Abstract
We previously reported the identification and development of novel inhibitors of streptokinase (SK) expression by Group A Streptococcus (GAS), originating from a high throughput cell-based phenotypic screen. Although phenotypic screening is well-suited to identifying compounds that exert desired biological effects in potentially novel ways, it requires follow-up experiments to determine the macromolecular target(s) of active compounds. We therefore designed and synthesized several classes of chemical probes for target identification studies, guided by previously established structure-activity relationships. The probes were designed to first irreversibly photolabel target proteins in the intact bacteria, followed by cell lysis and click ligation with fluorescent tags to allow for visualization on SDS-PAGE gels. This stepwise, “tag-free” approach allows for a significant reduction in molecular weight and polar surface area compared to full-length fluorescent or biotinylated probes, potentially enhancing membrane permeability and the maintenance of activity. Of the seven probes produced, the three most biologically active were employed in preliminary target identification trials. Despite the potent activity of these probes, specific labeling events were not conclusively observed due to a considerable degree of nonspecific protein binding. Nevertheless, the successful synthesis of potent biologically active probe molecules will serve as a starting point for initiating more sensitive methods of probe-based target identification.
Keywords: Group A Streptococcus, virulence inhibitor, streptokinase, antibiotic, phenotypic screening, target identification, tag-free photoprobes, photo-crosslinking, click chemistry
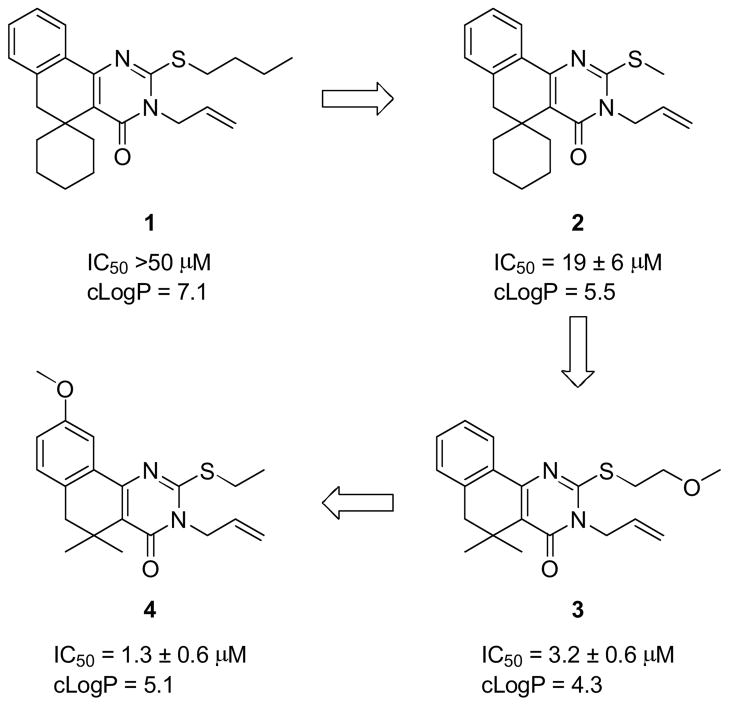
The growing threat of bacterial resistance to antibiotic treatment necessitates the development of new antibacterial agents with novel mechanisms of action.1 Bacterial pathogens express genes known as virulence factors that do not contribute to the growth and maintenance of the cell, but are critical for infection in the host. As a result, virulence-attenuating therapeutics have emerged as a potential mechanism for managing bacterial infection without selecting for mutants that are resistant to treatment.2 Our group has applied this paradigm toward the development of a new class of anti-virulence antibiotics that suppress the expression of streptokinase (SK), a bacterial activator of human plasmin, that plays a direct role in enhancing Group A Streptococcus (GAS) virulence.3 The lead compound of this series (1, Scheme 1) was identified via a cell-based high-throughput screen (HTS) for compounds that reduce ska gene transcription without inhibiting bacterial growth.4 Optimization of the lead compound through structure-activity relationship (SAR) studies5 led to a number of incremental improvements in activity and lipophilicity (2 and 3), eventually resulting in the discovery of potent analog 4 with a 35-fold greater potency and 2-log reduction in cLogP compared to lead 1.
Scheme 1.
Selected compounds from the SAR effort leading to potent analogs of screening hit 1.
Phenotype-based HTS strategies, like the one used to identify 1, return hit compounds with physicochemical properties sufficient for activity in whole cells and do not rely on a priori knowledge of the affected biological pathway, making them useful for discovering compounds with novel mechanisms of action.6,7 These advantages, taken together with the lack of antibiotic leads discovered via bacterial target-based screening,8 suggest phenotypic screening may be a potentially more fruitful tool for identifying novel antibacterial agents. However, phenotypic screening does not explicitly identify the molecular target of individual hit compounds, and therefore they must be established through subsequent studies. In the context of our project, identifying the target of this compound series would be important for (a) helping to establish a biochemical assay with which to improve the potency and specificity of the series, and (b) elucidating potentially novel virulence control pathways.
The potent activity of our compounds against SK expression, combined with RNA microarray data indicating the down-regulation of other important GAS virulence factors,4 suggests that their macromolecular target(s) are involved in the upstream regulation of GAS virulence mechanisms. Several proteins governing GAS virulence have been studied in detail,9 including Mga,10 Rgg,11,12 and CovR/CovS,13 but the genomic sequencing of several clinically relevant GAS serotypes has revealed multiple well-conserved virulence control elements that remain uncharacterized.14 Thus, identifying the target of this compound series has the potential to ascertain novel control mechanisms and further elucidate the complex nature of GAS virulence.
