Abstract
Upon muscle injury the high mobility group box 1 (HMGB1) protein is up-regulated and secreted to initiate reparative responses. Here we show that HMGB1 controls myogenesis both in vitro and in vivo, during development and after adult muscle injury. HMGB1 expression in muscle cells is regulated at the translational level: the miRNA miR-1192 inhibits HMGB1 translation and the RNA-binding protein HuR promotes it. HuR binds to a cis-element, HuRBS, located in the 3′UTR of the HMGB1 transcript, and at the same time miR-1192 is recruited to an adjacent seed element. The binding of HuR to the HuRBS prevents the recruitment of Argonaute 2 (Ago2), overriding miR-1192-mediated translation inhibition. Depleting HuR reduces myoblast fusion and silencing miR-1192 re-establishes the fusion potential of HuR-depleted cells. We propose that HuR promotes the commitment of myoblasts to myogenesis by enhancing the translation of HMGB1 and suppressing the translation inhibition mediated by miR-1192.
Introduction
The process leading to muscle fiber formation during embryonic development, also known as myogenesis, involves the fusion of mononucleated myoblasts to form multinucleated myofibers 1. Likewise, upon injury adult muscle tissues are repaired by satellite cells, which are quiescent mononucleated cells that coexist with myofibers 2. In response to injuries, satellite cells are activated; they first proliferate and then exit the cell cycle to fuse and form muscle fiber 3–5. During both embryonic and injury-induced myogenesis a cohort of intra- and extra-cellular factors act in concert.
HMGB1 (the high mobility group box 1) is a cytokine that is secreted by damaged muscle fibers and by infiltrating inflammatory cells after muscle injury. One of its main functions is to promote myogenesis by associating with the receptor for advanced glycation end products (RAGE), which is expressed on the surface of myoblasts, resulting in the activation of a signal transduction cascade that induces the expression of promyogenic factors such as MyoD and Myogenin 6–12. It is also known that while HMGB1 is highly expressed in myoblasts or satellite cells, its level in muscle fibers is significantly reduced 3,9. This suggests that maintaining a high expression level of HMGB1 during the early steps of myogenesis is required for the formation of functional myotubes. However, the mechanism controlling HMGB1 levels during myogenesis have never been investigated.
It has been shown that the 3′ untranslated region (3′UTR) of HMGB1 mRNA is very long and contains elements that are uridyl(U)-rich 13. U-rich elements in the 3′UTR are known to modulate posttranscriptional events such as the cellular movement, the turnover and the translation of many mRNAs 14,15. The expression of mRNAs encoding MyoD and Myogenin is regulated posttranscriptionally. These mRNAs harbour AU-rich elements (AREs) located in their 3′UTRs that mediate their association with RNA-binding proteins (RBPs) such as HuR. This association is crucial for the stability and the expression of these messages during myogenesis 16,17. Since HuR binds to MyoD and Myogenin mRNAs only during the transition state from myoblasts to myotubes but not at earlier stages 17, we concluded that HuR promotes myogenesis by stabilizing these mRNAs specifically at this later step during the myogenic process. However, knocking down the expression of HuR in undifferentiated muscle cells prevented their entry into the differentiation process 17. Thus, HuR-dependent promyogenic activities could also involve modulating the expression of mRNA targets during the early steps of myogenesis.
In this study, we show that HMGB1 is required for myogenesis and that its expression in muscle cells is controlled at the translational level. Both miR-1192 and HuR associate with a U-rich element in the 3′UTR of the HMGB1 mRNA. miR-1192 inhibits HMGB1 translation, but HuR promotes the translation of HMGB1 mRNA by preventing the formation of Ago2/miR-1192 complex. We propose that HuR promotes the commitment of myoblasts to myogenesis by enhancing the translation of HMGB1 and suppressing the translation inhibition mediated by miR-1192.
Results
The HuR-mediated expression of HMGB1 promotes myogenesis
HuR modulates the expression of MyoD and Myogenin mRNAs in an ARE-dependent manner during the transition state from myoblasts to myotubes, but not at earlier stages 16–18. To identify potential HuR mRNA targets during the early steps of myogenesis, we performed an immunoprecipitation (IP) experiment combined with cDNA microarray analysis on total extracts from undifferentiated C2C12 cells, a well-established murine myogenic cell line 19.
C2C12 cell extracts were immunoprecipitated with an anti-HuR or -IgG antibody. The RNAs associated with HuR were isolated and hybridized to mouse arrays. We revealed that HuR bound to 64 mRNAs in undifferentiated myoblasts (Supplementary Table S1). Among these messages, HMGB1 and the β-actin mRNAs are known to encode proteins that directly affect muscle cell differentiation 9,10,20. Since HuR associates with MyoD and Myogenin mRNAs only at later stages of the myogenic process 17,21 these messages were not on this list. While β-actin mRNA expression is known to depend on HuR22, nothing is known regarding the link between HMGB1 expression, its promyogenic function and HuR protein. Using IP coupled with quantitative (q) RT-PCR (RT-qPCR) we validated the association between HuR and HMGB1 mRNA in these cells (Supplementary Figs. S1a–b). Therefore, it is possible that HuR regulates HMGB1 expression during the early steps of myogenesis.
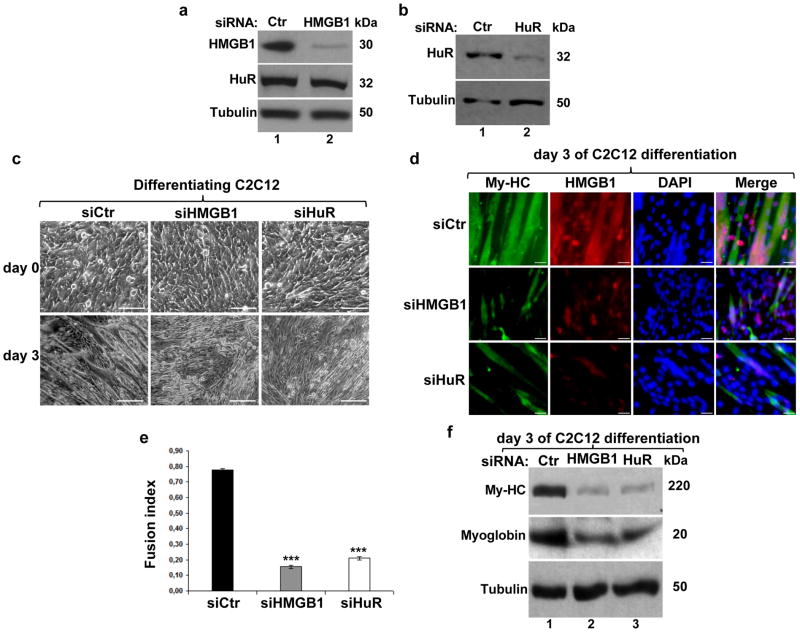
Several studies have suggested that the high expression level of HMGB1 in myoblasts is important for myogenesis 3,9. Indeed, we observed that while HMGB1 mRNA and protein are highly expressed during the early steps of muscle cell differentiation, their expression decreases at later steps (Supplementary Fig. S2). Despite this, the role of HMGB1 in the commitment of muscle cells to myogenesis is still elusive. To address this question, we depleted the expression of HMGB1 or HuR in C2C12 cells and the efficiency of myogenesis was determined by assessing cell morphology by phase contrast, the expression of the Myosin heavy chain (My-HC) by immunofluorescence (IF), and by determining the fusion index 17 (Figs. 1a–e). We observed that HMGB1 depletion reduced the efficiency of muscle fiber formation by >85% (Figs. 1c–e). As expected 17 a similar reduction in myogenesis was also observed in C2C12 depleted of HuR. Western blot analysis showed that the levels of My-HC and to a lesser extent myoglobin were reduced due to the knockdown of HMGB1 or HuR (Fig. 1f). Interestingly, exposing C2C12 cells depleted of endogenous HMGB1 to 400nM of recombinant HMGB1 (rHMGB1) re-established their ability to enter the myogenic process (Supplementary Figs. S2c–d). These observations demonstrate that HMGB1 promotes the early steps of myogenesis via its extracellular association with the RAGE receptor.
Figure 1. Knocking down HMGB1 expression in myoblasts prevents their entry into the myogenic process.
(a–b) HuR and HMGB1 knockdown were performed in C2C12 cells and total cell lysates were prepared 48 hours post-transfection. Western blotting was performed using antibodies against HMGB1, HuR and α-tubulin as a loading control. (c) Phase contrast pictures showing the morphology of C2C12 cells transfected with control (siCtr), siHMGB1 or siHuR, at the time of differentiation induction (day 0) and at day 3. Scale Bars, 50μm (d) Immunofluorescence with anti-My-HC, anti-HMGB1 antibodies and DAPI was performed to determine the differentiation status of the C2C12 cells treated with the indicated siRNAs. Scale Bars, 20 μm. Representative images from three independent experiments are shown. (e) The fusion index indicating the efficiency of C2C12 differentiation was determined by calculating the number of nuclei in cells with more than 2 nuclei (myotubes) in relation to the total number of nuclei in each microscopic field. Data are presented as +/− S.E.M. of three independent experiments. ***P<0.0001 (t test). (f) Total cell extracts were prepared at day 3 of differentiation. Western blotting was performed using antibodies against My-HC, myoglobin, and α-tubulin as a loading control. Blots shown in a-b-f are representative of three independent experiments.
