Abstract
We described the natural polymorphism of cytomegalovirus DNA polymerase in 42 unrelated isolates susceptible to ganciclovir, foscarnet, and cidofovir. All variations, including an eight-amino-acid deletion, were located between domains delta-C and II and between domains III and I, suggesting that these specific residues are not involved in enzymatic functions.
The DNA polymerase of cytomegalovirus (CMV) is the target of the three drugs, ganciclovir, foscarnet, and cidofovir, currently approved for the treatment of severe CMV disease. The 1,242-amino-acid DNA polymerase, encoded by the 3,729-bp gene UL 54, is related to the DNA polymerases of other herpes viruses through a series of eight conserved domains, named I to VII and delta-C, and it has both activities, polymerase and exonuclease (27, 29). Resistance to antiviral drugs occurs in immunosuppressed patients receiving prolonged therapy (3). Prior to the era of highly active antiretroviral therapy, in patients with AIDS the incidence of resistance had reached 27% after 9 months of ganciclovir therapy (5), 13% after 1 year of valganciclovir therapy (4), and 37% after 1 year of foscarnet therapy (33). In solid organ recipients, 7% of the CMV infections are resistant to ganciclovir within a median delay of 10 months (25), with lung transplant recipients having the highest incidence of resistance (26). Resistance is most prevalent in the population of CMV-seronegative transplant recipients of CMV-seropositive organs (28).
Ganciclovir resistance results mostly from changes in the UL97 phosphotransferase (7, 30) responsible for the primary phosphorylation of ganciclovir. Mutations in the UL 54 polymerase gene appear after prolonged ganciclovir therapy. They contribute to a high level of resistance to ganciclovir and induce cross-resistance to cidofovir (23, 29). All foscarnet- and cidofovir-resistance mutations map to the UL 54 gene. Resistance mutations are mainly located in the conserved domains of the polymerase, and their location can often predict the resistance patterns of the strains (15, 20).
As CMV resistance to antiviral drugs is a factor in therapeutic failure and disease progression (1, 3, 24), accurate and early resistance detection is needed. Phenotypic susceptibility assays require a long turnaround time in order to identify drug-resistant CMV isolates. The most convenient means of laboratory diagnosis of a drug-resistant virus is genotypic testing which shortens the delay of results, avoids bias by cell culture selection, and detects the resistance earlier (17, 19, 21, 22). The interpretation of genetic assays requires that resistance-associated mutations to natural variation be distinguished. Some of the DNA polymerase changes have been validated as resistance markers by a process of marker transfer (2, 8, 9, 11, 12, 13, 14, 15, 18, 29), but others have not. In the latter situation the role of such changes in inducing resistance is questionable.
To date, only one study which listed changes in 40 ganciclovir- and foscarnet-susceptible CMV isolates from the United States has been performed to define natural variations (10).
The aim of the multicenter study in France was to describe DNA polymerase polymorphisms in the region spanning domain IV to domain V.
Forty-two unrelated patients who had not received prior anti-CMV treatment (29 infants with congenital infection, 11 transplant recipients, and 2 human immunodeficiency virus-infected patients) were included. The following were collected: 13 amniotic fluid samples, 15 urine samples, 12 peripheral blood leukocyte samples, 1 bronchoalveolar fluid sample, and 1 lung biopsy sample. Also studied were four reference CMV strains, AD169, Towne, Davis (ATCC strains VR-538, VR-977, and VR-807, respectively), and Toledo (kindly provided by S. Michelson).
Phenotypic susceptibility of the isolates to ganciclovir, foscarnet, and cidofovir was verified by measuring the drug concentration required to reduce the number of plaques by 50% (50% inhibitory concentration [IC50]) compared to controls according to the Agence Nationale de Recherches sur le SIDA consensus method (16). An isolate was considered sensitive if the ganciclovir sensitivity index (IC50 of the isolate/IC50 of strain AD169) was <3, the foscarnet IC50 was <400 μM, and the cidofovir IC50 was <2 μM. According to these criteria, all the isolates were susceptible to the three drugs.
