Abstract
Sequential pulses of the steroid hormone ecdysone regulate the major developmental transitions in Drosophila, and the duration of each developmental stage is determined by the length of time between ecdysone pulses. Ecdysone regulates biological responses by directly initiating target gene transcription. In turn, these transcriptional responses are known to be self-limiting, with mechanisms in place to ensure regression of hormone-dependent transcription. However, the biological significance of these transcriptional repression mechanisms remains unclear. Here we show that the chromatin remodeling protein ino80 facilitates transcriptional repression of ecdysone-regulated genes during prepupal development. In ino80 mutant animals, inefficient repression of transcriptional responses to the late larval ecdysone pulse delays the onset of the subsequent prepupal ecdysone pulse, resulting in a significantly longer prepupal stage. Conversely, increased expression of ino80 is sufficient to shorten the prepupal stage by increasing the rate of transcriptional repression. Furthermore, we demonstrate that enhancing the rate of regression of the mid-prepupal competence factor βFTZ-F1 is sufficient to determine the timing of head eversion and thus the duration of prepupal development. Although ino80 is conserved from yeast to humans, this study represents the first characterization of a bona fide ino80 mutation in any metazoan, raising the possibility that the functions of ino80 in transcriptional repression and developmental timing are evolutionary conserved.
Keywords: developmental timing, chromatin remodeler, ecdysone, transcriptional repression
Introduction
In all metazoans, the life cycle is divided into discrete stages, including embryonic development, a juvenile growth phase, sexual maturation, and reproductive adulthood. In Drosophila, these stages correspond to embryogenesis, three instars of larval development, metamorphosis with prepupal and pupal stages, and adulthood. The transition between each of these developmental stages is regulated by the steroid hormone 20-hydroxyecdysone (Riddiford, 1993), henceforth referred to as ecdysone. At the onset of each transition, a systemic pulse of ecdysone triggers the appropriate morphological and behavioral changes, causing the animal to progress forward in development toward adulthood. After a developmental transition has occurred, the ecdysone pulse recedes, only to be triggered again when the animal reaches the next developmental transition. Therefore, the duration of each developmental stage is determined by the timing of ecdysone pulses.
At the onset of metamorphosis, two sequential pulses of ecdysone regulate the transformation of a crawling larva into an immature adult. These two pulses outline the boundaries of prepupal development, an intermediate stage during which adult tissues undergo morphogenesis and obsolete larval tissues are destroyed (Robertson, 1936). The first pulse, referred to as the late larval pulse, triggers puparium formation, signaling the transition from larval development to prepupal development. The second pulse, referred to as the prepupal pulse, triggers head eversion, signaling the end of prepupal development and the start of pupal development. The duration of prepupal development is one of the most tightly regulated stages during the fly life cycle, with most animals in a population completing the stage within an hour of one another (Bainbridge and Bownes, 1981).
Ecdysone, like all steroid hormones, acts through its nuclear receptor to directly regulate transcription of target genes. These transcriptional responses not only initiate stage- and tissue-specific biological responses, but also play a critical role in regulating the onset of the subsequent pulse of ecdysone, particularly during prepupal development (Thummel, 1996). Following the late larval ecdysone pulse, the early gene E74A is directly induced by ecdysone (Burtis et al., 1990). The early-late gene DHR3 is also regulated by ecdysone signaling, but additional factors are required for maximal expression, resulting in delayed induction compared to early genes like E74A (Horner et al., 1995; Huet et al., 1995). DHR3 then induces expression of the mid-prepupal competence factor βFTZ-F1 (Lam et al., 1999; 1997; White et al., 1997). In turn, βFTZ-F1 expression is required to initiate the prepupal pulse of ecdysone (Broadus et al., 1999; Woodard et al., 1994), whereupon E74A is induced again. In this manner, the late larval pulse of ecdysone initiates a sequential wave of gene activation that determines the timing of the prepupal ecdysone pulse and the duration of prepupal development.