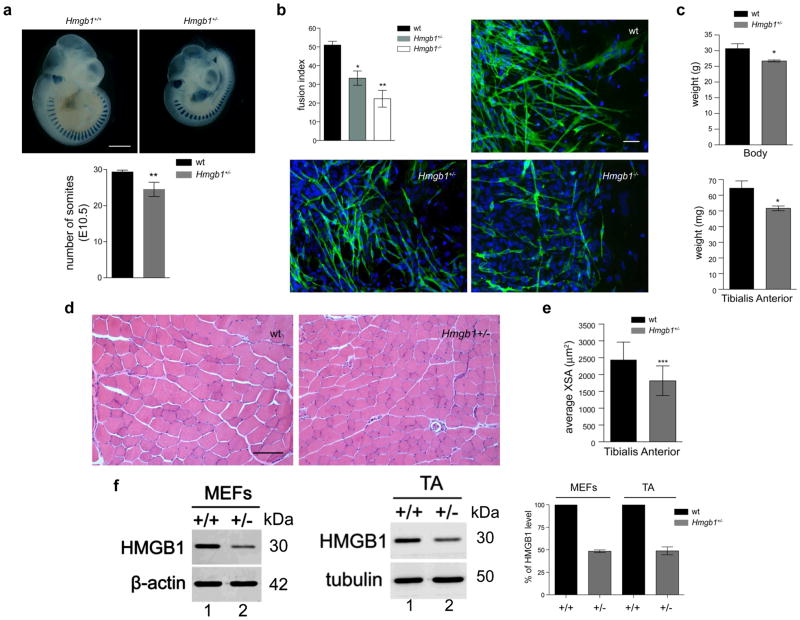
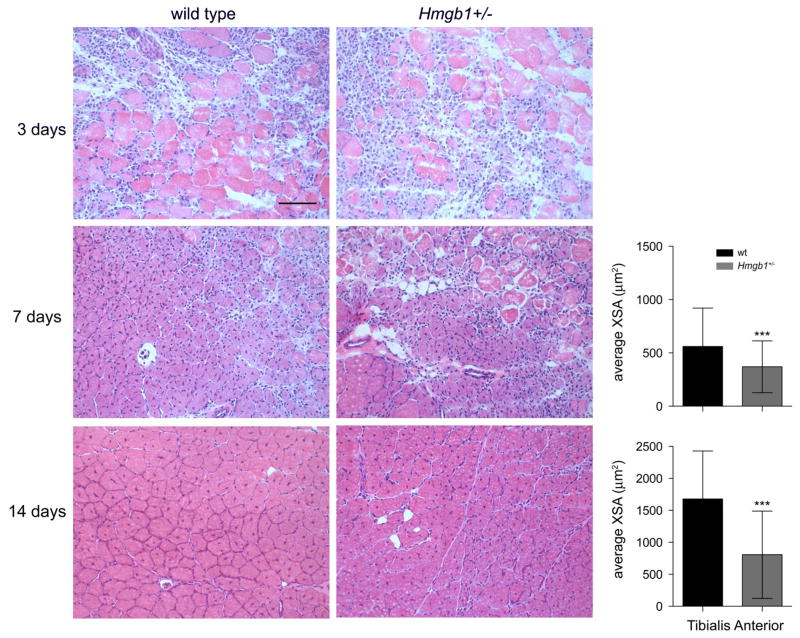
Next, we assessed the impact of HMGB1 on muscle development and growth in vivo using Hmgb1 wild type (+/+), +/− and −/− mice. During vertebrate embryogenesis, skeletal muscle in the limb develops from progenitor cells originating in the somites 23. Mice where the Hmgb1 gene has been deleted die perinatally 24. To assess the role of HMGB1 in mouse myogenesis, we therefore crossed Hmgb1+/− mice with homozygous MLC1/3F-nlacZ transgenic mice (wt for Hmgb1) 25 and collected the embryos at E10.5. As expected, half of the embryos were Hmgb1+/+, whereas the other half was Hmgb1+/−; all of them carried the MLC1/3F-nlacZ transgene and showed blue myonuclei when stained for X-gal (Fig. 2). We observed a significant reduction in the number of somites in Hmgb1+/− embryos when compared to wt controls (Fig. 2a). We then collected explants from wt, Hmgb1+/− and Hmgb1−/− embryos (E9.5), cultured them for 4 days 26 and determined the differentiation efficiency of myoblasts. We observed that Hmgb1−/− and +/− myoblasts formed significantly fewer myotubes than wt myoblasts (Fig. 2b). We next analyzed one-year old Hmgb1+/− mice to score for any reduction in muscle mass and cellularity when compared to wt mice. Hmgb1+/− mice showed a significant reduction in both total body weight and the mass of the Tibialis Anterior (TA) muscle (Fig. 2c). The myofibers in the TA muscle of Hmgb1+/− mice had a smaller cross-sectional area (XSA) than those of control mice (Figs. 2d–e). We also confirmed that mouse embryonic fibroblasts and muscle tissue from Hmgb1+/− mice express 50% less HMGB1 than their Hmgb1+/+counterparts (Fig. 2f). Additionally, a muscle injury experiment showed a marked delay in the Hmgb1+/− mice in the regeneration process after injury as evidenced by the significant reduction in the XSA of regenerating fibers at both 7 and 14 days after injury (Fig. 3). Together, these data demonstrate that maintaining a high expression level of HMGB1 in myoblasts is required for embryonal myogenesis and muscle regeneration after acute injury because of its release from myoblasts and/or damaged myofibers10,12,27.
Figure 2. HMGB1 is required for proper muscle fiber formation in vivo.
(a) Top panel: whole-mount X-Gal staining of control heterozygote MLC1/3F-nlacZ and double heterozygote MLC1/3F-nlacZ-Hmgb1+/− embryos collected at E10.5. Scale Bars, 1mm. Bottom panel: Histogram showing the average number of somites expressing MLC1/3F-nlacZ in control and double heterozygotes embryos at E10.5. **P <0.01 indicates a significant reduction in the number of transgene-expressing somites in Hmgb1+/− embryos compared to Hmgb1+/+ (Data are presented as +/− SD, t test, n=6). (b) Presomitic mesoderm (PSM) explants were derived from wt, Hmgb1+/− and Hmgb1−/− embryos (E9.5), cultured for 4 days and analyzed by immunofluorescence with a specific antibody against My-HC (green) and DAPI counterstaining (blue). Scale bar, 20 μm. Fusion index was calculated as the number of nuclei in My-HC positive cells with more than 2 nuclei (myotubes) in relation to the total number of nuclei in each microscopic field. Data are presented as +/− S.E.M. of three independent experiments. *P<0.05 and **P<0.01 (t test, n=3). (c) Body and Tibialis Anterior weights of 1-year old wt and Hmgb1+/− mice. Three animals were analyzed per group. Error bars represent SD. **P<0.01 (t test). (d) Histology of TA muscle. Representative images of H&E stained sections of TA muscles of 3-month old wt and Hmgb1+/− mice. Scale bar, 100 μm. (e) Mean cross-section area (XSA) of TA muscle fibers from 1-year old wt and Hmgb1+/− mice. Error bars represent SD. Nine hundred fibers were analyzed for each group. ***P<0.001 vs wt (ANOVA). (f) Western blots for HMGB1 were performed on equal amounts of total extracts from wt (+/+) and heterozygous (+/−) Mouse Embryonic Fibroblasts (MEFs) and adult Tibialis Anterior (TA). β-actin and tubulin are shown as loading controls. The blots shown are representative of three independent experiments. Western blot signals were quantified with ImageQuant software (GE Healthcare) and plotted +/− SD from three independent experiments.
Figure 3. HMGB1 is required for normal muscle regeneration after injury.
Representative sections of TA muscles from wt and Hmgb1+/− mice stained with H&E 3, 7 and 14 days after CTX injury. Scale bar, 100 μm. The cross-sectional area (XSA) of regenerating TA myofibers was decreased in Hmgb1+/− mice at both 7 and 14 days following injury. Nine hundred fibers were analyzed for each group (3 animals per group). Error bars in the histograms represent SD ***P<0.001 vs wt (ANOVA).
HuR promotes the translation of the HMGB1 mRNA
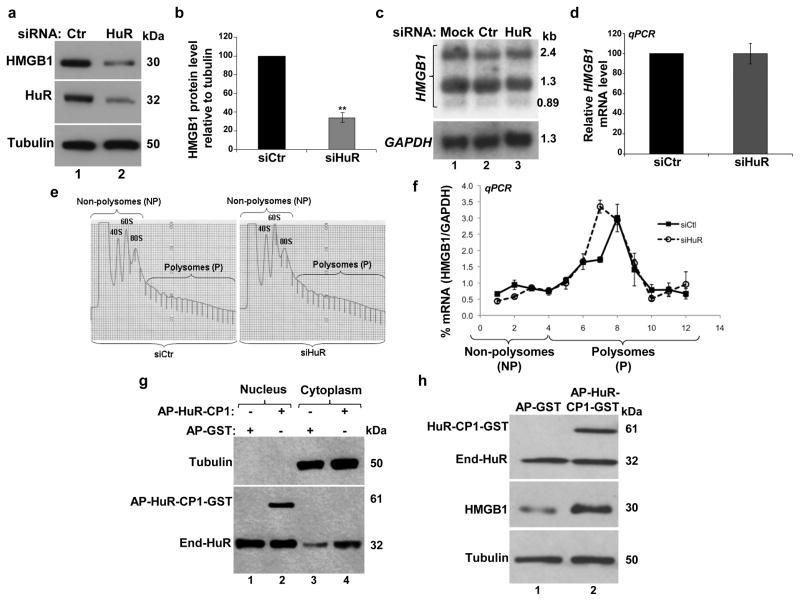
The HuR protein is known to modulate the export, stability and/or the translation of its target mRNAs 14,28. The fact that HuR plays a key role during the early steps of myogenesis 17 and associates with HMGB1 mRNA in muscle cells (Supplementary Fig. S1) suggested that HuR promotes muscle differentiation by regulating HMGB1 expression posttranscriptionally. Hence, we first determined whether HuR is required for the expression of HMGB1 in C2C12 cells. We observed that knocking down HuR in myoblasts reduced the levels of the HMGB1 protein (by >65%), but not its three mRNA isoforms (Figs. 4a–d). Pulse-chase mRNA stability experiments 22, RNA fluorescence in situ hybridization (FISH) 29 and cellular fractionation, indicated that HuR does not affect the cellular movement or the stability of HMGB1 messages (Supplementary Figs. S3–4). These observations therefore suggest that HuR regulates the translation of the HMGB1 mRNA. Sucrose fractionation experiments were performed to assess this possibility. We observed that while the depletion of HuR had no significant effect on the general distribution profile of polysomes (Fig. 4e), knocking down HuR resulted in a shift in the distribution of HMGB1 mRNA towards lighter polysome fractions when compared to siCtr-treated cells. These results indicated that the HMGB1 transcript is less translated in the absence of HuR (Fig. 4f). Since HuR-mediated effects on target mRNAs in muscle cells have been linked to its ability to accumulate in the cytoplasm 30, we tested whether this could also be the case for HMGB1 mRNA. To do this, we treated undifferentiated C2C12 cells with the HuR cleavage product 1 (HuR-CP1), which promotes the cytoplasmic accumulation of HuR during myogenesis 30. We observed that HuR-CP1 increased HuR level in the cytoplasm of muscle cells and promoted the expression of the HMGB1 protein (Figs. 4g–h). Taken together, these results show that HuR maintains the high expression level of HMGB1 in muscle cells by promoting the translation of HMGB1 mRNA.