The use of chemical probes is a proven strategy for successfully establishing the protein targets of small molecules.15,16,17,18 We chose to pursue a tandem photolabeling-bioorthogonal conjugation strategy that has become widespread since the development of click chemistry.19,20,21 In this approach, a small-molecule analog of a potent compound possessing a photoreactive group and a terminal alkyne is covalently crosslinked to target proteins in the intact cellular milieu with UV light. After cell lysis, an azide-modified fluorescent or biotin-derived moiety can then be appended to the alkyne-functionalized protein(s) via copper(I)-mediated click chemistry, resulting in target proteins with covalently attached tags for visualization or selective purification. The lower molecular weight and topological polar surface area (TPSA) of these “tag-free”20 compounds compared to traditional biotinylated probes increases their likelihood of being cell-permeable, allowing them to be used in whole-cell systems rather than lysates. Cell-permeable affinity probes are advantageous in that their biological activity can be confirmed in phenotypic assays before beginning target identification studies. The probes also have access to all proteins in their native cellular conformations.
We envisioned the design of several tag-free photoprobes based on structural insights gained from our SAR studies on this scaffold.5 Although the maintenance of a high level of potency was a primary concern, the nature and positioning of the UV-active and terminal alkyne groups were equally important to us as they are crucial for ensuring a compatible orientation for labeling.22,23 A number of different photolabile groups have been successfully employed in the literature, varying in their stability, reactivity, and preference for carbon-carbon or carbon-heteroatom bond formation.24 The unknown nature of the binding site ultimately determines the structural features necessary for a functional probe, so we set out to synthesize a diverse series of compounds with one of three photolabile groups (benzophenone, diazirine, aryl azide) at different points on the scaffold to maximize the probability of identifying the target.
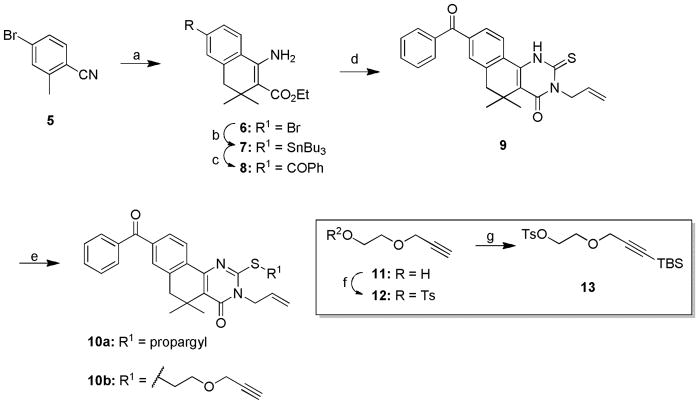
The chemistry to install benzophenone, diazirine, and aryl azide functionality onto the core scaffold of this class of compounds required three distinct synthetic routes. Synthesis of the benzophenone-based probes (10a–b, Scheme 2) began with the LDA-promoted Michael addition of 4-bromo-2-methylbenzonitrile (5) to ethyl 3,3-dimethyl acrylate and subsequent ZnI2-mediated cyclization to afford β-aminoester 6.25 Stannylation via halogen-metal exchange26 followed by carbonylative Stille coupling27 successfully delivered benzophenone 8. Acid-catalyzed addition of the amine to allyl isothiocyanate and subsequent cyclization generated 2-thioxopyrimidinone 9 that underwent S-alkylation with propargyl bromide or 12 (generated via tosylation of commercially available propynol ethoxylate 11) under mildly basic conditions to generate probes 10a and 10b.
Scheme 2.
Synthesis of benzophenone-functionalized tag-free probes 10a–b.a
aReagents and conditions. a) LDA, 1h; then ethyl 3,3-dimethyl acrylate, ZnI2, diglyme, −78°C – RT, 2h, 50%; b) (Bu3Sn)2, Pd(PPh3)4, toluene, reflux, 16h, 37%; c) 1 atm CO, PdCl2(PPh3)2, iodobenzene, DMF, 60°C, 4h, 58%; d) allyl isothiocyanate, AcOH, EtOH, 70°C, 16h, 27%; e) propargyl bromide or 12, Cs2CO3, 2-butanone, 70°C, 16h, 69–70%; f) Ts-Cl, pyridine, 0–4°C, 24h, 72%; g) nBuLi, −78°C, THF, 2h, then TBS-OTf, −78°C-RT, 1h, 63%.