The sequential transcriptional responses during prepupal development are subject to elaborate mechanisms that ensure timely repression of hormone-dependent gene expression. Many of the protein products of these ecdysone-induced genes repress their own transcription. For example, at the prepupal pulse of ecdysone, E74A protein binds the E74A genomic locus and inhibits ecdysone-dependent transcription, resulting in a sharp and self-limiting peak of E74A mRNA expression (Ihry et al., 2012; Urness and Thummel, 1990). βFTZ-F1 is thought to have a similar auto-inhibitory property (Woodard et al., 1994). In addition to auto-inhibitory regulation, ecdysone-regulated genes exhibit cross-inhibitory regulation. For example, DHR3 protein represses E74A mRNA transcription after the late larval ecdysone pulse (Lam et al., 1997), ensuring that E74A levels decrease as DHR3 levels increase. Together, these transcriptional repression mechanisms help generate the sharp peaks and proper sequence of transcription in response to ecdysone pulses. However, despite ample evidence for mechanisms regulating repression of ecdysone-induced transcription, the biological significance of these mechanisms remains unclear.
The INO80 complex is one of the most highly conserved chromatin remodelers (Clapier and Cairns, 2009). The INO80 protein contains two functional domains: an N-terminal helicase-SANT-associated/post-HSA (HSA/PTH) domain and a Snf2 ATPase domain, which is split by a spacer region into N-terminal and C-terminal regions (Watanabe and Peterson, 2010). Each of these domains is required for binding to specific proteins that together comprise the INO80 chromatin remodeling complex, the composition of which is highly conserved from yeast to humans (R. C. Conaway and J. W. Conaway, 2009). INO80, together with its protein partners, facilitates ATP-dependent nucleosome sliding (Jin et al., 2005; Shen et al., 2000). Studies in yeast reveal that ino80 is required for proper transcriptional regulation of many target genes (Jónsson et al., 2004; Shimada et al., 2008), and in vitro biochemical studies in yeast reveal that ino80 tends to relocate nucleosomes from the edges of DNA fragments toward the center (Shen et al., 2003). Recent biochemical studies in a Drosophila cell culture system reveal that ino80 primarily functions by increasing nucleosome density at its target loci (Moshkin et al., 2012), likely helping to establish repressive chromatin signatures and inhibit transcription of target genes. Despite a high level of evolutionary conservation, studies of INO80 function have been largely limited to biochemical analysis, as mutant alleles have only been characterized in yeast and Arabidopsis (Fritsch et al., 2004; Shen et al., 2000).
Here, we report the identification and characterization of the first metazoan mutant of ino80. We demonstrate that ino80 function is essential for development, with mutant animals arresting during metamorphosis. This lethality can be rescued by ectopic expression of ino80 from a transgenic construct. By focusing on prepupal development, we show that mutation of ino80 results in defects in the timely repression of ecdysone-induced transcription. Importantly, the biological consequence of these repression defects is an extended duration of prepupal development. Moreover, increased expression of ino80 reduces the duration of prepupal development, suggesting that ino80-dependent regression is critical for developmental timing. We also show that the rate of regression of the mid-prepupal competence factor βFTZ-F1 is critical to determine the timing of head eversion and the duration of prepupal development. These results provide the first characterization of ino80 function in a metazoan organism, and also provide novel insights linking transcriptional repression mechanisms with regulation of developmental timing.
Materials And Methods
Stocks and recombination mapping
The following stocks were obtained from the Bloomington Drosophila Stock Center: Df(3R)ED5942, Df(3R)DI-BX12, Df(3R)BSC475, P{EPgy2}Ino80EY09251, Δ2-3 transposase, tubulin-Gal4, UAS-Dcr-2, Sgs3-GFP. ino80 RNAi KK102202 was obtained from the Vienna Drosophila RNAi Center. The ino80psg25 mutation was generated in an EMS mutagenesis screen (Wang et al., 2008). C. Thummel (University of Utah, Salt Lake City, UT) kindly provided the hs-E74A and hs-βFTZ-F1 stocks. The dominant marker recombination mapping method used to identify ino80psg25 is described elsewhere (Sapiro et al., 2013). Experiments with hemizygous ino80psg25 mutant animals used the Df(3R)ED5942 chromosomal deficiency.