Figure 4. HuR promotes the translation of HMGB1 in C2C12 cells.
(a) Total extracts from C2C12 cells treated with siHuR or siCtr were used for western blots with antibodies against HMGB1, HuR and α-tubulin as a loading control. (b) The HMGB1 protein level relative to α-tubulin for each treatment was plotted as the percentage relative to siCtr-treated sample +/− the S.E.M. of three independent experiments. **P<0.001 (t test). (c) Northern blot of total RNA from C2C12 cells treated with siHuR or siCtr. A representative blot of three independent experiments is shown. (d) Total RNAs from siHuR or siCtr treated C2C12 cells were subjected to RT-qPCR analysis using specific primers for HMGB1 and GAPDH mRNAs and plotted +/− the S.E.M from three independent experiments. (e) Sucrose gradient (15–50%) polysome fractionation of extracts from exponentially growing C2C12 cells that were treated with siHuR or siCtr. The profile of polysome distribution did not differ between C2C12 cells treated with siHuR or siCtr. (f) Quantitative RT-PCR was performed on the sucrose fractions using specific primers for HMGB1 and GAPDH mRNAs. Error bars represent S.E.M. from four independent experiments. (g–h) C2C12 cells were treated with HuR-CP1 as described 30. The nuclear and cytoplasmic fractions (g) or total extracts (h) were prepared and used for western blot analysis with anti-HuR, -HMGB1 and α-tubulin antibodies. All blots shown in g and h are representative of three independent experiments.
HuR prevents miR-1192-mediated repression of HMGB1 translation
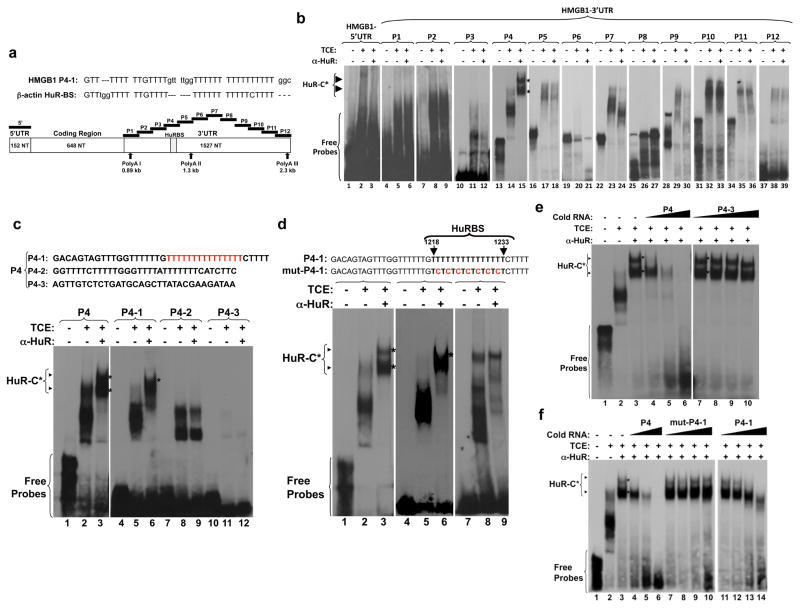
HuR affects the translation of target messages by associating with HuR binding sites (HuRBS) located at either the 5′ or 3′UTRs 14. Sequence analysis revealed the presence of a U-rich element in the 3′UTR of the HMGB1 mRNA that is similar to a HuRBS in the β-actin mRNA 22 (Fig. 5a). To assess whether this and/or other elements could mediate HuR binding, we performed RNA electromobility shift assays (REMSAs) using total extracts from C2C12 cells and thirteen radiolabeled cRNA probes that covered the entire 5′ and 3′ UTRs of HMGB1 mRNA (Fig. 5a). All the probes except P6 and P8 formed RNA-protein complexes when incubated with total cell extract (Fig. 5b). However, an anti-HuR antibody only shifted RNA-complexes (HuR-C) containing the P4 but not the other probes (Fig. 5b). A pull-down experiment confirmed the specificity of HuR binding: biotinylated P4 associated with HuR but not with CUGBP1, an RNA-binding protein known to modulate muscle differentiation 31 (Supplementary Fig. S5a).
Figure 5. HuR specifically binds to a U-rich element within the HMGB1 3′UTR.
(a) Upper panel: alignment of the U-rich HuR binding sites (HuRBS) within the HMGB1 and β-actin 3′UTRs. Lower panel: schematic representation of the HMGB1 mRNA sequence. The elements covering the 5′ and the 3′ (P1-P12) UTRs of HMGB1 used to generate radiolabeled RNA probes for RNA electromobility shift assays are indicated. The accession number in the NCBI database of the HMGB1 mRNA sequence used to generate these probes is NM_010439. (b–f) Representative gels of supershifts carried out by incubating total C2C12 cell extracts (TCE) with radiolabelled cRNA probes and anti-HuR antibody. Supershifted complexes (HuR-C) contain HuR protein. (b) Gel-shift assay performed using radiolabeled 5′UTR and the 12 probes (P1 to P12) depicted in (a). (c) Nucleotide sequence of the probes P4-1, P4-2 and P4-3 that were generated to localize HuR binding site (upper panel). These probes were used for gel-shift assays (lower panel). (d) Nucleotide sequences of probe P4-1 (HuRBS) and mut-P4-1, showing the T->C changes in mut-P4-1 (upper panel). The arrowhead shows HuR-C (lower panel). (e–f) Gel-shift competition was performed with radiolabeled probe P4 and 1X, 10X and 100X excess of the indicated unlabeled probes. All blots shown in b-f are representative of three independent experiments.
To delineate the HuRBS within P4, we divided this element into radiolabeled cRNA probes P4-1, -2 and -3 (Fig. 5c). A supershift containing the HuR-C was only generated with P4-1. A P4-1 mutant probe (mut-P4-1) in which every second U in the U-rich element (U15) was changed to a C (Fig. 5d) also failed to form a complex when incubated with extracts in the presence of the anti-HuR-antibody. P4-HuR complexes were gradually competed away in the presence of an excess of unlabeled P-4 or P4-1 probes but not in the presence of an excess of unlabeled P4-3 or mut-P4-1 probes (Figs. 5e–f). These observations, together with the fact that recombinant GST-HuR but not GST alone was able to form an HuR-C (Supplementary Fig. S5b), demonstrate that the U15 element located between nucleotides (nt) 1218 and 1233 comprises the HuRBS in the HMGB1-3′UTR.
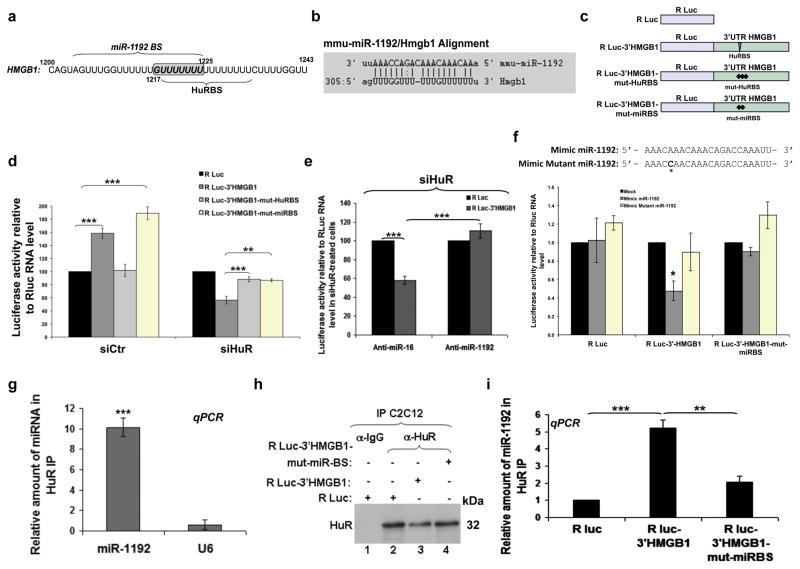
Previous studies have indicated that HuR either competes or collaborates with miRNAs in order to regulate the translation of some of its mRNA targets 32,33. We investigated whether this could also be the case for HMGB1 expression in muscle cells. As a first step, we identified miRNAs that associate with HuR in muscle cells. C2C12 cell extracts were immunoprecipitated with an anti-HuR antibody as described above and the miRNAs associated with HuR were isolated and identified using miRNA arrays (Exiqon, USA). We identified 20 miRNAs that were immunoprecipitated two folds or more with the anti-HuR antibody when compared to the IgG control (Supplementary Table S2). Using the MSKCC (http://www.microrna.org/microrna/releaseNotes.do), the BiBiSer (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) and the miRmap (http://mirmap.ezlab.org/app/) websites, we identified that among these 20 HuR-miRNAs only miR-1192 is predicted to target the HMGB1 mRNA. miR-1192 stood out since the first seven Us (nt 1218-1225) of its “seed” sequence (miRNA binding site, miRBS) are part of the U-rich element containing the HuRBS (Figs. 6a–b). Additionally, our screening (RT-qPCR and sequencing) indicated that miR-1192 is expressed in C2C12 cells as well as in muscle and heart tissues, but not in lung, spleen, MEFs, or HeLa cells (Supplementary Fig. S6 and Table S3). Therefore, it was possible that this miRNA could contribute to the modulation of HMGB1 expression in muscle cells.