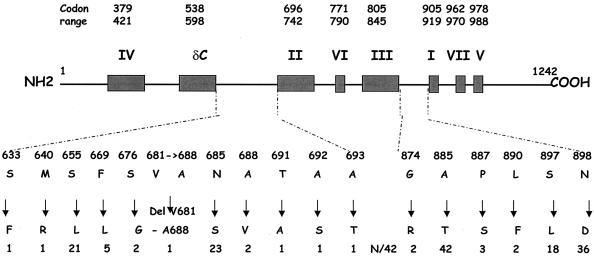
CMV DNA was extracted from clinical samples or infected fibroblasts (isolates and reference strains) by means of a Qia-Amp DNA blood kit (QIAGEN, Hilden, Germany). The 2,019-bp region from codon 349 to codon 1022 of the UL 54 gene was amplified by using a high fidelity polymerase (LA PCR kit; Takara, Shiga, Japan) and the forward primer 5′-ATC TCT TTA CGA TCG GCA CC-3′ and reverse primer 5′-ATC CTC AAA GAG CAG GGA GAG-3′. Sequences were determined either directly from initial samples (18 cases) or from isolates (24 cases) if amplification from the initial sample failed. PCR products were sequenced in forward and reverse directions by using internal primers that produced overlapping sequences (1,810 bps, codons 370 to 990), encompassing the conserved domains. Sequencing reactions were performed by Big Dye assay (Applied Biosystems, Warrington, United Kingdom) and analyzed with an automated sequencer (ABI 3100 Genetic Analyser; Applied Biosystems). Nucleotide and amino acid sequences were compared to the AD169 sequences with AutoAssembler and sequence navigator software. All interstrain changes including single-amino-acid substitutions and an eight-amino-acid deletion were clustered in two nonconserved regions located between domains delta-C and II and between domains III and I, respectively (Fig. 1).
FIG. 1.
Human CMV DNA polymerase natural polymorphism map. Shown at the top are the conserved functional domains and their codon ranges (boxed). Below the boxes are the loci of amino acid changes. These changes are then shown mapped to the natural polymorphism in the clinical isolates. The number (N) of isolates (total, 42) harboring each change is shown at the bottom of the figure.
Phylogenetic analysis was performed by using CLUSTAL W version 6.1 and PHYLIP software programs. The alignment of sequences showed 98% nucleotide homology. A phylogenetic tree, including the sequences of our 42 CMV strains, 19 strains from the United States published by Chou et al. (10), and reference strains AD169, Towne, Davis, and Toledo, was constructed by the neighbor-joining method and was generated with 100 bootstrap values. It showed no distinctive clustering based on geographical origin.
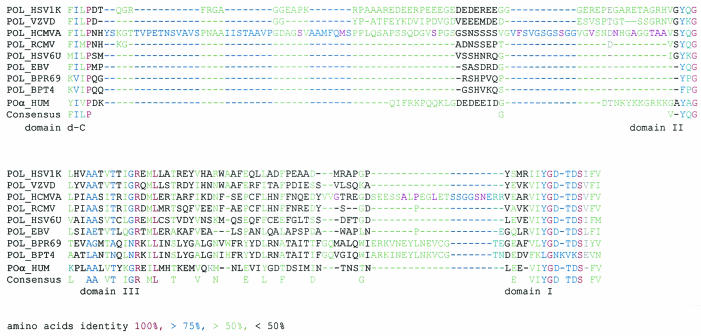
These results confirm the very weak variability of the CMV DNA polymerase gene as reported previously (10). All the previously reported polymorphisms, except two changes located between domains IV and delta-C and one in the conserved domain II (10), were located in the regions between domains delta-C and II and between domains III and I (29, 33, 35). Amino acid alignments of the human herpes virus polymerases, the human polymerase α (6), and the polymerases of bacteriophages RB69 (32) and T4 (31) showed the following features: (i) the counterparts of the region between domains delta-C and II were either absent or significantly shorter in the other polymerases, and (ii) when present, they were highly divergent. The whole region between domains delta-C and II was rich in glycine (G) and serine (S), amino acids known to be present in flexible regions of proteins, as was a short segment close to domain I (Fig. 2) between domains III and I, where most of interstrain changes were located. Indeed, the region between domains delta-C and II of herpes simplex DNA polymerase was also likely to be flexible, according to proteolytic studies (34). The combination of these characteristics suggests that these specific residues are not involved in the enzymatic functions.
FIG. 2.
Amino acid sequence alignment of selected polymerases of the B family in the regions located between domains delta-C and II (top) and between domains III and I (bottom) of polymerase. HSV1K, herpes simplex virus 1 strain kos; VZVD, varicella-zoster virus strain Dumas; HCMVA, human CMV strain AD169; RHCMV, rat CMV; HSV6U, human herpes virus 6 strain GS from Uganda; EBV, Epstein-Barr virus; PBR69, bacteriophage RB 69; BPT4, bacteriophage T4; and HUM, human polymerase α.
