WI]. The column was washed with 10 column volumes of buffer, and proteins were eluted by addition of cation exchange elution buffer [10 mM sodium acetate (pH 5.2), 500 mM NaCl]. Eluted chitinases were concentrated by addition of ammonium sulfate to either 40% saturation (232 g/L; full-length ChitAs) or 50% (314 g/L; truncated ChitAs) followed by centrifugation (22,500g). Precipitated proteins were resuspended in 0.5 mL 10 mM sodium acetate, pH 5.2 and dialyzed against like buffer.
Chitinase assays
For oligosaccharide substrates, chitinase assays and MALDI-TOF mass spectrometry analysis were performed as previously described.19 For insoluble chitin, assays were performed as described.35 Reactions were incubated for 1 h at 37°C in 0.4 mL McIlvaine's buffer (pH 5.0) and contained 50 mg purified chitinase and 1.5 mg chitin. Synthetic p-methoxyphenyl TMG-chitotriomycin (TMG-pMP), an enzyme inhibitor specific for insect and fungal β-N-acetylglucosaminidases was obtained from Dr. Qing Yang, Dalian University of Technology, Dalian, China. Tetra-N-acetylchitopentaose oligosaccharide (GlcN-GlcNAc4)23 was obtained from Drs. Sami Halila and Eric Samain, Centre de Recherches sur les Macromolécules Végétales (CERMAV), Grenoble. France.
Chitin binding assays
Heterologous strains of P. pastoris that express ChitA, ChitA-EQ, and ChitA ΔN-EQ were inoculated as 40 mL cultures of buffered glycerol complex medium and grown at 25°C in a shake incubator to an optical density of 10. Cells were pelleted, media removed, and cells were resuspended in 40 mL of buffered methanol complex medium. Cultures were incubated for 24 h to allow expression and secretion of recombinant chitinases (25°C, 250 RPM). Cells were then removed by centrifugation. Proteins were precipitated from cell-free media by addition of ammonium sulphate (390 g/L) followed by centrifugation. Protein pellets were resuspended in 20 mM sodium bicarbonate (pH 8.4) and dialyzed overnight against the same buffer.
Dialyzed protein solutions were combined with 10 mg of chitin (Sigma-Aldrich, St. Louis,MO) or 20 mL of chitin agarose (New England Biolabs, Ipswich, MA). After 30 min of incubation, the protein solution was removed and retained for SDS-PAGE analysis. Chitin/chitin agarose was washed twice with 1 mL buffer (discarded). SDS-PAGE loading buffer was added directly to the chitin/chitin agarose; samples were heated in boiling water bath for 1 min to elute bound proteins. Results were assessed by SDS-PAGE.
Crystallization
ChitA ΔN-EQ protein sample was dialyzed into sodium acetate buffer, pH 5.2, prior to crystallization experiments. Crystals were obtained at room temperature by the vapor diffusion method with sitting drops using 1 μL of reservoir and 1 μL of protein (26mg/mL). ChitA ΔN-EQ crystallized in the presence of 1.5M ammonium sulfate, 0.1M Tris, pH 8.5, and 0.16M potassium chloride, with crystals observed after 4 weeks of incubation. Crystals were cryoprotected by soaking in CryoOil (Mitegen) for 10 seconds, and flash-cooled.
Data collection
Data were collected at a temperature of 100K on a home source with a Rigaku Micromax x-ray generator. Diffraction data were processed with Structure Studio and HKL2000 software.36 Diffraction for ChitA ΔN-EQ was to 1.45 Å and observed to be in the space group P212121 with cell dimensions of a=37.15 Å, b=65.17 Å and c=72.33 Å. Data collection statistics are reported in Table I.
Table I.
Data Collection and Refinement Statistics for ChitA ΔN-EQ PDB Code 4MCK
| Data collection statistics | |
| Wavelength (Å) | 1.54178 |
| Space group | P 21 21 21 |
| Unit cell (a,b,c) (Å) | 37.152, 62.167, 72.334 |
| Unit Cell (α,β,γ) (°) | 90,90,90 |
| # Molecules in the Asymmetric Unit | 1 |
| Mosaicity (°) | 0.5 |
| Resolution range (Å) | 25.91-1.5 |
| Reflections (total/unique) | 292,194/27,880 |
| Completeness (%) | 96.67 (71.79) |
| Mean I/sigma(I) | 27.72 (3.31) |
| Wilson B-factor | 16.34 |
| Average Redundancy | 10.5 |
| Refinement statistics | |
| Rwork/Rfree | 0.1784/0.2031 |
| Average B-factor | 39.8 |
| Number of Protein Atoms | 201 |
| RMS Bonds(°) | 0.007 |
| RMS Angles (Å) | 1.25 |
| Validation statistics | |
| Ramachandran favored (%) | 95 |
| Ramachandran outliers (%) | 0.51 |
| Clashscore | 5.96 |
Structure determination and refinement
Analysis of the unit-cell content suggested one protein molecule in the asymmetric unit. Molecular replacement was conducted on preliminary data for ChitA ΔN using the model of the Norway spruce chitinases structure (3HBD)21 in Phenix Phaser-MR.37 A 56% amino acid similarity was observed between ChitA ΔN and 3HBD. The final model was refined in Phenix Refine (Table I).37 Molecular modeling and visualization were performed in Coot.28
Acknowledgments
The authors thank Dr. Qing Yang, Dalian University of Technology, Dalian, China for the sample of synthetic p-methoxyphenyl TMG-chitotriomycin (TMG-pMP), and Drs. Sami Halila and Eric Samain, Centre de Recherches sur les Macromolécules Végétales (CERMAV), Grenoble, France for the modified tetra-N-acetylchitopentaose oligosaccharide (GlcN-GlcNAc4).