Figure 6. HuR binding to a U8 element within HuRBS is sufficient to prevent the miR-1192-mediated inhibition of HMGB1 translation.
(a) Schematic representation of the HuRBS and the seed element of miR-1192 (miR-1192 BS). (b) Diagram illustrating the bioinformatic approach used to predict miR-1192 as a putative miRNA targeting HMGB1 3′UTR. Shown here are complementarities of miR-1192 with HMGB1 3′UTR. (c) Schematic diagrams of luciferase constructs. (d-g) Effects of HuR, miRNAs, antagomirs, and mimics. Error bars represent S.E.M of three independent experiments. ***P<0.0001, **P<0.001, *P<0.05 (Student’s t test). The amount of reporter RNA expressed in cells was determined by qPCR and used to normalize Renilla luciferase activities for each treatment. (d) Exponentially growing C2C12 cells were treated with siCtr or siHuR and 24 h later luciferase constructs were introduced. Luciferase activity of R Luc construct was considered as 100%. (e) miR-1192 and miR-16 antagomirs were transfected in siHuR-treated C2C12 cells previously transfected either with R luc alone or R luc-3′HMGB1. (f) Upper panel: Schematic representation of the Mimic miR-1192 and Mimic mutant miR-1192. The asterisk (*) indicates the A to C mutation in the Mimic mutant miR-1192. Lower panel: HeLa cells were transfected with Mimic miR-1192 or Mimic mutant miR-1192 and 24 h later luciferase constructs were transfected. The luciferase activity of the mock treated cells was set as reference. (g) Immunoprecipitation experiments were performed using a monoclonal HuR antibody, or IgG as a control, on total cell lysates from exponentially growing C2C12 cells. RNA was isolated from the immunoprecipitate, and quantitative RT-PCR was performed using primers specific to miR-1192, miR-16 and U6 RNAs. The levels of miR-1192 and U6 RNAs in each IP were normalized against the level of miR-16. Error bars represent S.E.M of three independent experiments. t test was used for statistical analysis. (h–i) Immunoprecipitation experiments were performed as in (g) on total cell lysates from C2C12 cells transfected with either R-luc alone, R-luc-3′HMGB1 or R-luc-3′HMGB1-mut-miRBS. (h) Western blot was performed using an HuR antibody. This blot is a representation of three independent experiments. (i) The levels of miR-1192 in each IP were determined and are plotted with the S.E.M. of three independent experiments. ***P<0.0001, **P<0.001 (Student’s t test).
We first determined the impact of the miRBS and HuRBS on the translation of a reporter mRNA containing the HMGB1 3′UTR. For these experiments we used a Renilla luciferase (R Luc) reporter containing either a wt (R Luc-3′HMGB1) or a mutated 3′UTR with mutations in the seed element of miR-1192 (R Luc-3′HMGB1-mut-miRBS), or the HuRBS (R Luc-3′HMGB1-mut-HuRBS) (Fig. 6c). Of note, since all the 15 Us of HuRBS were mutated in R Luc-3′HMGB1-mut-HuRBS, this reporter should be unable to recruit both HuR and miR-1192 to the HMGB1-3′UTR. We transfected each one of the constructs in C2C12 cells depleted or not of HuR and measured luciferase activity 34 in comparison to the baseline activity of R Luc alone (Fig. 6d). In cells expressing endogenous HuR the luciferase activity of R Luc-3′HMGB1 was >60% higher than R Luc alone (Fig. 6d). Mutating the HuRBS (R Luc-3′HMGB1-mut-HuRBS) prevented this increase, confirming that the binding of HuR to the HMGB1-3′UTR actively promotes translation. Mutating the seed element of miR-1192 (R Luc-3′HMGB1-mut-miRBS) however, did not prevent the translation increase mediated by the HMGB1-3′UTR. Indeed, REMSA experiments demonstrated that HuR can bind to the HuRBS despite mutations in miR-1192 seed element (Supplementary Fig. S7). These results clearly indicate that the recruitment of HuR to the HuRBS is sufficient to promote the translation of HMGB1 in muscle cells.
Since the inhibition of HMGB1 translation occurs only in cells depleted of HuR (Fig. 4), we repeated the experiments described above in HuR-knockdown C2C12 cells. We observed that the R Luc-3′HMGB1 reporter, unlike the R Luc-3′HMGB1-mut-miRBS or R Luc-3′HMGB1-mut-HuRBS constructs, displayed a ~50% decrease in luciferase activity when compared to its control R Luc counterpart (Fig. 6d). Additionally, silencing miR-1192 by a miRNA silencer (antagomir anti-miR-1192) abrogated the decrease in the luciferase activity mediated by the HMGB1-3′UTR. No such effect was however seen in cells treated with a control antagomir against miR-16 (Anti-miR-16) (Fig. 6e and Supplementary Fig. S8a). Importantly, the concentration of antagomir used in these experiments did not affect the viability of C2C12 cells and the steady state levels of endogenous HMGB1 mRNA (Supplementary Fig. S8b–d). We next generated mimics of miR-1192 with or without an A-to-C mutation in its 5th nucleotide (Fig. 6f). In HeLa cells, which do not express endogenous miR-1192 (Supplementary Fig. S6), the mimic miR-1192 reduced the luciferase activity of R Luc-3′HMGB1 by ~50%. The mutant miR-1192 mimic however, had no effect on this activity (Fig. 6f). The luciferase activity of the R Luc-3′HMGB1-mut-miRBS was not affected by the Mimic miR-1192, indicating that inhibition of R Luc-3′HMGB1 translation occurs via a direct binding to the miRBS. Collectively, these results demonstrate that in muscle cells miR-1192 directly inhibits the translation of HMGB1 mRNA via the miRBS element, but this effect is overcome by the binding of HuR to its HuRBS.
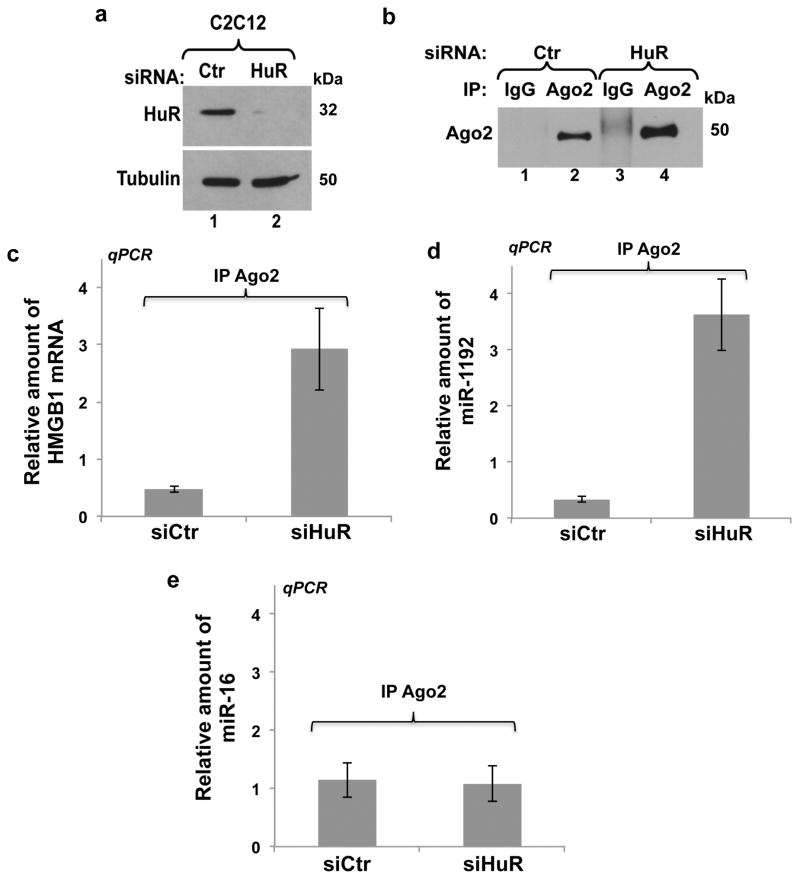
Our IP combined to miRNA microarray data (Supplementary Table S2), which were validated by performing IP/RT-qPCR experiments (Fig. 6g) indicated that HuR and miR-1192 can coexist in the same complex in C2C12 cells. To determine whether this complex is indeed assembled on the HMGB1-3′UTR, we performed the same IP/RT-qPCR experiment on C2C12 cells expressing either R Luc alone, R Luc-3′HMGB1 or the R Luc-3′HMGB1-mut-miRBS mRNAs. We observed that the level of miR-1192 in the HuR complex was five fold greater in cells expressing the R Luc-3′HMGB1 mRNA when compared to cells expressing mRNAs deficient in their ability to bind miR-1192 (R Luc or R Luc-3′HMGB1-mut-miRBS) (Figs. 6h–I). Therefore, these observations clearly indicate that HuR and miR-1192 can bind simultaneously the 3′UTR of HMGB1 mRNA. We next verified mechanistically how HuR could negate the ability of miR-1192 to repress the translation of the HMGB1 mRNA. miRNAs are known to translationally repress mRNAs via the recruitment of the Ago2, a component of the RNA-induced silencing complex (RISC) 35. We thus verified if HuR prevents the miR-1192 mediated recruitment of Ago2 to the HMGB1-miRBS. Our data indicate that although both HuR and miR-1192 simultaneously bind to the HMGB1 mRNA 3′UTR, the association of Ago2 to both miR-1192 and the HMGB1 mRNA is increased when HuR is depleted from C2C12 cells (Fig. 7). These experiments, therefore, indicate that HuR prevents the miR-1192 mediated translational repression of the HMGB1 mRNA by interfering with the recruitment of Ago2.