The five most frequent changes from the AD169 sequence (A885T, N898D, S655L, N685S, S897L) in our study were also detected by Chou et al. (10). As summarized in Table 1, T885 was rarely present either in field isolates or in the other reference strains. The four other mutations were hot spot sites of polymorphism as already described (29, 33, 35). Each of the four reference strains had a specific pattern concerning these five positions. AD169 is a good reference strain because, on the one hand, it is the reference for IC50 determination and, on the other hand, its sequence is well known.
TABLE 1.
Amino acid changes at five positions of the polymerase protein in comparison to reference strain AD169
| Position and amino acida | No. of isolates
|
Reference strain
|
||||
|---|---|---|---|---|---|---|
| This study (n = 42) | Chou et al. (n = 40)b | % of total (n = 82) | Towne | Davis | Toledo | |
| 885 | ||||||
| A | 0 | 3 | ||||
| T | 42 | 37 | 96 | + | + | + |
| 898 | ||||||
| N | 5 | 5 | + | |||
| D | 37 | 35 | 87.8 | + | + | |
| 655 | ||||||
| S | 21 | 20 | + | + | + | |
| L | 21 | 20 | 50 | |||
| 685 | ||||||
| N | 19 | 15 | + | + | ||
| S | 23 | 25 | 58.5 | + | ||
| 897 | ||||||
| S | 24 | 29 | + | + | + | |
| L | 18 | 11 | 35.3 | |||
Amino acids harbored by reference strain AD169 are in bold.
Reference 10.
In conclusion, although the natural polymorphism of CMV DNA polymerase is weak, knowledge of the polymorphism improves the interpretation of the sequencing results in patients with antiviral treatment failure. Indeed, the location of the mutations in the regions between domains delta-C and II and between domains III and I argues strongly in favor of a natural polymorphism. However, wild isolates should be further studied in order to draw a precise map of a polymorphism and, in particular, the location of a rare mutation.
Nucleotide sequence accession numbers.
Sequences determined in this study have been deposited in the GenBank database under accession numbers AY422355 through AY422377.
Acknowledgments
This work was supported by the National Agency on AIDS ANRS.
We thank Benoit Barrou for providing clinical information concerning kidney transplant recipients and Nathalie Dhédin for information concerning bone marrow recipients. We appreciate the advice provided by Constance Delaugerrre on phylogenetic studies and by Stephane Duquerroy on sequence alignment.
REFERENCES
- 1.Alain, S., P. Honderlick, D. Grenet, M. Stern, C. Vadam, M. J. Sanson-Le-Pors, and M. C. Mazeron. 1997. Failure of ganciclovir treatment associated with the selection of a ganciclovir-resistant strain in a lung transplant recipient. Transplantation 63:1533-1536. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti, F., M. R. Underwood, S. C. Stanat, K. K. Biron, S. Chou, A. Sarasini, E. Silini, and G. Gerna. 1996. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J. Virol. 70:1390-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldanti, F., and G. Gerna. 2003. Human cytomegalovirus resistance to antiviral drugs: diagnosis, monitoring and clinical impact. J. Antimicrob. Chemother. 52:324-330. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., C. Gilbert, A. Gaudreau, I. Greenfield, R. Sudlow, and N. A. Roberts. 2001. Rate of emergence of cytomegalovirus (CMV) mutations in leukocytes of patients with acquired immunodeficiency syndrome who are receiving valganciclovir as induction and maintenance therapy for CMV retinitis. J. Infect. Dis. 184:1598-1602. [DOI] [PubMed] [Google Scholar]
- 5.Bowen, E. F., V. C. Emery, P. Wilson, M. A. Johnson, C. C. Davey, C. A. Sabin, D. Farmer, and P. D. Griffiths. 1998. Cytomegalovirus polymerase chain reaction viraemia in patients receiving ganciclovir maintenance therapy for retinitis. AIDS 12:605-611. [DOI] [PubMed] [Google Scholar]