Glossary
- BMGY
buffered glycerol complex medium
- CBD
carbohydrate binding domain
- dp6
chitohexaose
- dp2
chitobiose
- dp3
chitotriose
- dp4
chitotetraose
- dp5
chitopentaose
- GH19
glycoside hydrolase family 19
- GlcNAc
N-acetylglucosamine
- PC4
plant class IV
- TMG-pMP
p-methoxyphenyl TMG-chitotriomycin.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Koehler B. Corn ear rots in Illinois. Agric Exp Stn Publ. 1951:639. [Google Scholar]
- 2.Wild CP, Gong YY. Mycotoxins and human disease: a largely ignored global health issue. Carcinogenesis. 2010;31:71–82. doi: 10.1093/carcin/bgp264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akande KE, Abubaker MM, Adegbola TA, Bogoro SE. Nutritional and health implications of mycotoxins in animals feeds: a review. Pakistan J Nutr. 2006;5:398–403. [Google Scholar]
- 4.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 5.Parijs JVan, Broekaert WF, Goldstein IJ, Peumans WJ. Hevein: an antifungal protein from rubber-tree (Hevea brasiliensis) latex. Planta. 1991;183:258–264. doi: 10.1007/BF00197797. [DOI] [PubMed] [Google Scholar]
- 6.Huynh QK, Hironaka CM, Levine EB, Smith CE, Borgmeyer JR, Shah DM. Antifungal proteins from plants. Purification, molecular cloning and antifungal properties of chitinases from Maize seeds. J Biol Chem. 1992;267:6635–6640. [PubMed] [Google Scholar]
- 7.Naumann TA. Modification of recombinant maize ChitA chitinase by fungal chitinase-modifying proteins. Mol Plant Pathol. 2011;12:365–372. doi: 10.1111/j.1364-3703.2010.00677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naumann TA, Wicklow DT, Price NPJ. Identification of a chitinase-modifying protein from Fusarium verticillioides: truncation of a host resistance protein by a fungalysin metalloprotease. J Biol Chem. 2011;286:35358–35366. doi: 10.1074/jbc.M111.279646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naumann TA, Wicklow DT. Allozyme-specific modification of a maize seed chitinase by a protein secreted by the fungal pathogen Stenocarpella maydis. Phytopathology. 2010;100:645–654. doi: 10.1094/PHYTO-100-7-0645. [DOI] [PubMed] [Google Scholar]
- 10.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-active enzymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K. Plant chitinases. Plant J. 1993;3:31–40. doi: 10.1046/j.1365-313x.1993.t01-1-00999.x. [DOI] [PubMed] [Google Scholar]
- 12.Honda Y, Fukamizo T. Substrate binding subsites of chitinase from barley seeds and lysozyme from goose egg white. Biochim Biophys Acta. 1998;1388:53–65. doi: 10.1016/s0167-4838(98)00153-8. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki C, Itoh Y, Takehara H, Kuhara S, Fukamizo T. Family 19 chitinase from rice (Oryza sativa L.): substrate-binding subsites demonstrated by kinetic and molecular modeling studies. Plant Mol Biol. 2003;52:43–52. doi: 10.1023/a:1023972007681. [DOI] [PubMed] [Google Scholar]
- 14.Mizuno R, Fukamizo T, Sugiyama S, Nishizawa Y, Kezuka Y, Nonaka T, Suzuki K, Watanabe T. Role of the loop structure of the catalytic domain in rice class I chitinase. J Biochem. 2008;143:487–495. doi: 10.1093/jb/mvn004. [DOI] [PubMed] [Google Scholar]
- 15.Hollis T, Honda Y, Fukamizo T, Marcotte E, Day PJ, Robertus JD. Kinetic analysis of barley chitinase. Arch Biochem Biophys. 1997;344:335–342. doi: 10.1006/abbi.1997.0225. [DOI] [PubMed] [Google Scholar]
- 16.Iseli B, Armand S, Boller T, Neuhaus JM, Henrissat B. Plant chitinases use two different hydrolytic mechanisms. FEBS Lett. 1996;382:186–188. doi: 10.1016/0014-5793(96)00174-3. [DOI] [PubMed] [Google Scholar]
- 17.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 18.Hoell IA, Dalhus B, Heggset EB, Aspmo SI, Eijsink VGH. Crystal structure and enzymatic properties of a bacterial family 19 chitinase reveal differences from plant enzymes. FEBS J. 2006;273:4889–4900. doi: 10.1111/j.1742-4658.2006.05487.x. [DOI] [PubMed] [Google Scholar]