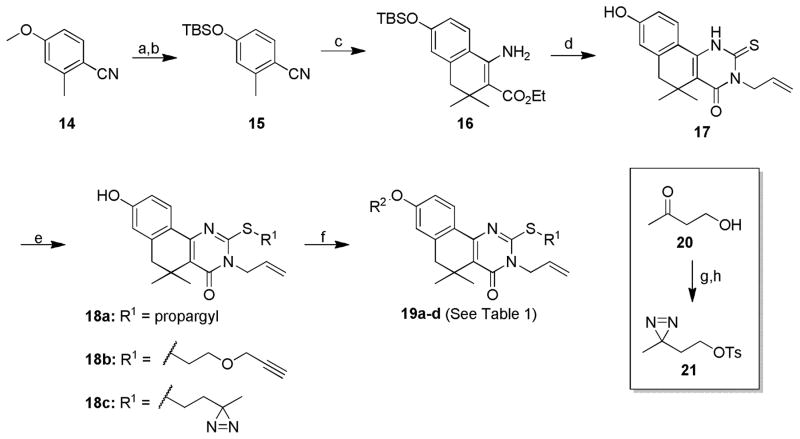
Diazirine probes 19a–d were synthesized via an analogous route beginning from aryl methyl ether 14 (Scheme 3). Demethylation and protection of the resulting phenol as a silyl ether resulted in 15, which could be employed in the synthesis of β-aminoester 16 in a manner similar to the one used to generate 6 in Scheme 3. In the course of our investigation, we found that cyclization with allyl isothiocyanate in the presence of cesium carbonate installed the desired 2-thioxopyrimidinone ring system while simultaneously cleaving the silyl ether protecting group, affording common intermediate 17. Successive chemoselective alkylations at sulfur and oxygen were exploited to install the diazirine and alkyne linker moieties in four different arrangements, yielding probes 19a–d. Diazirine-containing alkylating agent 21 was synthesized from 4-hydroxy-2-butanone (20) via diaziridination with 7N methanolic ammonia and hydroxylamine O-sulfonic acid,28 followed by oxidation and tosylation.29
Scheme 3.
Synthesis of diazirine-functionalized tag-free probes 19a–d.a
aReagents and conditions. a) BBr3, DCM, 0°C-RT, 16h; b) TBS-Cl, imidazole, DCM, 0°C-RT, 16h, 96% over 2 steps; c) LDA, then ethyl 3,3-dimethyl acrylate, ZnI2, diglyme, −78°C-RT, 3h, 63%; d) allyl isothiocyanate, Cs2CO3, EtOH, 70°C, 16h, 57%; e) 12, 21, or propargyl bromide, NaHCO3, 70°C, DMF, 3–6h, 45–71%; f) 12, 21, or propargyl bromide, Cs2CO3, 70°C, DMF, 3h, 16–64%; g) 7N NH3:MeOH, hydroxylamine-O-sulfonic acid, 0°C-RT, 16h; then I2, 0°C, 30 min; h) Ts-Cl, pyridine, 0–4°C, 24h, 15% over 3 steps.
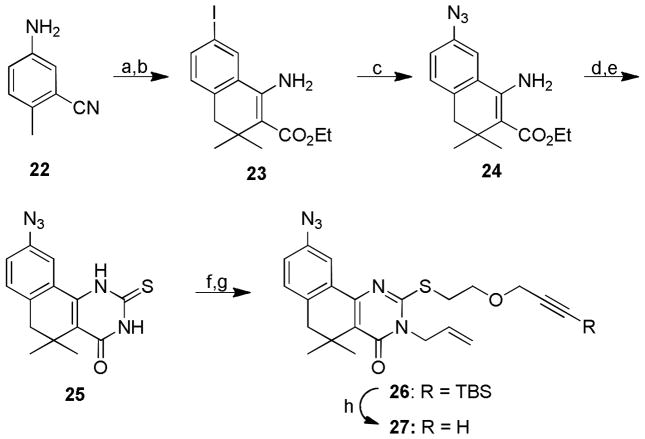
Synthesis of the aryl azide probe 27 began from commercially available 2-cyano-3-methylaniline 22 (Scheme 4). Masking the aniline functionality of 22 as a mono- or bis-Boc carbamate, diphenyl imine, or azide proved incompatible with the LDA/ZnI2 cyclization conditions. Finally, it was found that modified Sandmeyer conditions to transform the aniline to the aryl iodide30 permitted the formation of β-aminoester 23 in sufficient yields. Conversion to the corresponding aryl azide 24 via Cu(I)-mediated SNAr proceeded in excellent yields.31 Stepwise generation of the unsubstituted 2-thioxopyrimidinone with benzoyl isothiocyanate and potassium hydroxide afforded 25,32 which was then selectively S-alkylated with TBS-protected alkyne-containing unit 13. Alkyne protection was necessary to minimize cross-reactivity during the subsequent N-allylation under basic conditions to furnish 26. Finally, fluoride ion-promoted removal of the silyl group yielded the desired aryl azide probe 27.
Scheme 4.
Synthesis of aryl azide-functionalized tag-free probe 27.a
aReagents and conditions. a) NaNO2, HCl, then KI, H2O, 0°C; b) LDA, then ethyl 3,3-dimethyl acrylate, ZnI2, diglyme, −78°C-RT, 3h, 16% over 2 steps; c) NaN3, CuI, N,N-dimethyl ethylene diamine, Na-ascorbate, DMSO:H2O (5:1), RT, 3h, 85%; d) benzoyl isothiocyanate, EtOH, 70°C, 3h; e) KOH, EtOH:H2O, 2:1, 70°C, 3h, 54% over 2 steps; f) 13 (Scheme 2), NaHCO3, 70°C, DMF, 16h; g) NaOMe, allyl bromide, EtOH, 70°C, 3h, 42% over 2 steps; h) TBAF, THF, 0°C, 1h, 65%.
All completed tag-free affinity probes were assessed for their inhibitory effects on streptokinase expression using our previously described assay.4 Briefly, GAS culture supernatant was mixed with human plasma, and the resulting plasmin activity was assayed by turnover of a plasmin-specific chromogenic substrate by monitoring absorbance at 405 nm. Activity data are reported in Table 1 as the ratio of plasmin activity in test cultures divided by plasmin activity in a control culture treated with DMSO alone. Since retaining high potency is important for target identification, 50% streptokinase expression inhibition at 5 μM was used as the cutoff value for further evaluating probe compounds. Compounds meeting this threshold were more completely characterized with full IC50 determinations. Results are summarized in Table 1.