Figure 7. HuR binding to the HuRBS prevents the recruitment of Ago2 to the HMGB1 mRNA.
(a) Western blot analysis was performed using antibodies against HuR and α-tubulin as a loading control on total cell extracts obtained from exponentially growing C2C12 cells treated with a control (Ctr) or HuR specific (siHuR) siRNA. (b) Immunoprecipitation experiments were performed using a monoclonal Ago2 antibody, or anti-IgG antibody as a control, on the total cell lysates described in (a). The immunoprecipitation of Ago2 was then assessed by western blotting using an anti-Ago2 antibody. (c–e) RNA was isolated from the immunoprecipitate described above, and quantitative RT-PCR was performed using primers specific to (c) HMGB1 (d) miR-1192 (e) miR-16. The levels of HMGB1 mRNA, miR-1192 and miR-16 in each IP, relative to those in the IgG IP, were respectively normalized against the GAPDH mRNA and U6 levels. Error bars represent S.E.M of three independent experiments.
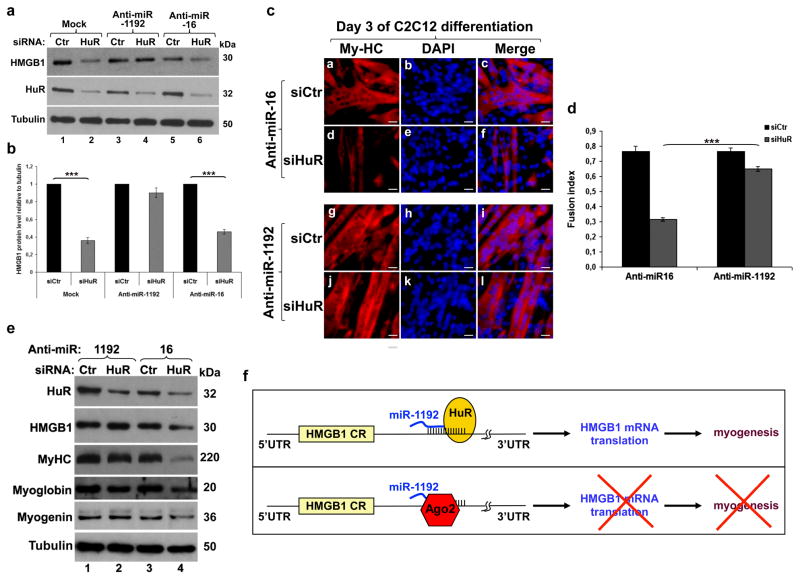
Our data show that HMGB1 is one of the mRNA targets through which HuR promotes myogenesis. Specifically, the results outlined in Figure 6 suggest that HuR-depleted muscle cells fail to enter myogenesis because in the absence of HuR miR-1192 inhibits HMGB1 translation. Hence, silencing miR-1192 should rescue HMGB1 expression in HuR-depleted muscle cells and should also re-establish their myogenic potential. Our experiments showed that silencing miR-1192 but not miR-16 prevented the inhibition of HMGB1 translation in HuR-depleted C2C12 cells (Figs. 8a–b and Supplementary Figs. S9a). In the presence of endogenous HuR, however, Anti-miR-1192 had no effect on HMGB1 translation (Figs. 8a–b and Supplementary Figs. S9a). This result was further confirmed by the fact that while the depletion of HuR shifted the distribution of HMGB1 mRNA towards lighter polysome fractions (Fig. 4f), silencing miR-1192 reversed this effect (Supplementary Fig. S9b). Next, we assessed whether the Anti-miR-1192-mediated rescue of HMGB1 translation is sufficient to promote the myogenic potential of HuR-knockdown cells. Indeed, silencing miR-1192 but not miR-16 reestablished the myogenic potential of HuR-depleted cells (Figs. 8c–d) and the expression levels of My-HC, myoglobin and myogenin (Fig. 8e).
Figure 8. Silencing miR-1192 reestablishes HMGB1 translation and rescues the myogenic potential of HuR depleted muscle cells.
(a) Exponentially growing C2C12 were treated with siCtr or siHuR and were transfected 5 h later with antagomirs to miR-1192 or miR-16. Extracts from these cells were harvested and used for western blotting with anti-HMGB1, -HuR and -α-tubulin antibodies. (b) The expression level of HMGB1 protein relative to α-tubulin. Error bars represent S.E.M. of three independent experiments. ***P<0.0001 (student’s t test). (c) C2C12 treated as in (A) were grown to confluency, induced to differentiate for three days and stained with anti-My-HC antibody and DAPI. Scale bars, 20 μm. The images shown are representative fields for each cell treatment from three independent experiments. (d) Fusion index of cells in (c). Error bars represent S.E.M. of three independent experiments. ***P<0.0001 (student’s t test). (e) Total extracts from cells treated as described above were blotted and probed with antibodies against HuR, HMGB1, My-HC, myoglobin, myogenin and α-tubulin. A representative blot of two independent experiments is shown. (f) Schematic model illustrating how HuR promotes the myogenesis in undifferentiated muscle cells. HuR recognizes an U8 element in the HMGB1-3′UTR adjacent to the seed element of miR-1192, and promotes HMGB1 translation and myogenesis. HuR mediates this effect even if miR-1192 is bound to its seed element (upper panel). In the absence of HuR however, miR-1192 recruits Ago2 thus inhibiting HMGB1 translation and preventing myogenesis (lower panel).
It was previously shown that miRNAs such as miR-519 and miR-16 inhibit HuR expression in various cell lines 36–38. However, since HuR levels does not significantly change during the early steps of myogenesis in vitro 16,17,39 and increase during muscle regeneration in vivo 30 (Supplementary Fig. S10), miRNA-mediated modulation of HuR expression in muscle cells seems to be ineffective in these conditions. Our experiments show that while the expression level of miR-16 increases during myogenesis both in vitro and in vivo (Supplementary Figs. S11a–b), it is the depletion but not the increased expression of miR-16 that modestly reduces the expression level of HuR, with limited effects on HMGB1 as well as My-HC and Myoglobin (Fig. 8e). In fact, a marked reduction of HMGB1 expression and the inhibition of myogenesis is observed only when HuR level is further reduced by siRNA (Fig. 8e). These observations indicate that miR-1192 is active only when HuR level is below a critical threshold. All together, our data support a model whereby HuR helps undifferentiated muscle cells to enter myogenesis by supporting the translation of HMGB1 transcripts and preventing the action of Ago2/miR-1192 (Fig. 8f).
Discussion
In this study we show that HMGB1 is required for muscle fiber formation both in vitro and in vivo and uncover a novel mechanism by which muscle cells modulate HMGB1 expression. A general reduction in skeletal muscle tissues was previously observed in Hmgb1−/− mice, among several other defects 24. Here, we show that a precise level of HMGB1 expression is required for proper muscle fiber formation, since a 50% reduction in HMGB1 expression in embryos significantly decreases the efficiency of myogenesis. Thus, a tight control of HMGB1 expression levels appears essential for myogenesis.
More than a decade ago it was discovered that HMGB1 mRNA harbours a long 3′UTR with U-rich elements 40–42 suggesting that posttranscriptional events could regulate HMGB1 expression. Surprisingly, this possibility was never explored. Our data clearly establish that a posttranscriptional mechanism, via HuR, plays a key role in promoting HMGB1 expression in muscle cells. HuR, one of the well known posttranscriptional regulators, affects the expression of its target mRNAs by binding to specific U-rich-elements in their 3′UTRs 14,15. We show that the 3′UTR of HMGB1 mRNA harbours such an element, the HuRBS, through which HuR promotes the translation of HMGB1 mRNA. In the absence of HuR, however, the microRNA miR-1192 is able to inhibit the expression of HMGB1. To our knowledge, this is the first report of the impact of a miRNA on HMGB1 expression. Incidentally, this is also the first report on the biological activity of miR-1192. Using several different techniques including miRNA microarray analysis, RT-qPCR with specific primers for miR-1192, and the sequencing of the PCR product that was generated by these primers, our data indicate that miR-1192 is expressed in muscle cells. Previous reports using RNA sequencing (RNA-seq), however, have failed to detect miR-1192 in the whole mouse embryo and muscle tissues43,44. As demonstrated by several recent studies 45,46, technical biases during capture and preparation of the small RNA library could impact the detection of small RNA molecules. Despite potential technical differences in methods used to precisely quantify the expression level of miR-1192 in muscle cells, our data clearly show that this miRNA is expressed in muscle cells and is functionally relevant in modulating HMGB1 expression.
Since HuR, HMGB1 and miR-1192 are highly expressed in muscle cells and during muscle regeneration 30 (Supplementary Figures S10, S11) and both HuR and miR-1192 can simultaneously associate with the HMGB1-3′UTR, our data support a model whereby the binding of HuR to its HuRBS is sufficient to prevent miR-1192-mediated inhibition. Interestingly, the association of miR-1192 with Ago2, a key player in the RISC complex 35,47, is dramatically enhanced in muscle cells depleted of HuR. This suggests that HuR prevents the action of miR-1192 by interfering with the recruitment of Ago2 to the HMGB1 3′UTR. These data are consistent with previous observations showing that HuR promotes the translation of target messages by preventing the assembly of an active RISC complex on the Let-7 miRNA seed element 48 even if its binding site is far away from the miRNA seed element. Therefore, these observations with our data suggest that HuR promotes the translation of target mRNAs by interfering with the formation of an active RISC complex regardless of the distance of its binding site from the seed element. This mechanism of action is different from the one described for the dead end 1 protein (Dnd1), another RBP, which promotes translation of the the cyclin-kinase inhibitor p27cip by preventing the recruitment of miR-221/222 to their seed elements 49. Therefore, our findings not only add another example to the few studies on the cross talk between RBPs and miRNAs, but also uncover a novel way by which cells, via proteins such as HuR, control miRNA-mediated effects. Our data demonstrate that via such a mechanism HuR induces the translation of HMGB1, which in turn promotes the entry of muscle cells into myogenesis.