Developmental staging and lethal phase analysis
Animals were raised on standard cornmeal molasses medium in the presence of yeast in an environmental chamber set to 25°C. For developmental staging during metamorphosis, animals were synchronized either at puparium formation (white prepupa) or pupation (head eversion), placed on grape agar plates, and aged for the appropriate time at 25°C. Staging prior to head eversion was based on the average duration of prepupal development (puparium formation to head eversion) of each respective genotype. Standard “blue food” technique was used to stage late third instar larvae (Andres and Thummel, 1994). Additional developmental stages prior to metamorphosis were aged at 25°C from egg lay. For heat-shock treatments, animals at the appropriate stage were placed on grape agar plates sealed with Parafilm and submerged in a 37.5°C water bath for 30 minutes. Bainbridge and Bownes staging criteria was used to assign lethal phases: P1-P3 for prepupae (PP), P4-P9 for pupae (P), P10-15 for pharate adults (PA), and eclosed adults (A) (Bainbridge and Bownes, 1981).
Developmental timing analysis
For head eversion timing, white prepupae were placed on grape agar plates and allowed to age for an appropriate time at 25°C. Aging on agar plates allowed for proper control of humidity. Animals were then scored for completed head eversion in 15 minute intervals. For eclosion timing, adult flies of the appropriate genotype were allowed to mate for two days, then transferred to fresh vials each day for three consecutive days. To determine the timing of eclosion, the vials were examined for empty pupal casings every 12 hours until all animals eclosed.
Generation of the ino80 transgene
The ino80 sequence was amplified from cDNA clone SD02886 (BDGP) with the following primers: 5′ GAAATTAATACGACTCACTATAGGGAGACCGG 3′ and 5′ CGTCGACGTTAGAATTCGGCTACAAT 3′. The resulting amplicon was digested with EcoRI, ligated into pCASPER-hs, and sequence-verified. Transgenic flies were generated via standard protocols (Genetic Services, Inc.).
Imprecise Excision
To generate excision events within the ino80 locus, virgin females containing the viable P{EPgy2}Ino80EY09251 P-element were crossed to males with the Δ2-3 transposase. Progeny that lost the mini-white (w+mC) marker on the P-element were selected, indicating that an excision event occurred, and stable stocks were generated from these animals. To identify imprecise excision events, each stock was screened for lethality. Complementation tests were carried out on the lethal excisions with Df(3R)ED5942 and ino80psg25 to identify potential ino80 alleles. Standard PCR on a genomic DNA template was used to identify the deleted region in ino80ex64.
A previously published report characterizes 2 excision alleles generated from mobilization of the same P-element we used (P{EPgy2}Ino80EY09251), resulting in a 3 or 4 kilobase deletion, respectively, in the vicinity of the P-element (Bhatia et al., 2010). These alleles, unlike our ino80 alleles, have a lethal phase early in development. However, these reported excision alleles were not tested for complementation with the ino80-containing chromosomal deficiency. Moreover, the specific location of the deletions was not directly characterized. Given that the P-element resides within a ∼10 kilobase intron, it is unclear if these excision events disrupt ino80 coding sequences. Importantly, in our excision scheme, most (27 of 28) of the excision events recovered were generated from a single locus residing outside the ino80-containing chromosomal deficiency. Each of the 27 lethal excision events of this complementation group has a lethal phase early in development. Thus, our results suggest that the original stock may have a second P-element, and the majority of lethal excision events affect that currently unidentified locus.
Quantitative PCR
qPCR was performed as previously described (Ihry et al., 2012). RNA was isolated from appropriately-staged whole animals or dissected salivary glands using the RNeasy Plus Mini Kit (Qiagen). cDNA was synthesized from 100-400 ng of total RNA using the SuperScript III First-Strand Synthesis System with oligo(dT)20 primers (Invitrogen). qPCR was performed using LightCycler 480 SYBR Green I Master Mix (Roche) on a Roche 480 LightCycler. For each experiment, target genes of interest and the reference gene rp49 were run simultaneously on three independently-isolated control and experimental biological samples. 3-4 serial dilutions of pooled cDNA derived from samples spanning development were used to determine the amplification efficiency of the primers in each experiment. Roche LightCycler 480 Software (Version 1.5) was used to calculate cycle threshold values and melting curves for each reaction. Relative expression and standard error were calculated using Relative Expression Software Tool (REST) (Pfaffl et al., 2002). Standard error calculated by REST reflects asymmetrical tendencies in the data. Primer sequences are listed in Table S1. Primers were designed with A plasmid Editor (ApE), GenScript Real-time PCR Primer Design tool, or QuantPrime (Arvidsson et al., 2008).