Table 1.
Activity data for tag-free photoprobes 10, 19, and 27.
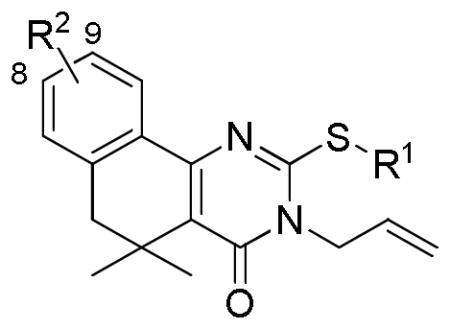
| |||||
|---|---|---|---|---|---|
| Cmpd | R1 | R2 | T/C (5 μM)a | Growth T/C (50 μM)b | IC50 (μM)c |
| 10a | propargyl | 8-Bz | 0.77 ± 0.34 | 1.09 ± 0.04 | nd |
| 10b |
|
8-Bz | 0.51 ± 0.07 | 0.94 ± 0.18 | 3.8 ± 0.6 |
| 19a | propargyl |
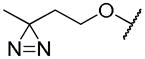
|
1.07 ± 0.16 | 0.96 ± 0.09 | nd |
| 19b |
|
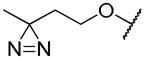
|
0.49 ± 0.34 | 1.07 ± 0.02 | 0.33 ± 0.70 |
| 19c |
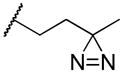
|
|
0.71 ± 0.40 | 1.01 ± 0.09 | nd |
| 19d |
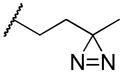
|
8-O-propargyl | 0.89 ± 0.16 | 1.02 ± 0.01 | nd |
| 27 |
|
9-N3 | 0.30 ± 0.08 | 0.98 ± 0.04 | 0.19 ± 0.26 |
| 4 | Et | 9-OMe | 0.10 ± 0.06 | 0.73 ± 0.07 | 1.3 ± 0.6 |
Amount of SK activity in GAS culture measured via colorimetric assay (A405) after incubation with 5 μM test compound for 24 hours divided by SK activity measured in GAS culture 24 hours after treatment with DMSO control, +/− standard deviation.
Growth inhibition as measured by OD600 of GAS culture 24 hours after treatment with 50 μM test compound divided by OD600 of control GAS culture treated with DMSO control after 24 hours, +/− standard deviation.
Concentration at which 50% maximal inhibitory effect against SK expression was achieved +/− standard deviation. Each data point is derived from the statistical treatment of at least 3 replicates. nd = not determined.
Benzophenone 10b, with its S-propargyloxyethyl click ligation sidechain, was found to be more potent than the corresponding S-propargyl analog 10a. This trend was also evident in diazirine probes 19a and 19b. Disappointingly, reversing the positioning of the alkyne and diazirine in probes 19c and 19d resulted in complete loss of activity. Azide probe 27, on the other hand, possessed potency slightly greater than lead compound 4, the most potent compound derived from our prior SAR studies. Fortuitously, we were able to identify a sufficiently active probe containing each of the selected photolabile groups (10b, 19b, and 27) for use in target identification studies.
Representative probe 19b was subjected to crosslinking conditions in methanol to assess whether the core scaffold would exhibit undesired photo-induced reactivity. Exposure of a methanolic solution of 19b to 365 nm UV light led to conversion of the parent compound into 2 new isolable compounds within 30 minutes (Scheme 5). NMR and ESI-MS analysis of the purified products identified the major product as 28, consistent with carbene insertion into the O-H bond of methanol (43% yield). The most prominent minor product consisted of alkene regioisomers that resulted from the two possible 1,2 hydride shifts (29, 27% combined yield) common for carbenes adjacent to saturated carbons.33 A previously reported study involving methanol capture by aliphatic diazirines observed analogous insertion and rearrangement products in similar ratios.34 Importantly, this study established that the remainder of the template is stable to degradation by UV irradiation.
Scheme 5.
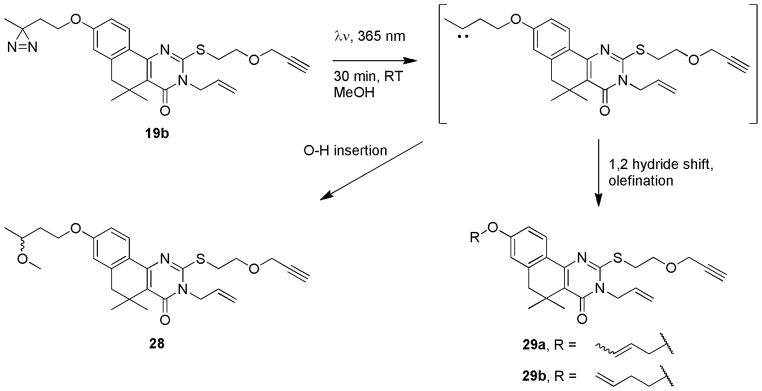
UV-induced reactivity of probe compound 19b in methanol.