By linking the interplay between HuR and miR-1192 to the translation modulation of HMGB1, we found a posttranscriptional mechanism that controls the expression of the main “alarmin” in the organism 50,51. HMGB1 is a damage-associated molecular pattern (DAMP) molecule that upon severe injury activates the innate immune system to recognize tissue damage and initiate reparative responses 52,53. Typically, the increase in HMGB1 levels in stressed or activated inflammatory cells precedes the upregulation of HMGB1 transcript levels by several hours 53. Our results suggest that posttranscriptional events might be responsible for this increase since the HMGB1 mRNAs are both abundant 40,41 and stable (Supplementary Figure S2). Therefore, our study may open the door to new strategies to manipulate HMGB1 levels under various conditions. Such a strategy could help promote the beneficial effects of HMGB1 (e.g. the activation of muscle regeneration and wound healing) while limiting/preventing the deleterious outcomes of its overproduction during life-threatening assaults (e.g. ischemia, burn, infection, or sepsis).
Methods
Plasmid construction
The pCMV-SPORT6 plasmid containing the full-length HMGB1 cDNA (Accession Number: BC008565) was purchased from Open Biosystems (Catalogue Number: MMM1013-64094). pRL-luc-3′HMGB1-mut-miRBS was generated by Norclone Biotech Laboratories, Kingston, ON, Canada. The full-length 3′UTR of mouse HMGB1 was subcloned into a pRL-SV40 vector (Promega) by performing PCR amplification using the following primers: forward 5′-TTG GTT CTA GCG CAG TTT TT-3′ and reverse 5′-TCA TCC AGG ACT CAT GTT CAG-3′. The pRL-SV40 vector was digested by Xba1 restriction enzyme (New England Biolabs), followed by a treatment with the T4 DNA polymerase, and then dephosphorylated. The PCR insert was ligated into the plasmid using the Quick Ligase enzyme (Promega) according to the manufacturer’s instructions. pRL-luc-3′HMGB1-mutHuRBS was generated using a specific primer containing the mutations and Quickchange site-directed mutagenesis kit (Stratagene). The MISSION shRNA plasmid (siHMGB1; Sigma-Aldrich) was used to knock down HMGB1. MISSION pLKO.1-puro vector encoding scrambled shRNA (siCtr; SHC001) was used as a control.
Cell culture and transfection
C2C12 muscle cells (ATCC) were grown in media containing 20% fetal bovine serum (FBS, Invitrogen) in DMEM (Dulbecco’s modified eagle medium from Invitrogen). In order to induce muscle cell differentiation, cells were switched to a media containing DMEM, 2% horse serum, penicillin/streptomycin antibiotics (Invitrogen), and 50mM HEPES, pH 7.4 (Invitrogen) when their confluency reached 100%17,39. Hmgb1+/+ and Hmgb1−/− mouse embryonic fibroblasts (MEFs) (HMGBiotech, Milan, Italy) were cultured in DMEM supplemented with Penicillin/streptomycin and 10% FBS (Sigma-Aldrich). Transfections with siRNAs specific for HuR or plasmids were performed using Lipofectamine and Plus reagents (Invitrogen) or jetPEI (Polyplus Transfection) according to the manufacturer’s instructions. The same transfection protocol was used to treat C2C12 cells with 200 nM miR-1192 or miR-16 antagomirs (Dharmacon).
HMGB1 rescue experiments
Transient transfection of C2C12 cells was carried out using jetPEI™ (Polyplus Transfection), as recommended by the manufacturer. Briefly, C2C12 cells were transfected with the MISSION shRNA plasmid (Sigma-Aldrich) to knock down HMGB1 or with the MISSION pLKO.1-puro vector encoding a scrambled shRNA (SHC001) as a control. 48 h post-transfection, the cells were switched to differentiation media and cultured for additional 48 h in the absence or presence of recombinant HMGB1 (400nM).
Preparation of cell extracts and immunoblotting
Total cell extracts were prepared by incubating undifferentiated or differentiated C2C12 cells on ice for 15 min with lysis buffer (50 mM HEPES pH 7.0, 150 mM NaCl, 10% glycerol, 1% Triton, 10 mM pyrophosphate sodium, 100 mM NaF, 1 mM EGTA, 1,5 mM MgCl2, 1X protease inhibitor (Roche), 0,1 M orthovanadate, 0,2 M PMSF), then centrifuge at 12 000 rpm at 4°C and the supernatant was kept17. The extracts were run on an SDS-PAGE and transferred to nitrocellulose membranes (BIORAD). The samples were analyzed by western blotting 54 with antibodies against HuR (3A2,1:10000) 54, HMGB1 (Abcam,1:1000), My-HC (MY32;Sigma,1:1000), Myogenin (F5D, Developmental studies Hybridoma Bank, 1:250) α-tubulin (Developmental studies Hybridoma Bank, 1:1000), hnRNPA1 (Cell Signalling, 1:1000) 22, β-actin (Sigma,1:500), myoglobin (DAKO, 1:500) and Ago2 (Cell Signalling,1:1000). Full blots are provided in Supplementary Figure S12.
Immunofluorescence
Immunofluorescence was performed using undifferentiated or differentiated C2C12 cells that were grown to sub-confluency in DMEM (Dulbecco’s modified eagle medium from Invitrogen)17. After the appropriate experimental treatments, cells were rinsed twice in PBS, fixed in 3% phosphate-buffered paraformaldehyde (Sigma), and permeabilized in 0.5% PBS-goat serum with Triton. After permeabilization, cells were incubated with primary antibodies [against My-HC (MF-20, Developmental studies Hybridoma Bank, 1:1000), HuR (3A2,1:1000) or HMGB1 (Abcam,1:1000)] for 1 h at room temperature and then incubated with goat anti-mouse secondary antibodies conjugated with Rhodamine (red) or FITC (green) from Molecular Probes (Eugene, OR). To visualize the nucleus, cells were stained with DAPI (Molecular Probes). Microscopic analyses were performed using an AXIOVERT 200M (Zeiss).
β-Galactosidase Staining
MLC1/3F-nlacZ homozygous transgenic mice 25 were mated with Hmgb1+/− heterozygotes. Pregnant females were sacrified by cervical dislocation at E10.5. Embryos were isolated in phosphate-buffered saline (PBS), fixed for 2 hours in 4% paraformaldehyde pH 7.4 at 4°C, washed in PBS for 20 min and equilibrated in 20% sucrose PBS. Fixed embryos were then stained for 1 hr at 37°C in 1mg/ml X-gal solution of PBS, also containing 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 2 mM MgCl2, 0.2% NP40 PBS. Hmgb1+/− embryos were identified by PCR analysis of yolk sac DNA using the following primers: HMGB1for GCA GGC TTC GTT GTT TTC ATA CAG and HMGB1rev TCA AAG AGT AAT ACT GCC ACC TTC for the wt allele; NEOfor TGG TTT GCA GTG TTC TGC CTA GC and NEOrev CCC AGT CAT AGC CGA ATA GCC for the targeted allele. These animal studies were approved by the San Raffaele University Animal Care Committee.
Histology and morphometric analysis
Tibialis anterior (TA) muscles were dissected from 1-year old wild type and Hmgb1+/− mice and frozen in liquid-nitrogen-cooled isopentane. Serial sections (8 μm thick) were stained with hematoxylin and eosin (H&E). Morphometric analyses were performed on sections of TA using ImageJ to determine the cross-sectional area of 900 fibers for each group.
Fusion index
Explants of presomitic mesoderm (PSM)55 were dissected from wt, Hmgb1+/− and Hmgb1−/− embryos at E9.5, plated on gelatin-coated dishes and cultured for four days in DMEM supplemented with 20% FCS and 50μg/ml gentamycin. On day 4, cultures were processed for immunoflorescence with an antibody specific for MyHC (MF20, Developmental Studies Hybridoma Bank, 1:20 dilution). Fusion index was determined as the number of nuclei in sarcomeric myosin-expressing cells with more than two nuclei versus the total number of nuclei.
Regeneration assay
Muscle injury was performed on the TA of 3-mo-old wt and Hmgb1+/− mice by injecting 50 μl of 10 μM CTX (three animals per group). Mice were sacrificed 3, 7 or 14 d after CTX injection, and the TA muscles were dissected and frozen in liquid N2–cooled isopentane. Ten-μm serial muscle sections were stained with H&E. Alternatively, injury of C57BL/6 (Charles River) mouse muscles was performed by BaCl2 injection in the TA muscle of 8-wk old mice, under zolazepam/tiletamine anesthesia 56. These animal studies were approved by the San Raffaele University Animal Care Committee.
RNA electromobility shift assays
The HMGB1 cRNA probes were produced by in vitro transcription using a T7 RNA polymerase 57. All HMGB1 probes (5′UTR and P1 to P12) were generated by PCR amplification using a forward primer fused to the T7 promoter as well as pCMV-HMGB1 expression vector as the template (see Supplementary Table S4). For smaller probes (P4-1 to P4-3), oligonucleotide sense and anti-sense were directly annealed and used for in vitro transcription. The RNA binding assays were performed22 by incubating either 10 μg total cell extracts (TCE) or 300 ng purified recombinant protein (GST or GST-HuR) with 50 000 cpm of 32P-labelled cRNAs in a total volume of 20 μl EBMK buffer (25 mM HEPES pH 7.6, 1.5 mM KCl, 5 mM MgCl2, 75 mM NaCl, 6% sucrose and protease inhibitors) at RT for 15 minutes. For competition assays, 0.01X, 0.1X, 1X, 10X and 100X excess unlabeled specific or unspecific transcripts were incubated with the TCE for 15 minutes at RT before the 32P-labelled probes were added to the reaction. 2 μl of a 50 mg/ml heparin sulphate stock solution were then added to the reaction for an additional 15 minutes at RT. In supershift experiments, 5 μg of a purified monoclonal anti-HuR antibody were then added to the reaction for an additional 15 minutes at RT. Samples were then loaded on a 4% polyacrylamide gel containing 0.05% NP-40.