- 6.Braithwaite, D. K., and J. Ito. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases.Nucleic Acid Res. 21:787-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, S., A. Erice, M. C. Jordan, G. M. Vercellotti, K. R. Michels, C. L. Talarico, S. C. Stanat, and K. K. Biron. 1995. Analysis of the UL97 phosphotransferase coding sequence in clinical cytomegalovirus isolates and identification of mutations conferring ganciclovir resistance. J. Infect. Dis. 171:576-583. [DOI] [PubMed] [Google Scholar]
- 8.Chou, S., G. Marousek, S. Guentzel, S. E. Follansbee, M. E. Poscher, J. P. Lalezari, R. C. Miner, and W. L. Drew. 1997. Evolution of mutations conferring multidrug resistance during prophylaxis and therapy for cytomegalovirus disease. J. Infect. Dis. 176:786-789. [DOI] [PubMed] [Google Scholar]
- 9.Chou, S., G. Marousek, D. M. Parenti, S. M. Gordon, A. G. LaVoy, J. G. Ross, R. C. Miner, and W. L. Drew. 1998. Mutation in region III of the DNA polymerase gene conferring foscarnet resistance in cytomegalovirus isolates from 3 subjects receiving prolonged antiviral therapy. J. Infect. Dis. 178:526-530. [DOI] [PubMed] [Google Scholar]
- 10.Chou, S., N. S. Lurain, A. Weinberg, G. Y. Cai, P. L. Sharma, C. S. Crumpacker, and adults AIDS clinical trials group CMV laboratories. 1999. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Antimicrob. Agents Chemother. 43:1500-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou, S., R. C. Miner, and W. L. Drew. 2000. A deletion mutation in region V of the cytomegalovirus DNA polymerase sequence confers multidrug resistance. J. Infect. Dis. 182:1765-1768. [DOI] [PubMed] [Google Scholar]
- 12.Chou, S., R. H. Waldemer, A. E. Senters, K. S. Michels, G. W. Kemble, R. C. Miner, and W. L. Drew. 2002. Cytomegalovirus UL97 phosphotransferase mutations that affect susceptibility to ganciclovir. J. Infect. Dis. 185:162-169. [DOI] [PubMed] [Google Scholar]
- 13.Chou, S., N. S. Lurain, K. D. Thompson, R. C. Miner, and W. L. Drew. 2003. Viral DNA polymerase associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32-39. [DOI] [PubMed] [Google Scholar]
- 14.Cihlar, T., M. D. Fuller, A. S. Mulato, and J. M. Cherrington. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382-393. [DOI] [PubMed] [Google Scholar]
- 15.Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ducancelle, A., S. Belloc, S. Alain, C. Scieux, F. Petit, M. J. Sanson Le Pors, and M. C. Mazeron. 2004. Comparison of sequential cytomegalovirus isolates in a patient with lymphoma failing antiviral therapy. J. Clin. Virol. 29: 241-247. [DOI] [PubMed]
- 17.Ecle, T., L. Prix, G. Jahn, T. Klingebiel, R. Handgretinger, B. Selle, and K. Hamprecht. 2000. Drug-resistant human cytomegalovirus infection in children after allogeneic stem cell transplantation may have different clinical outcomes. Blood 96:3286-3289. [PubMed] [Google Scholar]
- 18.Erice, A., C. Gil-Roda, J. L. Perez, H. H. Balfour Jr., K. J. Sannerud, M. N. Hanson, G. Boivin, and S. Chou. 1997. Antiviral susceptibilities and analysis of UL97 and DNA polymerase sequences of clinical cytomegalovirus isolates from immunocompromised patients. J. Infect. Dis. 175:1087-1092. [DOI] [PubMed] [Google Scholar]
- 19.Erice, A., N. Borrell, W. Li, W. J. Miller, and H. H. Balfour, Jr. 1998. Ganciclovir susceptibilities and analysis of UL97 region in cytomegalovirus (CMV) isolates from bone marrow recipients with CMV disease after antiviral prophylaxis. J. Infect. Dis. 178:531-534. [DOI] [PubMed] [Google Scholar]
- 20.Erice, A. 1999. Resistance of human cytomegalovirus to antiviral drugs. Clin. Microbiol. Rev. 12:286-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert, C., and G. Boivin. 2003. Discordant phenotypes and genotypes of cytomegalovirus (CMV) in patients with AIDS and relapsing CMV retinitis. AIDS 17:337-341. [DOI] [PubMed] [Google Scholar]