Having confirmed the photostability of our probe scaffold, we began photo-crosslinking35 and fluorescent tagging assays. Briefly, GAS cultures were incubated in the presence of probe compounds for 30 minutes, then exposed to long-wave UV radiation for 5 (19b, 27) or 10 (10b) minutes to promote crosslinking. The cells were lysed and pelleted, and the resulting lysates were subjected to click chemistry conditions with Alexa Fluor 488 azide. Proteins were then precipitated via methanol/chloroform/water solution, separated via SDS-PAGE gel, and fluorescently visualized. Total protein levels were subsequently visualized by staining with Sypro Ruby (see Supplemental data for more detailed protocol).
Benzophenone probe 10b appeared to crosslink to a distinct subpopulation of proteins when separated on a polyacrylamide gel and fluorescently imaged (Figure 1). Tagging was found to be dose-dependent, evidenced by lower fluorescent signal in protein bands from samples tagged with lower probe concentrations. An identically treated control omitting the UV crosslinking step (Lane 6) did not fluorescently tag any proteins. Staining the gel to visualize total protein content with Sypro Ruby stain confirmed an abundance of protein in the sample, indicating that only a small subset of proteins were crosslinked to 10b and tagged with Alexa Fluor 488. Although the intensity of several protein bands appears to have been diminished by the introduction of soluble competitor 3, these findings could not be reliably reproduced in biotin pulldown experiments (data not shown). The presence of several other protein bands on the gel that did not lose intensity in the presence of 3 implies some degree of nonspecific protein binding by 10b.
Figure 1. Crosslinking trials using probe 10b.
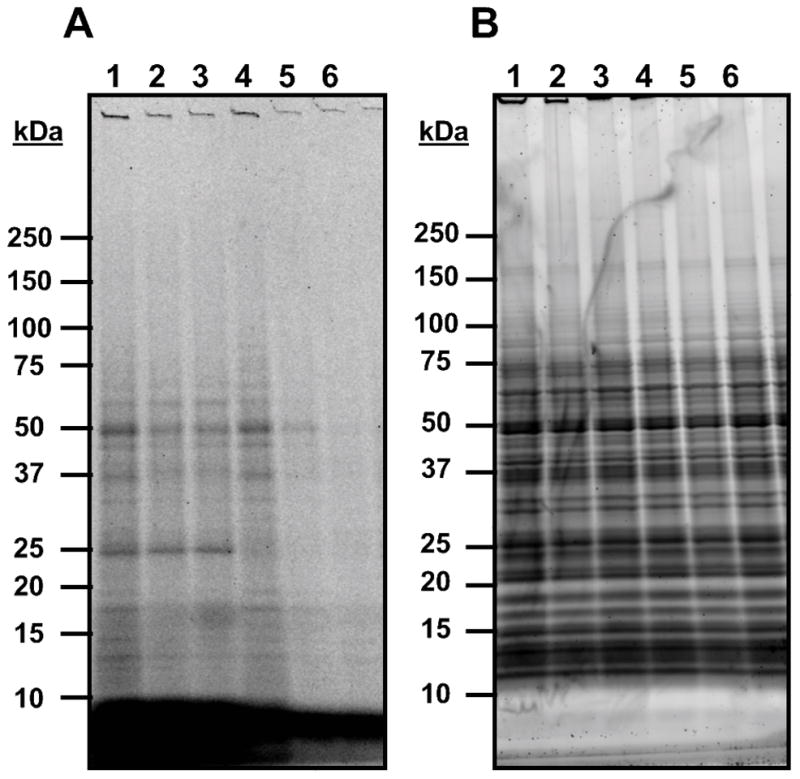
GAS cultures were grown in the presence of 10b with or without competitor 3, then subjected to UV crosslinking conditions. The cells were lysed and a fluorescent Alexa Fluor 488 moiety was appended to probe-protein adducts via click chemistry, then proteins were isolated and separated via SDS-PAGE. A: Fluorescent visualization of proteins from crosslinking and competition experiments with 10b on PAGE gel. B: Total protein stain of gel with Sypro Ruby stain. Lane 1: 100 μM 10b; Lane 2: 100 μM 10b + 500 μM 3; Lane 3: 100 μM 10b + 1 mM 3; Lane 4: 10 μM 10b; Lane 5: 1 μM 10b; Lane 6: 100 μM 10b, no UV crosslinking.
Crosslinking experiments undertaken with diazirine 19b and azide 27 (Figures 2 and 3) showed similar trends to 10b. In each case the intensity of fluorescently-tagged proteins was dependent on both the concentration of the probe compound and the exposure to UV light. Qualitative comparison of the fluorescent and total protein stains suggests that the fluorescent intensity was generally proportional to protein abundance, with a few exceptions. Bands at approximately 30K and 60K (probe 19b, Figure 2) and 18K and 110K (probe 27, Figure 3), for example, appear to be disproportionately labeled compared to their relative abundance, but the lack of competition-induced signal reduction suggests these are due to nonspecific interactions as well.
Figure 2. Crosslinking trials using diazirine probe 19b.