Quantitative RT-PCR
One μg of total RNA was reverse transcribed using the M-MuLV Reverse Transcriptase (New England Biolabs) according to the manufacturer’s protocol. 1/80 dilution of cDNA was used to detect HMGB1, GAPDH and R luc mRNAs using SsoFast™ EvaGreen® Supermix (Bio-Rad). Expression of HMGB1 and R luc was standardized using GAPDH as a reference, and relative levels of expression were quantified by calculating 2−ΔΔCT, where ΔΔCT is the difference in CT between target and reference. For miRNA detection, 200 ng of total RNA was reverse transcribed using the Universal cDNA synthesis kit (Exiqon) and the presence of miR-1192, miR-16 and U6 was assessed by qPCR using the SyBr Green Master Mix (Exiqon).
Northern blot analysis and Actinomycin D pulse-chase experiments
The extraction of total RNA was performed using Trizol reagent (Invitrogen) 22. For the isolation of miRNAs, twice as much isopropanol was used in the RNA purification protocol. Northern blot analysis was performed using 10 μg total RNA 22. After transferring to a Hybond-N membrane (Amersham) and UV-cross-linking, the blot was hybridized with probes specific for MyoD 17, GAPDH 17, 18S 58, 5.8S 34 and HMGB1 mRNAs were generated using the PCR Purification Kit (GE Healthcare) with primers described below. The probes were radiolabeled with α–32P dCTP using Ready-to-Go DNA labeling beads (GE Healthcare) according to the manufacturer’s instructions. The stability of HMGB1 mRNA was assessed by the addition of the general transcriptional inhibitor actinomycin D (5 μg/ml)58 for the indicated periods of time. Total RNA was isolated from the cells after 0, 4, 6, 8, 10 and 12 hours following ActD treatment using TRIzol reagent (Invitrogen), and analyzed by Northern blotting.
For miRNA detection, 25 μg of total RNA was separated on a 12% denaturating urea polyacrylamide gel. RNA was subsequently transferred to a Hybond-N+ membrane (Amersham Biosciences) and cross-linked to the membrane. Hybridization was carried out by using ULTRAHybOligo solution according to the manufacturer’s instructions (Ambion). The probe sequences were complementary to the mature forms of miR-16 or U6 RNA and were labeled using StarFire system (Integrated DNA Technologies) according to the manufacturer’s protocol.
Polysome fractionation
Forty million myoblasts were grown and treated with siRNAs as described above and polysome fractionation experiments were performed. Briefly, the cytoplasmic extracts obtained from lysed myoblast cells were centrifuged at 130,000 x g for 2 h on a sucrose gradient (10–50% w/v)59,60. Polysomal (P) or non-polysomal (NP) fractions were pooled and RNA was extracted using Trizol LS (Invitrogen). RNA samples were then analyzed on an agarose gel. The levels of HMGB1 and GAPDH mRNAs were determined using quantitative RT-PCR.
Luciferase activity
The activity of Renilla luciferase was measured using a Renilla luciferase assay system (Promega) with a luminometer following the manufacturer’s instructions.
Fluorescence in situ Hybridization
The fluorescence in situ hybridization experiments were performed29 using a DNA fragment of ~500 bp corresponding to the coding region of mouse HMGB1. The fragment was amplified by PCR using the following primers fused to either a T7 or T3 minimal promoter sequence: HMGB1 forward, 5′-AAA AAG CCG AGA GGC AAA AT-3′, and HMGB1 reverse, 5′-CTT TTT CGC TGC ATC AGG TT-3′. The PCR product was used as the template for in vitro transcription of the HMGB1 probe needed for fluorescence in situ hybridization. The antisense (T3) and sense (T7) probes were prepared using digoxigenin-RNA labeling mix from Roche Diagnostics. The RNA probes were quantified, denatured and incubated with permeabilized cells at 37°C overnight in the hybridization buffer (50% formamide, 5X SSC, 50 mM phosphate buffer, pH=7.4, 5X Denhardt’s, 1 mM EDTA and 250 ng/μl of salmon sperm DNA). After the hybridization, the cells were used for immunofluorescence to detect the HMGB1 mRNA and HuR protein29. Finally, the cells were incubated with secondary goat anti-rabbit antibody and anti-DIG antibody for immunofluorescence.
Preparation of mRNA complexes and analysis with RT-PCR
Total cell extracts were prepared in lysis buffer (50 mM Tris, pH 8; 0.5% Triton X-100; 150 mM NaCl; complete protease inhibitor from Roche). 25 μl of the anti-HuR (3A2) or IgG (Jackson Immunoresearch Laboratories) antibodies were incubated with 200 μl of pre-swelled proteinA-Sepharose beads for 4 h at 4°C. Alternatively, C2C12 cells depleted or not of HuR were incubated with an anti-Ago2 (Cell Signalling) or IgG control antibody. After three washes in lysis buffer, 1.5 mg of cell extract was added overnight at 4°C. The final dilution of the antibodies during the incubation with lysates is 1/18. Beads were washed three times with cell lysis buffer, incubated with Proteinase K and the RNA isolated by phenol/cloroform extraction followed by precipitation overnight at −20°C with isopropanol 17,61. Purified RNA was resuspended in 12 μl of water, and 2 μl was reverse transcribed using the M-MuLV Reverse Transcriptase (New England Biolabs) according to the manufacturer’s protocol. Association of HMGB1 and β-actin mRNAs with HuR as well as HMGB1 with Ago2 was defined using quantitative RT-PCR. Ten μl of purified RNA was reverse transcribed using the Universal cDNA synthesis kit (Exiqon) and the presence of miR-1192, miR-16 and U6 was assessed by qPCR.
cDNA array analysis
Microarray experiments were performed using mouse array, which contain 17,000 probe sets of known and unknown expressed sequence tags 62. HuR was immunoprecipitated from exponentially growing C2C12 cells using an anti-HuR monoclonal antibody (3A2) and an anti-IgG monoclonal antibody as a negative control. The final dilution of the antibodies during the overnight incubation with lysates is 1/18. Then the samples were spun 5min at 3000 RPM and washed with TSE1 (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl pH: 8.1, 150mM NaCl), TSE2 (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl pH: 8.1, 500mM NaCl) and TSE3 (0.25M, LiCl, 1% NP-40, 1% deoxycholate, 1mM EDTA, 10mM Tris-HCl pH: 8.1) buffers. The immunoprecipitate were then incubated for 30 min at 55°C in 100ml NT2 buffer (50mM Tris pH:7.4, 150mM NaCl, 1mM MgCl2, 0.05% NP-40) with0.1% SDS and 30mg proteinase K. The associated RNAs were extracted by adding ~15ml of 2M NaOAc pH: 4.0, 150 μl of water saturated phenol and ~30 μl chloroform followed by 15 min incubation on ice. After spinning (12000g for 20min) the aqueous phase is mixed with glycogen and isopropanol and incubated overnight at −20°C. The isolated RNAs were then resuspended in water and hybridized on cDNA arrays. The data were processed using the Array Pro software (Media Cybernetics, Inc.), then normalized by Z-score transformation 63 and used to calculate differences in signal intensities. Significant values were tested using a two-tailed Z-test and a P of ≤0.01. The data were calculated from two independent experiments.
Subcellular Fractionation
Subcellular fractionation was performed using the PARIS kit (Ambion) (Austin, TX) according the manufacturer’s instructions. An equal number of cells per sample were used.
Biotin pull-down assay
The P4 fragment of the 3′UTR of mouse HMGB1 was subcloned into a pGEM-Teasy vector (Promega) according to the manufacturer’s instructions by performing PCR amplification using the following primers: forward 5′-GCC ACT AAC CTT GCC TGG TA-3′ and reverse 5′-TCG TAT AAG CTG CAT CAG AGA CA-3′. The transcript is transcribed using the T7RiboMAX™ Express large scale RNA production system (Promega) according to the manufacturer’s and the NotI digested PGEM-Teasy-P3P4 vector as a template. For preparing protein extracts, 30-40 millions of cells were washed with ice-cold phosphate-buffered saline (PBS) and resuspended in 1mL of EBMK buffer (25mM Hepes pH 7,6; 5mM MgCl2; 1,5mM KCl; 75mM NaCl; 175mM Sucrose) containing 0,5% NP-40, complete protease inhibitor without EDTA (Roche). Lysates were sonicated 10s 3 times at 200W and cleared by centrifugation at 12,000 rpm for 15 min. The protein concentration was determined (Bio-Rad microassay) and 1mg were used for each assay. Biotin pull-down assay is performed using Miltenyi Biotech mMACS streptavidin kit according to the manufacturer’s instructions.
Primers used to prepare probes for Northern blot analysis
mHMGB1: For: 5′-GCA TCC TGG CTT ATC CAT TG-3′; Rev: 5′-TGC TCT TTT CAG CCT TGA CC-3′
mGAPDH: For: 5′-AAG GTC ATC CCA GAG CTG AA-3′; rev: 5′-AGG AGA CAA CCT GGT CCT CA-3′
Statistical analyses
The Statistical analyses in this study were performed using the Graphpad Prism5 software to determine significance (two tailed, Student’s t-test).
Supplementary Material
Acknowledgments
We offer our sincere thanks to Dr. C. von Roretz for critical review of this manuscript and insightful discussion. We wish to thank Heikki Rauvala (Helsinki, Finland) for providing recombinant HMGB1. VDR is a Research Fellow of the Terry Fox foundation through an award from the Canadian Cancer Society (former National Cancer Institute of Canada). This work was supported by a CIHR (MOP-89798) operating grant and a NSERC discovery grant (RGPIN/371668-200) to I.G, a EU FP7 grant (Endostem) to M.E.B. and S.B. IG is a recipient of a TierII Canada Research Chair.