Immunofluorescence and Image Capture
Salivary glands from appropriately-staged animals were dissected, fixed, and immunostained using standard techniques (Yin et al., 2007). Primary antibodies used were rabbit α-cleaved caspase-3 (1:200; Cell Signaling) to detect activity of the caspase Nc (Fan and Bergmann, 2010) and mouse α-Lamin (1:50; Developmental Studies Hybridoma Bank) to detect breakdown of a target of caspases. Secondary antibodies used were a-rabbit Cy3 (1:200; Jackson Immuno-Research Labs) and α-mouse AlexaFluor 488 (1:200; Invitrogen). DAPI was used to counterstain DNA (1:1,000; Invitrogen). Stained tissues were mounted on slides in Vectashield (Vector Laboratories). Immunofluorescence images were captured using an Olympus FLUOVIEW FV1000 confocal microscope with FV10-ASW software. Persistent salivary gland and lethal phase images were captured using an Olympus SZX16 stereomicroscope coupled to an Olympus DP72 camera with Olympus DPX2-BSW software.
Western Blotting
Western blots were performed using standard methods as previously described (Ihry et al., 2012). 10 whole animals at the appropriate stage were homogenized in 75 μl of hi-salt lysis buffer, and total protein concentration was calculated using the Bradford assay. Transferred membranes were blocked with 5% BSA in PBST and then incubated with properly diluted primary antibodies overnight at 4°C. Primary antibodies used were mouse α-E74A (1:2,000; a gift from C. Thummel) and rabbit α-β-actin (1:1,000; Cell Signaling Technologies). Secondary antibodies used were α-rabbit IgG alkaline phosphatase-linked antibody (1:30,000; GE Healthcare) and α-mouse IgG alkaline phosphatase-linked antibody (1:30,000; Sigma). Membranes were developed for imaging with ECF substrate (GE Healthcare) and were imaged using a Storm 840 Scanner (Amersham Bioscience) with ImageQuant TL software version 7.0 (GE Healthcare).
Results
Identification of the first metazoan ino80 mutant
To gain new insights into the mechanisms regulating ecdysone signaling during development, we previously conducted an EMS-based mutagenesis screen for mutations that disrupt the ecdysone-triggered destruction of the larval salivary glands during metamorphosis (Wang et al., 2008). Here, we characterize one of the mutations identified in this screen: psg25. Recombination mapping with pairs of dominant markers, followed by complementation tests with chromosomal deficiencies, was used to identify the region containing the psg25 mutation (Sapiro et al., 2013). Additional complementation tests with deficiencies mapped psg25 to a small region containing 20 genes (Fig. S1). psg25 complemented all available lethals within the region; however, Sanger sequencing of psg25 revealed a point mutation disrupting a donor splice site in the chromatin remodeler, ino80. Sanger sequencing of psg25 cDNA confirmed that disruption of the donor splice site after the fourth exon results in the retention of a 26 base pair intron fragment, causing a frameshift leading to a nonsense mutation (Fig. 1A). When translated, psg25 mRNA would produce a truncated INO80 protein, with loss of the C-terminal region of the Snf2 ATPase domain (Fig. 1B).
Figure 1. Identification of a novel allele of ino80.