- 19.Price NPJ, Naumann TA. A high-throughput matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry-based assay of chitinase activity. Anal Biochem. 2011;411:94–99. doi: 10.1016/j.ab.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Liu T, Yang Y, Wu Q, Yang Q, Yu B. Synthesis, evaluation, and mechanism of N,N,N-trimethyl-d-glucosamine-(1→4)-chitooligosaccharides as selective inhibitors of glycosyl hydrolase family 20 β-N-acetyl-d-hexosaminidases. Chembiochem. 2011;12:457–467. doi: 10.1002/cbic.201000561. [DOI] [PubMed] [Google Scholar]
- 21.Ubhayasekera W, Rawat R, Ho SWT, Wiweger M, Von Arnold S, Chye M-L, Mowbray SL. The first crystal structures of a family 19 class IV chitinase: the enzyme from Norway spruce. Plant Mol Biol. 2009;71:277–289. doi: 10.1007/s11103-009-9523-9. [DOI] [PubMed] [Google Scholar]
- 22.Ubhayasekera W, Tang CM, Ho SWT, Berglund G, Bergfors T, Chye M-L, Mowbray SL. Crystal structures of a family 19 chitinase from Brassica juncea show flexibility of binding cleft loops. FEBS J. 2007;274:3695–3703. doi: 10.1111/j.1742-4658.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 23.Hart PJ, Ready MP, Robertus JD. Crystallization of an endochitinase from Hordeum vulgare L. seeds. J Mol Biol. 1992;225:565–567. doi: 10.1016/0022-2836(92)90942-d. [DOI] [PubMed] [Google Scholar]
- 24.Hahn M, Hennig M, Schlesier B, Höhne W. Structure of jack bean chitinase. Acta Cryst. 2000;D56:1096–1099. doi: 10.1107/s090744490000857x. [DOI] [PubMed] [Google Scholar]
- 25.Huet J, Rucktooa P, Clantin B, Azarkan M, Looze Y, Villeret V, Wintjens R. X-ray structure of papaya chitinase reveals the substrate binding mode of glycosyl hydrolase family 19 chitinases. Biochemistry. 2008;47:8283–8291. doi: 10.1021/bi800655u. [DOI] [PubMed] [Google Scholar]
- 26.Tang CM, Chye M-L, Ramalingam S, Ouyang S-W, Zhao K-J, Ubhayasekera W, Mowbray SL. Functional analyses of the chitin-binding domains and the catalytic domain of Brassica juncea chitinase BjCHI1. Plant Mol Biol. 2004;56:285–298. doi: 10.1007/s11103-004-3382-1. [DOI] [PubMed] [Google Scholar]
- 27.Ohnishi T, Juffer AH, Tamoi M, Skriver K, Fukamizo T. 26 kDa endochitinase from barley seeds: an interaction of the ionizable side chains essential for catalysis. J Biochem. 2005;138:553–562. doi: 10.1093/jb/mvi154. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Brameld KA, Goddard WA. The role of enzyme distortion in the single displacement mechanism of family 19 chitinases. Proc Natl Acad Sci USA. 1998;95:4276–4281. doi: 10.1073/pnas.95.8.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersen MD, Jensen A, Robertus JD, Leah R, Skriver K. Heterologous expression and characterization of wild-type and mutant forms of a 26 kDa endochitinase from barley (Hordeum vulgare L.) Biochem J. 1997;822:815–822. doi: 10.1042/bj3220815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnuma T, Numata T, Osawa T, Inanaga H, Okazaki Y, Shinya S, Kondo K, Fukuda T, Fukamizo T. Crystal structure and chitin oligosaccharide-binding mode of a “loopful” family GH19 chitinase from rye, Secale cereale, seeds. FEBS J. 2012;279:3639–3651. doi: 10.1111/j.1742-4658.2012.08723.x. [DOI] [PubMed] [Google Scholar]
- 32.Verburg JG, Smith CE, Lisek CA, Huynh QK. Identification of an essential tyrosine residue in the catalytic site of a chitinase isolated from Zea mays that is selectively modified during inactivation with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide. J Biol Chem. 1992;267:3886–3893. [PubMed] [Google Scholar]
- 33.Verburg JG, Rangwala SH, Samac DA, Luckow VA, Huynh QK. Examination of the role of tyrosine-174 in the catalytic mechanism of the Arabidopsis thaliana chitinase: Comparison of variant chitinases generated by site-directed mutagenesis and expressed in insect cells using baculovirus vectors. Arch Biochem Biophys. 1993;300:223–230. doi: 10.1006/abbi.1993.1031. [DOI] [PubMed] [Google Scholar]
- 34.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 35.Bruce A, Srinivasaqa U, Stainesb HJ, Highley TL. Chitinase and laminarinase production in liquid culture by Trichoderma spp. and their role in biocontrol of wood decay fungi. Int Biodeterior Biodegrad. 1995:337–353. [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, AJ McCoy, NW Moriarty, R Oeffner, RJ Read, DC Richardson, JS Richardson, TC Terwilliger, PH Zwart. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