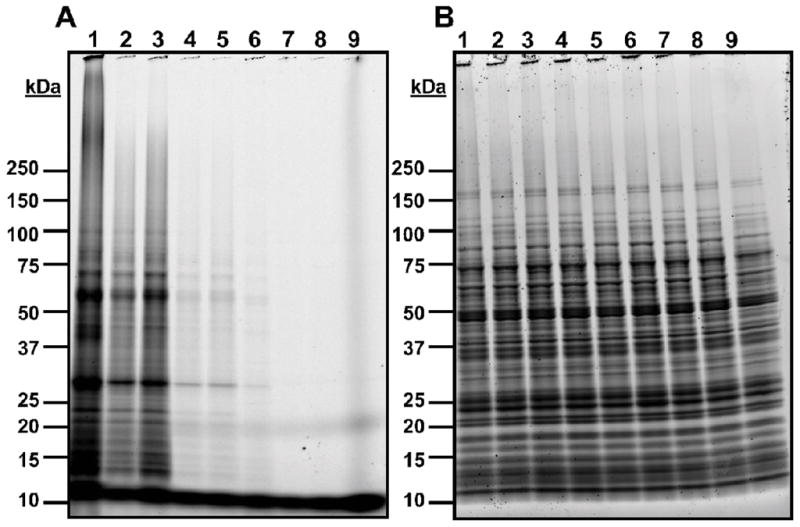
GAS cultures were grown in the presence of 19b with or without competitor 3, then subjected to UV crosslinking conditions. The cells were lysed and a fluorescent Alexa Fluor 488 moiety was appended to probe-protein adducts via click chemistry, then proteins were isolated and separated via SDS-PAGE. A: Fluorescent visualization of proteins from crosslinking and competition experiments with 19b on PAGE gel. B: Total protein stain of gel with Sypro Ruby stain. Lane 1: 100 μM 19b; Lane 2: 10 μM 19b; Lane 3: 10 μM 19b + 100 μM 3; Lane 4: 1 μM 19b; Lane 5: 1 μM 19b + 100 μM 3; Lane 6: 100 μM 19b, no UV crosslinking; Lane 7: 10 μM 19b, no UV crosslinking; Lane 8: 1 μM 19b, no UV crosslinking; Lane 9: no probe, no UV crosslinking.
Figure 3. Crosslinking trials using azide probe 27.
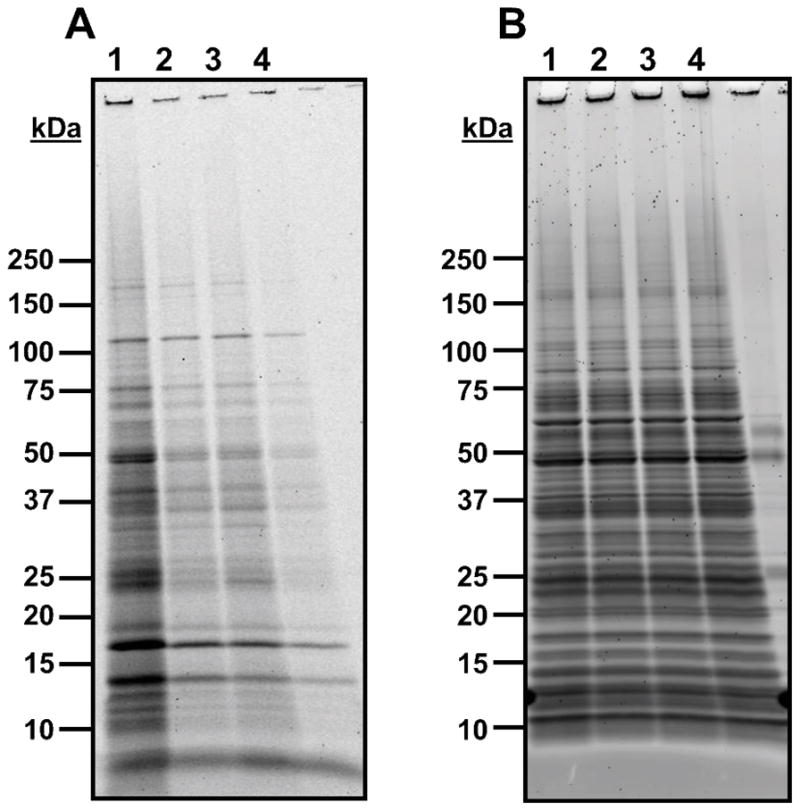
GAS cultures were grown in the presence of 27 with or without competitor 3, then subjected to UV crosslinking conditions. The cells were lysed and a fluorescent Alexa Fluor 488 moiety was appended to probe-protein adducts via click chemistry, then proteins were isolated and separated via SDS-PAGE. A: Fluorescent visualization of proteins from crosslinking and competition experiments with 27 on PAGE gel. B: Total protein stain of gel with Sypro Ruby stain. Lane 1: 100 μM 27; Lane 2: 10 μM 27; Lane 3: 10 μM 27 + 100 μM 3; Lane 4: 1 μM 27.
In summary, we report the synthesis of 7 tag-free photoaffinity probe compounds designed to determine the target of a novel class of Group A Streptococcal virulence inhibitors. Three probes, each with a different photolabile moiety, were found to be sufficiently active for use in target identification studies. After confirming adequate photoactivation of representative probe 19b without destruction of the overall template, we performed preliminary target identification studies. Crosslinking in whole cells, followed by cell lysis and covalent attachment of a fluorescent tag, led to relatively widespread fluorescent labeling that appears consistent with nonspecific labeling of a subset of total GAS proteins.