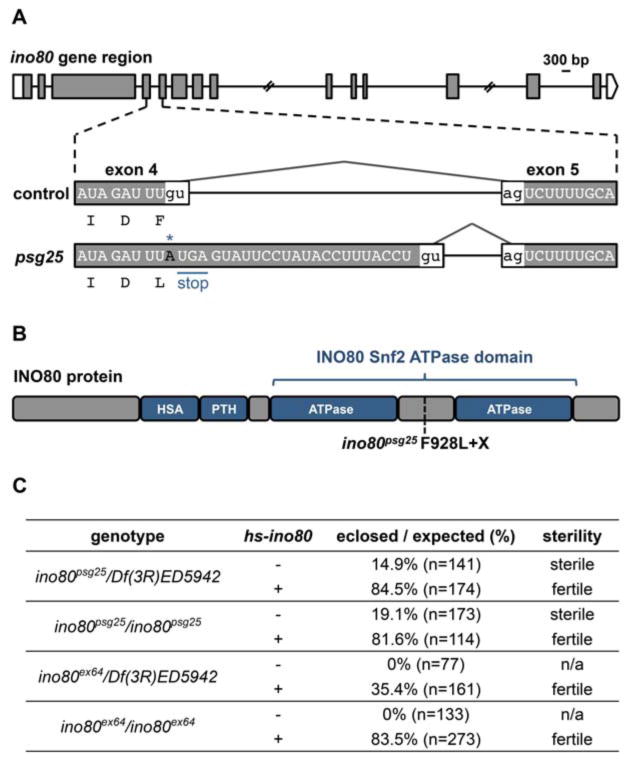
(A) Schematic describing the psg25 lesion in the ino80 locus. psg25 has a point mutation (G to A) that disrupts the donor splice site after the fourth exon, resulting in retention of an intron fragment and an in-frame stop codon immediately following the mutated donor splice site. Retention of the intron also changes a phenylalanine immediately before the donor splice site to a leucine. Boxes represent exons and shaded regions highlight coding sequences. Diagram to scale, except for two large introns, marked with slashes. Scale bar=300 base pairs (bp). (B) Schematic of the domains of the INO80 protein and the location of the psg25 nonsense mutation, indicated by the dotted line. (C) Leaky expression of ino80 from a hs-ino80 transgene rescues ino80psg25 and ino80ex64(+) or absence (−) of one copy of the hs-ino80 transgene. Presence of the hs-ino80 transgene also rescues the fertility of eclosing animals. Eclosion percentages expressed as observed eclosion percentage divided by expected eclosion percentage (33%).
We also generated an independent allele of ino80 via imprecise excision. We mobilized the P{EPgy2}Ino80EY09251 P-element, located in the 12th intron of the ino80 locus. After mobilization of the P-element, 163 flies with excision events were identified by loss of the mini-white marker (w+mC) on the transposable element; of these, 28 generated a lethal mutation on the excision chromosome (Fig. S2A). Of the 28 lethal mutations, only one failed to complement the ino80-containing chromosomal deficiency and resulted in sterile progeny when crossed to psg25. This allele, designated as ino80ex64, contains a large deletion to the right of the P-element (∼20 kilobases), excising most of the Snf2 ATPase domain and leaving a short stretch of P-element sequence within the ino80 locus (Fig. S2B). The excision event also deletes several other genes located within the introns of ino80. The remaining 27 lethal excisions comprise a single complementation group that has a lethal phase early in development, unlike ino80ex64 and ino80psg25, which arrest during metamorphosis (Fig. 2B, S2C). Additionally, the 27 early lethal mutations complement ino80psg25, ino80ex64, and the chromosomal deficiency that deletes the entire ino80 locus.
Figure 2. ino80is required for viability during metamorphosis.
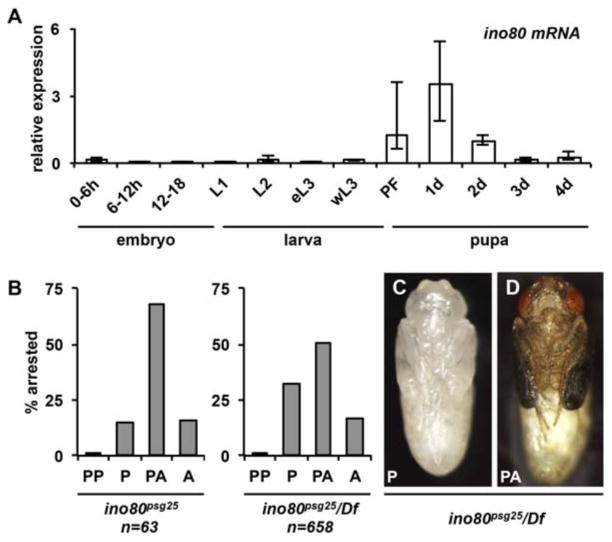
(A) qPCR analysis of ino80 mRNA expression throughout development. The highest expression levels are observed at the onset of metamorphosis, from puparium formation to 2 days after puparium formation (APF). Animals staged from egg lay for embryonic (0-6, 6-12 and 12-18 hours after egg lay (AEL)) and larval stages (L1: 30-42 AEL, L2: 54-66 AEL, eL3: 76-88 AEL). wL3 identified by robust expression of Sgs3-GFP. Stages during metamorphosis were synchronized at puparium formation (PF) and collected in 24-hour intervals. y-axis plots relative expression compared to 2 days APF; x-axis denotes the developmental stage analyzed. Three independently-isolated whole animal samples were run for each time point and target genes were normalized to rp49. (B) Lethal phase analysis of homozygous and hemizygous ino80psg25 mutant animals. Most homozygous and hemizygous ino80psg25 mutant animals die after head eversion, as either pupae or pharate adults. A smaller fraction eclose as adults. (C-D) Hemizygous ino80psg25 mutant animals that arrest as pupae (C) or pharate adults (D) have normal morphology, with head eversion and extension of wings and legs. Animals were dissected out of their pupal cases and imaged at their respective terminal stages. PF=puparium formation, PP=prepupa, P=pupa, PA=pharate adult, A=eclosed adult.
