Abstract
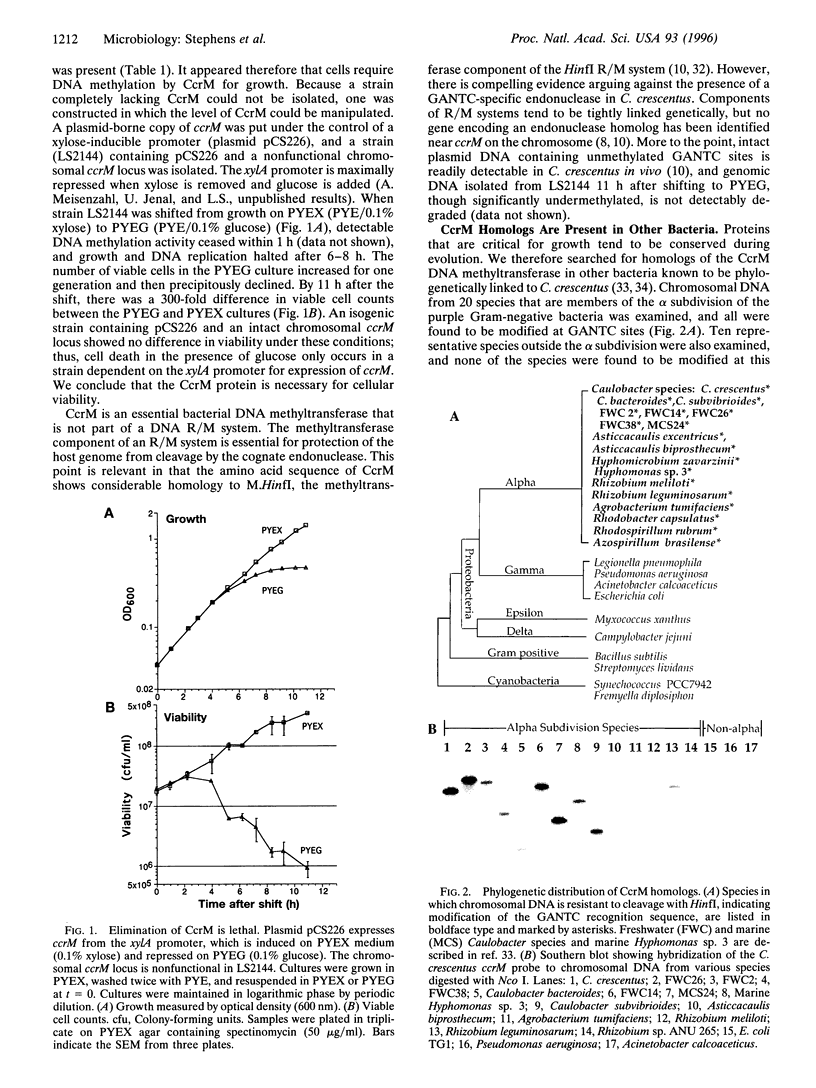
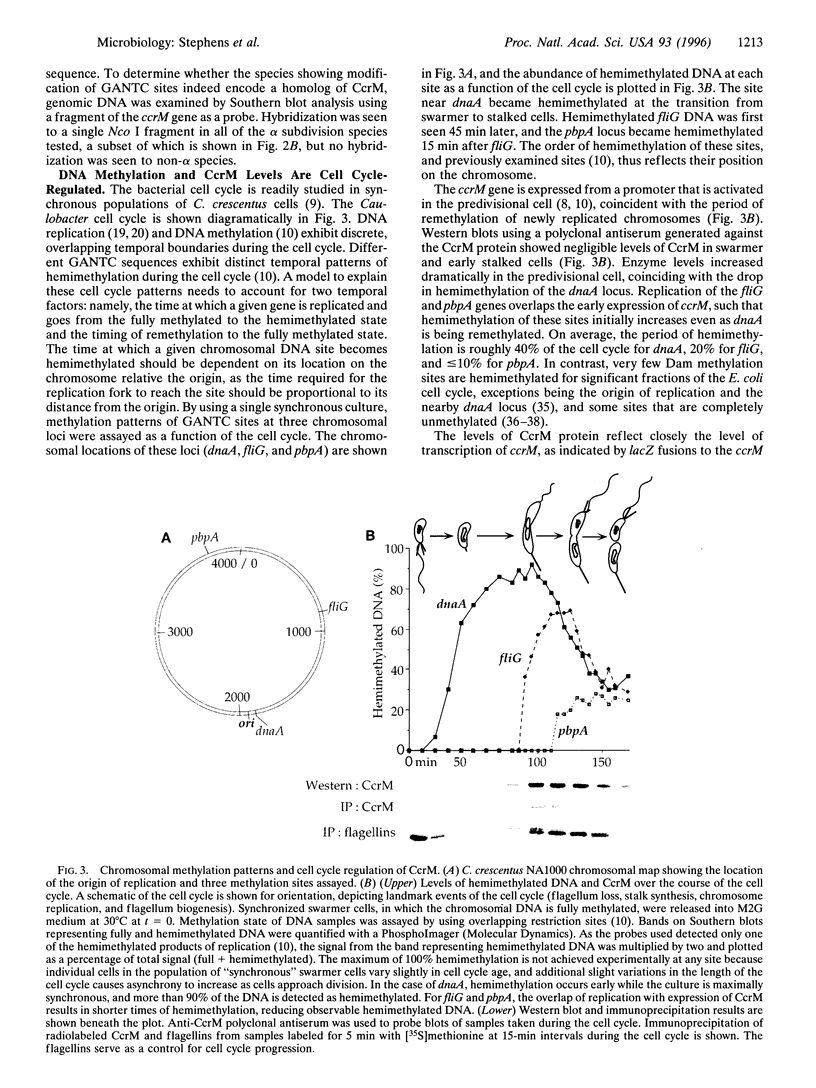
The CcrM adenine DNA methyltransferase, which specifically modifies GANTC sequences, is necessary for viability in Caulobacter crescentus. To our knowledge, this is the first example of an essential prokaryotic DNA methyltransferase that is not part of a DNA restriction/modification system. Homologs of CcrM are widespread in the alpha subdivision of the Proteobacteria, suggesting that methylation at GANTC sites may have important functions in other members of this diverse group as well. Temporal control of DNA methylation state has an important role in Caulobacter development, and we show that this organism utilizes an unusual mechanism for control of remethylation of newly replicated DNA. CcrM is synthesized de novo late in the cell cycle, coincident with full methylation of the chromosome, and is then subjected to proteolysis prior to cell division.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbeyron T., Kean K., Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J Bacteriol. 1984 Nov;160(2):586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E., Løbner-Olesen A. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990 Sep 7;62(5):981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- Boyes J., Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991 Mar 22;64(6):1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- Brun Y. V., Marczynski G., Shapiro L. The expression of asymmetry during Caulobacter cell differentiation. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Campbell J. L., Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990 Sep 7;62(5):967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- Chandrasegaran S., Lunnen K. D., Smith H. O., Wilson G. G. Cloning and sequencing the HinfI restriction and modification genes. Gene. 1988 Oct 30;70(2):387–392. doi: 10.1016/0378-1119(88)90210-7. [DOI] [PubMed] [Google Scholar]
- Degnen S. T., Newton A. Chromosome replication during development in Caulobacter crescentus. J Mol Biol. 1972 Mar 14;64(3):671–680. doi: 10.1016/0022-2836(72)90090-3. [DOI] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Evinger M., Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977 Oct;132(1):294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Claeys K. M., Selker E. U. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993 Dec 10;262(5140):1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985 Nov;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale W. B., van der Woude M. W., Low D. A. Analysis of nonmethylated GATC sites in the Escherichia coli chromosome and identification of sites that are differentially methylated in response to environmental stimuli. J Bacteriol. 1994 Jun;176(11):3438–3441. doi: 10.1128/jb.176.11.3438-3441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi-Ariga S. M., Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989 May;3(5):612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- Kakutani T., Jeddeloh J. A., Richards E. J. Characterization of an Arabidopsis thaliana DNA hypomethylation mutant. Nucleic Acids Res. 1995 Jan 11;23(1):130–137. doi: 10.1093/nar/23.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993 Nov 25;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Marczynski G. T., Dingwall A., Shapiro L. Plasmid and chromosomal DNA replication and partitioning during the Caulobacter crescentus cell cycle. J Mol Biol. 1990 Apr 20;212(4):709–722. doi: 10.1016/0022-2836(90)90232-B. [DOI] [PubMed] [Google Scholar]
- Modrich P. Methyl-directed DNA mismatch correction. J Biol Chem. 1989 Apr 25;264(12):6597–6600. [PubMed] [Google Scholar]
- Nelson M., Christ C., Schildkraut I. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 1984 Jul 11;12(13):5165–5173. doi: 10.1093/nar/12.13.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nou X., Skinner B., Braaten B., Blyn L., Hirsch D., Low D. Regulation of pyelonephritis-associated pili phase-variation in Escherichia coli: binding of the PapI and the Lrp regulatory proteins is controlled by DNA methylation. Mol Microbiol. 1993 Feb;7(4):545–553. doi: 10.1111/j.1365-2958.1993.tb01145.x. [DOI] [PubMed] [Google Scholar]
- Ogden G. B., Pratt M. J., Schaechter M. The replicative origin of the E. coli chromosome binds to cell membranes only when hemimethylated. Cell. 1988 Jul 1;54(1):127–135. doi: 10.1016/0092-8674(88)90186-9. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R., Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994 Jan;176(1):1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer B. R., Marinus M. G. The dam and dcm strains of Escherichia coli--a review. Gene. 1994 May 27;143(1):1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- Peterson K. R., Wertman K. F., Mount D. W., Marinus M. G. Viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol Gen Genet. 