While biotin/fluorescent tagging and SDS-PAGE separation have been used successfully in the past for target identification, the nature of affinity chromatography and PAGE gel visualization usually limits the effectiveness of this method to probes that potently interact with high-abundance proteins.36 The possibility of the target being a bacterial transcription factor, among the least abundant proteins in the cellular milieu,37 along with the high propensity for nonspecific binding exhibited by these compounds, suggests that visual inspection of SDS-PAGE gels may not be sufficiently sensitive to identify our target. Future studies will employ proteomic techniques that offer a far greater level of resolution, particularly stable isotope labeling by amino acids in cell culture (SILAC)36 and GAS genome phage display library screening.38 These techniques solve the issue of low abundance by vastly expanding the detection limit for specific interactions and expressing the entire proteome at equal levels, respectively. The development of more potent probes is also underway. Using probes with greater potency should enhance labeling efficiency and allow for the use of lower probe concentrations, reducing the likelihood of widespread nonspecific labeling.
Supplementary Material
Acknowledgments
This work was supported by a University of Michigan Life Sciences Institute Innovation Partnership grant to SDL, and an NIH Pharmacological Sciences Training Program fellowship (Grant T32 GM007767) to BDY.
Abbreviations
- SK
streptokinase
- GAS
Group A Streptococcus
- HTS
high-throughput screen
- SAR
structure-activity relationship
- TPSA
topological polar surface area
- cLogP
logarithm of the calculated octanol-water partition coefficient
- LDA
lithium diisopropylamide
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- SILAC
stable isotope labeling by amino acids in cell culture
- THY
Todd-Hewitt media with 0.2% yeast extract
- ESI-MS
electrospray ionization mass spectrometry
Footnotes
Methods for activity determination, target identification, and solvent capture studies, as well as detailed synthetic procedures and characterization data for all synthesized compounds, can be found accompanying the online version of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Otto M. Blocking the Spread of Resistance. Sci Trans Med. 2013;5(184):184fs17. doi: 10.1126/scitranslmed.3006128. [DOI] [PubMed] [Google Scholar]
- 2.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9(2):117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 3.Sun H, Ringdahl U, Homeister JW, Fay WP, Engleberg NC, Yang AY, Rozek LS, Wang X, Sjobring U, Ginsburg D. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305(5688):1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- 4.Sun H, Xu Y, Sitkiewicz I, Ma Y, Wang X, Yestrepsky BD, Huang Y, Lapadatescu MC, Larsen MJ, Larsen SD, Musser JM, Ginsburg D. Inhibitor of streptokinase gene expression improves survival after group A streptococcus infection in mice. PNAS. 2012;109(9):3469–3474. doi: 10.1073/pnas.1201031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yestrepsky BD, Xu Y, Breen ME, Li X, Rajeswaran WG, Ryu JG, Sorenson RJ, Tsume Y, Wilson MW, Zhang W, Sun D, Sun H, Larsen SD. Novel Inhibitors of Bacterial Virulence: Development of 5,6-dihydrobenzo[h]quinazolin-4(3H)-ones for the Inhibition of Group A Streptococcal Streptokinase Expression. Bioorg Med Chem. 2013;21:1880–1897. doi: 10.1016/j.bmc.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432(7019):846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10(7):507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 8.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6(1):29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 9.Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen–host interactions. Trends Microbiol. 2003;11(5):224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 10.McIver KS, Scott JR. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol. 1997;179(16):5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaussee MS, Ajdic D, Ferretti JJ. The rgg Gene of Streptococcus pyogenes NZ131 Positively Influences Extracellular SPE B Production. Infect Immun. 1999;67(4):1715–1722. doi: 10.1128/iai.67.4.1715-1722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaussee MS, Sylva GL, Sturdevant DE, Smoot LM, Graham MR, Watson RO, Musser JM. Rgg Influences the Expression of Multiple Regulatory Loci To Coregulate Virulence Factor Expression in Streptococcus pyogenes. Infect Immun. 2002;70(2):762–770. doi: 10.1128/iai.70.2.762-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heath A, DiRita VJ, Barg NL, Engleberg NC. A Two-Component Regulatory System, CsrR-CsrS, Represses Expression of Three Streptococcus pyogenes Virulence Factors, Hyaluronic Acid Capsule, Streptolysin S, and Pyrogenic Exotoxin B. Infect Immun. 1999;67(10):5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu M-Y, Smoot JC, Porcella SF, Parkins LD, Campbell DS, Smith TM, McCormick JK, Leung DYM, Schlievert PM, Musser JM. Genome sequence of a serotype M3 strain of group A Streptococcus: Phage-encoded toxins, the high-virulence phenotype, and clone emergence. PNAS. 2002;99(15):10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie BJ, Hergenrother PJ. Identification of the cellular targets of bioactive small organic molecules using affinity reagents. Chem Soc Rev. 2008;37(7):1347–1360. doi: 10.1039/b702942j. [DOI] [PubMed] [Google Scholar]