To confirm the gene identification of the ino80psg25 and ino80ex64 alleles, we conducted a rescue experiment. We generated a transgenic construct containing ino80 cDNA under control of the heat-shock promoter (hs-ino80), and placed this construct in the ino80psg25 mutant background. ino80psg25 hemizygous (ino80psg25/Df) and homozygous animals exhibit semi-lethality, with about 15% of the expected flies eclosing as viable, but sterile, adults (Fig. 1C). However, ubiquitous, leaky expression of ino80 from the hs-ino80 construct is able to rescue both the lethality and sterility observed in ino80psg25 hemizygous and homozygous animals. Additionally, hs-ino80 also rescues the lethality observed in hemizygous and homozygous ino80ex64, suggesting that the other genes disrupted by the excision event in ino80ex64 are not essential for viability. These results, together with the identical lethal phases of homozygous and hemizygous mutant animals (Fig. 2B, S2C), suggest that ino80psg25 and ino80ex64 are loss-of-function alleles of ino80. However, because ino80ex64 disrupts additional genes, we chose to continue our analysis of INO80 function with the EMS-induced ino80psg25 allele.
ino80 is primarily required at the onset of metamorphosis
To gain insights into the function of ino80, we first used quantitative reverse transcription real-time PCR (qPCR) to analyze the developmental profile of ino80 mRNA expression. RNA was isolated from whole animals spanning embryogenesis through metamorphosis, and qPCR was used to quantify ino80 mRNA levels in each developmental stage. ino80 mRNA is expressed at low levels throughout development; however, the highest levels are observed during metamorphosis, with a peak of expression from puparium formation to two days after puparium formation (Fig. 2A).
We next determined when ino80psg25 mutants arrest during development. Interestingly, all hemizygous mutant animals survive to enter metamorphosis, as the predicted percentage of expected progeny (one-third) is observed on the sides of vials when compared to balancer-containing siblings (33.4%, n=692). However, only a small percentage of these animals survive to reach adulthood. Most homozygous and hemizygous ino80psg25 mutant animals arrest during pupal or pharate adult stages (Fig. 2B), and the few that do eclose are sterile (Fig. 1C). A similar lethal phase is observed in hemizygous ino80ex64 mutant animals and in animals ubiquitously expressing ino80-RNAi (Fig. S2C). Although ino80psg25 mutant animals arrest during metamorphosis, they form normal pupae or pharate adults without visible abnormalities, demonstrating that morphogenetic processes are not disrupted by mutation of ino80 (Fig. 2C,D). Overall, the expression profile and mutant lethal phase suggest that ino80 plays a critical role during metamorphosis.