1985;201(1):14–19. doi: 10.1007/BF00397979. [DOI] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan G., Zhao J. L., Newton A. Multiple structural proteins are required for both transcriptional activation and negative autoregulation of Caulobacter crescentus flagellar genes. J Bacteriol. 1994 Dec;176(24):7587–7600. doi: 10.1128/jb.176.24.7587-7600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991 Sep;55(3):451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H. DNA methylation and genomic imprinting. Cell. 1994 May 20;77(4):473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- Ried J. L., Collmer A. An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene. 1987;57(2-3):239–246. doi: 10.1016/0378-1119(87)90127-2. [DOI] [PubMed] [Google Scholar]
- Ringquist S., Smith C. L. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4539–4543. doi: 10.1073/pnas.89.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Key R., Flesher B., Smit J. The phylogeny of marine and freshwater caulobacters reflects their habitat. J Bacteriol. 1992 Apr;174(7):2193–2198. doi: 10.1128/jb.174.7.2193-2198.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C. M., Shapiro L. An unusual promoter controls cell-cycle regulation and dependence on DNA replication of the Caulobacter fliLM early flagellar operon. Mol Microbiol. 1993 Sep;9(6):1169–1179. doi: 10.1111/j.1365-2958.1993.tb01246.x. [DOI] [PubMed] [Google Scholar]
- Stephens C. M., Zweiger G., Shapiro L. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol. 1995 Apr;177(7):1662–1669. doi: 10.1128/jb.177.7.1662-1669.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tilghman S. M. DNA methylation: a phoenix rises. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8761–8762. doi: 10.1073/pnas.90.19.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R. A., Richards E. J. Arabidopsis thaliana DNA methylation mutants. Science. 1993 Jun 25;260(5116):1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wang M. X., Church G. M. A whole genome approach to in vivo DNA-protein interactions in E. coli. Nature. 1992 Dec 10;360(6404):606–610. doi: 10.1038/360606a0. [DOI] [PubMed] [Google Scholar]
- Zweiger G., Marczynski G., Shapiro L. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994 Jan 14;235(2):472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- Zweiger G., Shapiro L. Expression of Caulobacter dnaA as a function of the cell cycle. J Bacteriol. 1994 Jan;176(2):401–408. doi: 10.1128/jb.176.2.401-408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]