- 22.Hamprecht, K., T. Eckle, L. Prix, C. Faul, H. Einsele, and G. Jahn. 2003. Ganciclovir-resistant cytomegalovirus disease after allogeneic stem cell transplantation: pitfalls of phenotypic diagnosis by in vitro selection of an UL 97 mutant strain. J. Infect. Dis. 187:139-143. [DOI] [PubMed] [Google Scholar]
- 23.Jabs, D. A., B. K. Martin, M. S. Forman, J. P. Dunn, J. L. Davis, D. V. Weinberg, H. K. Biron, F. Baldanti, and H. Hu for the Cytomegalovirus Retinitis and Viral Resistance Study Group. 2001. Longitudinal observations on mutations conferring ganciclovir resistance in patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis: the cytomegalovirus and viral resistance study group report number 8. Am. J. Ophthalmol. 132:700-710. [DOI] [PubMed] [Google Scholar]
- 24.Jabs, D. A., B. K. Martin, M. S. Forman, L. Hubbard, J. P. Dunn, J. H. Kempen, J. L. Davis, and D. V. Weinberg for the Cytomegalovirus Retinitis and Viral Resistance Study Group. 2003. Cytomegalovirus resistance to ganciclovir and clinical outcomes of patients with CMV retinits. Am. J. Ophthalmol. 135:26-34. [DOI] [PubMed] [Google Scholar]
- 25.Limaye, A. P., L. Corey, D. M. Koelle, C. L. Davis, and M Boeckh. 2000. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet 356:645-649. [DOI] [PubMed] [Google Scholar]
- 26.Limaye, A. P., G. Raghu, D. M. Koelle, J. Ferrenberg, M. L. Huang, and M. Boeckh. 2002. High incidence of ganciclovir-resistant cytomegalovirus infection among lung transplant recipients receiving preemptive therapy. J. Infect. Dis. 185:20-27. [DOI] [PubMed] [Google Scholar]
- 27.Lurain, N. S., K. D. Thompson, E. W. Holmes, and G. S. Read. 1992. Point mutations in the DNA polymerase gene of human cytomegalovirus that result in resistance to antiviral agents. J. Virol. 66:7146-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lurain, N. S., S. M. Bhorade, K. J. Pursell, R. K. Avery, V. V. Yeldandi, C. M. Isada, E. S. Robert, D. J. John, M. Q. Arens, E. R. Garrity, A. J. Taege, M. G. Mullen, K. M. Todd, J. W. Bremer, and B. Yen-Lieberman. 2002. Analysis and characterization of antiviral drug-resistant cytomegalovirus isolate from solid organ transplant recipients. J. Infect. Dis. 186:760-768. [DOI] [PubMed] [Google Scholar]
- 29.Smith, I. L., J. M. Cherrington, R. E. Jiles, M. D. Fuller, W. R. Freeman, and S. A. Spector. 1997. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J. Infect. Dis. 176:69-77. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan, V., C. L. Talarico, S. C. Stanat, M. Davis, D. M. Coen, and K. K. Biron. 1992. A protein kinase homologue controls phosphorylation of ganciclovir in human cytomegalovirus-infected cells. Nature 58:162-164. [DOI] [PubMed] [Google Scholar]
- 31.Wang, C. C., L. S. Yeh, and J. D. Karam. 1995. Modular organization of T4 DNA polymerase: evidence from phylogenetics. J. Biol. Chemistry 44:26558-26564. [DOI] [PubMed] [Google Scholar]
- 32.Wang, J., A. K. Sattar, C. C. Wang, J. D. Karam, W. H. Konigsberg, and T. A. Steitz. 1997. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 89:1087-1099. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg, A., D. A. Jabs, S. Chou, B. K. Martin, N. S. Lurain, M. S. Forman, and C. Crumpacker for the Cytomegalovirus Retinitis and Viral Resistance Study Group and the Adult AIDS Clinical Trials Group cytomegalovirus laboratories. 2003. Mutations conferring foscarnet resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J. Infect. Dis. 187:777-784. [DOI] [PubMed] [Google Scholar]
- 34.Weisshart K., A. A. Kuo, C. B. C. Hwang, K. Kumura, D. M. Coen. 1994. Structural and functional organization of herpes simplex virus DNA polymerase investigated by limited proteolysis. J. Biol. Chem. 269:22788-22796. [PubMed] [Google Scholar]
- 35.Wolf, D. G., N. S. Lurain, T. Zuckerman, R. Hoffman, J. Satinger, A. Honigman, N. Saleh, E. S. Robert, J. M. Rowe, Z. Kra-Oz. 2003. Emergence of late cytomegalovirus central nervous system disease in hematopoietic stem cell transplant recipients. Blood 101:463-465. [DOI] [PubMed] [Google Scholar]