Initiation of the ecdysone-dependent death response is delayed in ino80psg25 mutant salivary glands
To gain insights into the role of ino80 during metamorphosis, we first analyzed the function of this gene in the ecdysone-dependent destruction of the larval salivary glands. ino80psg25 mutant animals (ino80psg25/Df(3R)ED5942, unless otherwise indicated) exhibit a highly penetrant persistent salivary gland (PSG) phenotype (85.2%, n=54) at 24 h after puparium formation (APF) (Fig. S3A, B). The PSG phenotype is also observed in ino80ex64 hemizygous animals (83.7% PSG, n=37) (Fig. S3C). Importantly, other ecdysone-dependent processes, such as Sgs3 glue protein synthesis and secretion, occur normally (Fig. S3D-H), indicating that ino80psg25 mutant salivary glands have the ability to respond to ecdysone signals. To understand how ino80psg25 disrupts salivary gland destruction, we first analyzed induction of the death activators reaper (rpr) and hid involution defective (hid), which represents a point-of-no-return during the death response (Grether et al., 1995; White et al., 1996). We dissected salivary glands from control and ino80psg25 mutant animals staged relative to head eversion and analyzed their gene expression profiles using qPCR. As expected, control animals exhibit strong induction of both rpr and hid by 1.5 h after head eversion (AHE)(black line, Fig. 3A). In contrast, ino80psg25 salivary glands do not have maximal induction of rpr and hid until 3 AHE (blue line, Fig. 3A). Consistent with the observed delay in induction of rpr and hid, ino80psg25 salivary glands show a delay in caspase activation. At 1.5 AHE, control salivary glands have bright staining for cleaved-caspase-3 and dim staining for nuclear lamin, a substrate of caspases (Fig. 3B). In contrast, ino80psg25 mutant salivary glands at the same stage do not have staining for caspase activity and have bright staining for nuclear lamin (Fig. 3C). However, at 3 AHE, ino80psg25 mutant salivary glands exhibit caspase activity similar to that seen in control glands at 1.5 AHE (Fig. 3D). Overall, these results suggest that ino80psg25 delays, rather than disrupts, the ecdysone-triggered death response.
Figure 3. Destruction of the larval salivary glands is delayed in ino80psg25 mutant animals.
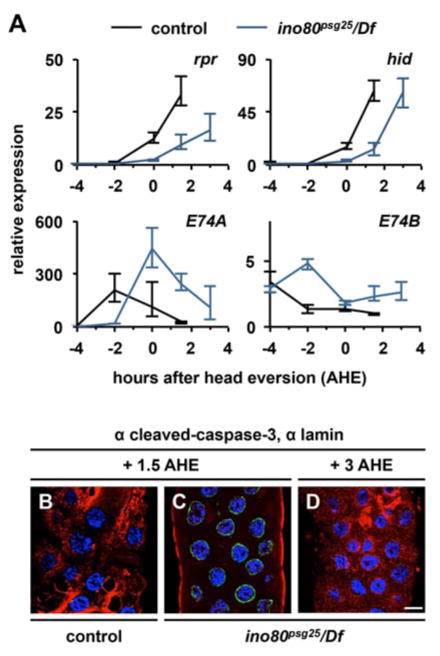
(A) qPCR analysis of target gene expression in control (black lines) and ino80psg25 (blue lines) mutant salivary glands staged relative to head eversion. Control glands have strong induction of rpr and hid by 1.5 h after head eversion (AHE), while ino80psg25 glands do not have maximal induction of rpr and hid until 3 AHE. Induction and regression of E74A and E74B is also delayed in ino80psg25 mutant salivary glands. y-axis represents relative expression compared to the lowest point in control animals; x-axis represents developmental stage relative to head eversion. Three independently-isolated samples were run for each timepoint; relative expression calculated by normalizing to rp49. (B-D) Activation of caspases detected by staining for cleaved-caspase-3 (red) and nuclear lamin (green) in control and ino80psg25 mutant salivary glands. DNA labeled with DAPI shown in blue. Control glands have ubiquitous caspase activation and strong loss of nuclear lamin staining by 1.5 AHE (B). In contrast, ino80psg25 glands have caspase activation similar to controls by 3AHE (D), but not at 1.5 AHE (C). Scale bar is 20μm. AHE= after head eversion.
Because the death response in salivary glands is triggered by the prepupal pulse of ecdysone (Jiang et al., 1997; 2000), we next used qPCR to measure expression of ecdysone response genes in ino80psg25 mutant salivary glands. The prepupal ecdysone pulse occurs two hours prior to head eversion, and, as expected, control glands exhibit strong induction of E74A, an early gene that is directly induced by ecdysone, at this timepoint (black line, Fig. 3A). However, at the same stage in ino80psg25 mutant salivary glands, E74A has yet to be induced (blue line, Fig. 3A), suggesting a delayed onset of the prepupal ecdysone pulse. Consistent with these results, expression of E74B, which is repressed by high ecdysone titers, shows a corresponding delayed repression in ino80psg25 mutant salivary glands (Fig. 3A). Taken together, these results suggest that the timing of the prepupal ecdysone pulse is likely delayed in ino80psg25 mutant animals.