- 16.Harding MW, Galat A, Uehling DE, Schreiber SL. A Receptor for the Immunosuppressant Fk506 Is a Cis-Trans Peptidyl-Prolyl Isomerase. Nature. 1989;341(6244):758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 17.Adam GC, Vanderwal CD, Sorensen EJ, Cravatt BF. (−)-FR182877 is a potent and selective inhibitor of carboxylesterase-1. Angew Chem Int Edit. 2003;42(44):5480–5484. doi: 10.1002/anie.200352576. [DOI] [PubMed] [Google Scholar]
- 18.Bach S, Knockaert M, Reinhardt J, Lozach O, Schmitt S, Baratte B, Koken M, Coburn SP, Tang L, Jiang T, Liang DC, Galons H, Dierick JF, Pinna LA, Meggio F, Totzke F, Schachtele C, Lerman AS, Carnero A, Wan YQ, Gray N, Meijer L. Roscovitine targets, protein kinases and pyridoxal kinase. J Biol Chem. 2005;280(35):31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 19.MacKinnon AL, Garrison JL, Hegde RS, Taunton J. Photo-leucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J Am Chem Soc. 2007;129(47):14560–14561. doi: 10.1021/ja076250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer T, Maier ME. Design and synthesis of a tag-free chemical probe for Photoaffinity Labeling. Eur J Org Chem. 2007;(28):4711–4720. [Google Scholar]
- 21.Lapinsky DJ. Tandem photoaffinity labeling–bioorthogonal conjugation in medicinal chemistry. Bioorg Med Chem. 2012;20(21):6237–6247. doi: 10.1016/j.bmc.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Fujii T, Manabe Y, Sugimoto T, Ueda M. Detection of 210 kDa receptor protein for a leaf-movement factor by using novel photoaffinity probes. Tetrahedron. 2005;61(33):7874–7893. [Google Scholar]
- 23.Salisbury CM, Cravatt BF. Click chemistry-led advances in high content functional proteomics. Qsar Comb Sci. 2007;26(11–12):1229–1238. [Google Scholar]
- 24.Fleming SA. Chemical reagents in photoaffinity labeling. Tetrahedron. 1995;51(46):12479–12520. [Google Scholar]
- 25.Kobayashi K, Uneda T, Takada K, Tanaka H, Kitamura T, Morikawa O, Konishi H. Efficient synthesis of 1-amino-2-naphthalenecarboxylic acid derivatives via a sequential Michael addition enolate-nitrile coupling route and its application to facile preparation of 9-amino analogues of arylnaphthofuranone lignans. J Org Chem. 1997;62(3):664–668. doi: 10.1021/jo961744r. [DOI] [PubMed] [Google Scholar]
- 26.Carato P, Moussavi Z, Sabaouni A, Lebegue N, Berthelot P, Yous S. Synthesis of 6- and 7-acyl-4H-benzothiazin-3-ones. Tetrahedron. 2006;62(38):9054–9058. [Google Scholar]
- 27.Moriello AS, Balas L, Ligresti A, Cascio MG, Durand T, Morera E, Ortar G, Di Marzo V. Development of the first potential covalent inhibitors of anandamide cellular uptake. J Med Chem. 2006;49(7):2320–2332. doi: 10.1021/jm051226l. [DOI] [PubMed] [Google Scholar]
- 28.Bond MR, Zhang HC, Vu PD, Kohler JJ. Photocrosslinking of glycoconjugates using metabolically incorporated diazirine-containing sugars. Nat Protoc. 2009;4(7):1044–1063. doi: 10.1038/nprot.2009.85. [DOI] [PubMed] [Google Scholar]
- 29.Church RFR, Weiss MJ. Diazirines .2. Synthesis and Properties of Small Functionalized Diazirine Molecules - Some Observations on Reaction of a Diaziridine with Iodine-Iodide Ion System. J Org Chem. 1970;35(8):2465. [Google Scholar]
- 30.Chae JY, Buchwald SL. Palladium-catalyzed regioselective hydrodebromination of dibromoindoles: Application to the enantioselective synthesis of indolodioxane U86192A. J Org Chem. 2004;69(10):3336–3339. doi: 10.1021/jo035819k. [DOI] [PubMed] [Google Scholar]
- 31.Andersen J, Madsen U, Bjorkling F, Liang XF. Rapid synthesis of aryl azides from aryl halides under mild conditions. Synlett. 2005;(14):2209–2213. [Google Scholar]
- 32.Markosyan AI, Kuroyan RA, Dilanyan SV, Aleksanyan MS, Karapetyan AA, Struchkov YT. Synthesis and structure of 2-methyl-6-oxo-7,8-dihydrospiro(benzo[h]triazolo[3,4-b]quinazoline-7,1 ′-cyclopentane) Khim Geterotsikl Soed. 2000;(5):658–662. [Google Scholar]
- 33.Nickon A. New perspectives on carbene rearrangements: migratory aptitudes, bystander assistance, and geminal efficiency. Acc Chem Res. 1993;26(3):84–89. [Google Scholar]
- 34.Kuhn C-S, Lehmann J, Steck J. Syntheses and properties of some photolabile β-thioglycosides. Potential photoaffinity reagents for β-glycoside hydrolases. Tetrahedron. 1990;46(9):3129–3134. [Google Scholar]
- 35.Colca JR, McDonald WG, Waldon DJ, Thomasco LM, Gadwood RC, Lund ET, Cavey GS, Mathews WR, Adams LD, Cecil ET, Pearson JD, Bock JH, Mott JE, Shinabarger DL, Xiong LQ, Mankin AS. Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J Biol Chem. 2003;278(24):21972–21979. doi: 10.1074/jbc.M302109200. [DOI] [PubMed] [Google Scholar]
- 36.Ong SE, Schenone M, Margolin AA, Li XY, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, Koehler AN, Marcaurelle LA, Golub TR, Gould RJ, Schreiber SL, Carr SA. Identifying the proteins to which small-molecule probes and drugs bind in cells. PNAS. 2009;106(12):4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghaemmaghami S, Huh W, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 38.Sche PP, McKenzie KM, White JD, Austin DJ. Display cloning: functional identification of natural product receptors using cDNA-phage display. Chem Biol. 1999;6(10):707–716. doi: 10.1016/s1074-5521(00)80018-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.