Footnotes
Author Contributions
V.D. and A.C. designed, carried out, analyzed and interpreted all of the in vitro experiments in the manuscript. B.C., S.B. and M.B. designed and carried out the in vivo experiments described in Figures 2, 3. X.J.L., K.V. and M.Z. assisted with some of the in vitro experiments. F.R., G.S. and R.D. R.D. designed and carried out the experiments described in Supplementary Figure S2 c,d as well as Supplementary Figure S10. R.D. helped writing part of the manuscript. B.W. provided technical expertise and helped in the analysis of microarray and sequencing experiments. S.D. helped in the design of several experiments in the manuscript. M.B. participated in writing and editing the manuscript. I.E.G. conceived and helped design all experiments in the manuscript, aided in the analysis of the data and wrote the manuscript.
Competing Financial Interests
M.E.B. is part owner of HMGBiotech, a company that provides reagents and services for HMGB1 research. The remaining authors declare no competing financial interests.
Accession codes
Microarray data has been deposited in Gene Expression Omnibus under accession code XXX.
References
- 1.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 2.Zhang K, Sha J, Harter ML. Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J Cell Biol. 2010;188:39–48. doi: 10.1083/jcb.200904144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Mori R, et al. Multiple effects of high mobility group box protein 1 in skeletal muscle regeneration. Arterioscler Thromb Vasc Biol. 2007;27:2377–2383. doi: 10.1161/ATVBAHA.107.153429. [DOI] [PubMed] [Google Scholar]
- 4.Filippin LI, Moreira AJ, Marroni NP, Xavier RM. Nitric oxide and repair of skeletal muscle injury. Nitric Oxide. 2009;21:157–163. doi: 10.1016/j.niox.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Palumbo R, et al. Extracellular HMGB1, a signal of tissue damage, induces mesoangioblast migration and proliferation. J Cell Biol. 2004;164:441–449. doi: 10.1083/jcb.200304135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takaesu G, et al. Activation of p38alpha/beta MAPK in myogenesis via binding of the scaffold protein JLP to the cell surface protein Cdo. J Cell Biol. 2006;175:383–388. doi: 10.1083/jcb.200608031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serra C, et al. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones NC, et al. The p38alpha/beta MAPK functions as a molecular switch to activate the quiescent satellite cell. J Cell Biol. 2005;169:105–116. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorci G, Riuzzi F, Arcuri C, Giambanco I, Donato R. Amphoterin stimulates myogenesis and counteracts the antimyogenic factors basic fibroblast growth factor and S100B via RAGE binding. Mol Cell Biol. 2004;24:4880–4894. doi: 10.1128/MCB.24.11.4880-4894.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riuzzi F, Sorci G, Donato R. RAGE expression in rhabdomyosarcoma cells results in myogenic differentiation and reduced proliferation, migration, invasiveness, and tumor growth. Am J Pathol. 2007;171:947–961. doi: 10.2353/ajpath.2007.070049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riuzzi F, Sorci G, Donato R. S100B stimulates myoblast proliferation and inhibits myoblast differentiation by independently stimulating ERK1/2 and inhibiting p38 MAPK. J Cell Physiol. 2006;207:461–470. doi: 10.1002/jcp.20580. [DOI] [PubMed] [Google Scholar]
- 12.Riuzzi F, Sorci G, Sagheddu R, Donato R. HMGB1-RAGE regulates muscle satellite cell homeostasis through p38-MAPK- and myogenin-dependent repression of Pax7 transcription. J Cell Sci. 2012;125:1440–1454. doi: 10.1242/jcs.092163. [DOI] [PubMed] [Google Scholar]
- 13.Begum N, Pash JM, Bhorjee JS. Expression and synthesis of high mobility group chromosomal proteins in different rat skeletal cell lines during myogenesis. J Biol Chem. 1990;265:11936–11941. [PubMed] [Google Scholar]
- 14.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Roretz C, Gallouzi IE. Decoding ARE-mediated decay: is microRNA part of the equation? J Cell Biol. 2008;181:189–194. doi: 10.1083/jcb.200712054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueroa A, et al. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Giessen K, Di-Marco S, Clair E, Gallouzi IE. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J Biol Chem. 2003;278:47119–47128. doi: 10.1074/jbc.M308889200. [DOI] [PubMed] [Google Scholar]
- 18.von Roretz C, Beauchamp P, Di Marco S, Gallouzi IE. HuR and myogenesis: being in the right place at the right time. Biochim Biophys Acta. 2011;1813:1663–1667. doi: 10.1016/j.bbamcr.2011.01.036. [DOI] [PubMed] [Google Scholar]
- 19.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 20.Conejo R, de Alvaro C, Benito M, Cuadrado A, Lorenzo M. Insulin restores differentiation of Ras-transformed C2C12 myoblasts by inducing NF-kappaB through an AKT/P70S6K/p38-MAPK pathway. Oncogene. 2002;21:3739–3753. doi: 10.1038/sj.onc.1205469. [DOI] [PubMed] [Google Scholar]
- 21.Li XL, Andersen JB, Ezelle HJ, Wilson GM, Hassel BA. Post-transcriptional regulation of RNase-L expression is mediated by the 3′-untranslated region of its mRNA. J Biol Chem. 2007;282:7950–7960. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- 22.Dormoy-Raclet V, et al. The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a U-rich-element-dependent manner. Mol Cell Biol. 2007;27:5365–5380. doi: 10.1128/MCB.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckingham M, et al. The formation of skeletal muscle: from somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calogero S, et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 25.Kelly R, Alonso S, Tajbakhsh S, Cossu G, Buckingham M. Myosin light chain 3F regulatory sequences confer regionalized cardiac and skeletal muscle expression in transgenic mice. J Cell Biol. 1995;129:383–396. doi: 10.1083/jcb.129.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cossu G, et al. Activation of different myogenic pathways: myf-5 is induced by the neural tube and MyoD by the dorsal ectoderm in mouse paraxial mesoderm. Development. 1996;122:429–437. doi: 10.1242/dev.122.2.429. [DOI] [PubMed] [Google Scholar]
- 27.Riuzzi F, Sorci G, Beccafico S, Donato R. S100B engages RAGE or bFGF/FGFR1 in myoblasts depending on its own concentration and myoblast density. Implications for muscle regeneration. PloS one. 2012;7:e28700. doi: 10.1371/journal.pone.0028700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- 29.Lian XJ, Gallouzi IE. Oxidative Stress Increases the Number of Stress Granules in Senescent Cells and Triggers a Rapid Decrease in p21waf1/cip1 Translation. J Biol Chem. 2009;284:8877–8887. doi: 10.1074/jbc.M806372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beauchamp P, et al. The cleavage of HuR interferes with its transportin-2-mediated nuclear import and promotes muscle fiber formation. Cell Death Differ. 2010 doi: 10.1038/cdd.2010.34. [DOI] [PubMed] [Google Scholar]
- 31.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PloS one. 2010;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HH, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009 doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Dormoy-Raclet V, Markovits J, Jacquemin-Sablon A, Jacquemin-Sablon H. Regulation of Unr expression by 5′- and 3′-untranslated regions of its mRNA through modulation of stability and IRES mediated translation. RNA Biol. 2005;2:e27–35. [PubMed] [Google Scholar]
- 35.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 36.Xu F, et al. Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma. J Cell Biochem. 2010;111:727–734. doi: 10.1002/jcb.22762. [DOI] [PubMed] [Google Scholar]
- 37.Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci U S A. 2008;105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marasa BS, et al. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany NY) 2010;2:333–343. doi: 10.18632/aging.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Giessen K, Gallouzi IE. Involvement of transportin 2-mediated HuR import in muscle cell differentiation. Mol Biol Cell. 2007;18:2619–2629. doi: 10.1091/mbc.E07-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrari S, Finelli P, Rocchi M, Bianchi ME. The active gene that encodes human high mobility group 1 protein (HMG1) contains introns and maps to chromosome 13. Genomics. 1996;35:367–371. doi: 10.1006/geno.1996.0369. [DOI] [PubMed] [Google Scholar]
- 41.Ferrari S, Ronfani L, Calogero S, Bianchi ME. The mouse gene coding for high mobility group 1 protein (HMG1) J Biol Chem. 1994;269:28803–28808. [PubMed] [Google Scholar]
- 42.Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes & Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuchen S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linsen SE, et al. Limitations and possibilities of small RNA digital gene expression profiling. Nature methods. 2009;6:474–476. doi: 10.1038/nmeth0709-474. [DOI] [PubMed] [Google Scholar]
- 46.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic acid Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 50.Andersson U, Tracey KJ. HMGB1 Is a Therapeutic Target for Sterile Inflammation and Infection. Annu Rev Immunol. 2010 doi: 10.1146/annurev-immunol-030409-101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 52.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 53.Bianchi ME, Manfredi AA. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol Rev. 2007;220:35–46. doi: 10.1111/j.1600-065X.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- 54.Gallouzi IE, et al. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cossu G, Tajbakhsh S, Buckingham M. How is myogenesis initiated in the embryo? Trends Genet. 1996;12:218–223. doi: 10.1016/0168-9525(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 56.Caldwell CJ, Mattey DL, Weller RO. Role of the basement membrane in the regeneration of skeletal muscle. Neuropathol Appl Neurobiol. 1990;16:225–238. doi: 10.1111/j.1365-2990.1990.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 57.Gallouzi IE, et al. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol Cell Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Di Marco S, et al. NF-(kappa)B-mediated MyoD decay during muscle wasting requires nitric oxide synthase mRNA stabilization, HuR protein, and nitric oxide release. Mol Cell Biol. 2005;25:6533–6545. doi: 10.1128/MCB.25.15.6533-6545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cammas A, et al. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol Biol Cell. 2007;18:5048–5059. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Di Marco S, et al. The translation inhibitor pateamine A prevents cachexia-induced muscle wasting in mice. Nat Commun. 2012;3:896. doi: 10.1038/ncomms1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 62.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lopez de Silanes I, et al. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bordeleau ME, et al. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem Biol. 2006;13:1287–1295. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 65.Dang Y, et al. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 66.Hood KA, West LM, Northcote PT, Berridge MV, Miller JH. Induction of apoptosis by the marine sponge (Mycale) metabolites, mycalamide A and pateamine. Apoptosis. 2001;6:207–219. doi: 10.1023/a:1011340827558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.