Regression of transcriptional responses is delayed during ino80psg25 prepupal development
To determine why the prepupal pulse of ecdysone is delayed in ino80psg25 mutant animals, we analyzed the sequence of expression of ecdysone-regulated genes during prepupal development. We collected control and ino80psg25 mutant whole animals staged relative to puparium formation and analyzed the expression profiles of ecdysone-regulated genes using qPCR. The samples collected span two pulses of ecdysone: the late larval pulse, which triggers puparium formation, and the prepupal pulse, which triggers head eversion and pupation. In ino80psg25 mutant animals, induction of ecdysone-target genes in response to the late larval pulse occurs normally (Fig. 4A). The early gene E74A is induced to maximal levels at -4 APF, followed by induction of the early-late gene DHR3, which peaks at 2 APF. βFTZ-F1 also reaches maximal expression levels on time, with a transcriptional peak at 8 APF. All of these genes are induced at the proper time, intensity, and sequence following the late larval pulse of ecdysone in ino80psg25 mutant animals, indicating that induction of target genes is unaffected by mutation of ino80.
Figure 4. ino80 is required for efficient regression of ecdysone-regulated genes during prepupal development.
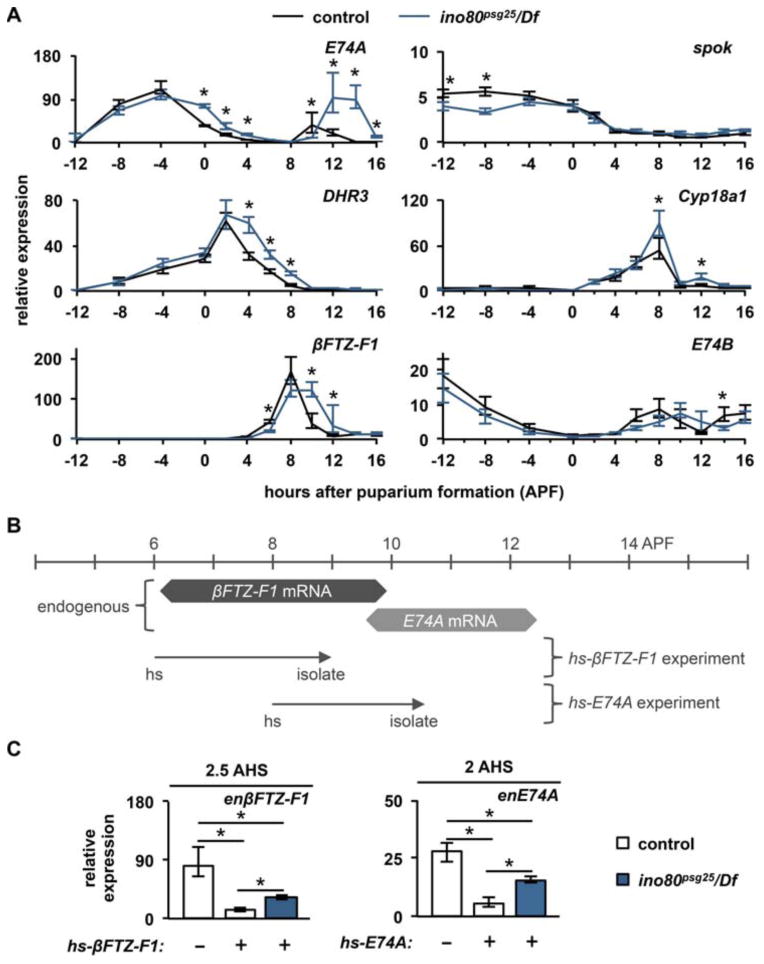
(A) qPCR analysis of target genes during prepupal development in control (black lines) and ino80psg25 (blue lines) mutant whole animals. The ecdysone-regulated genes E74A, DHR3, and βFTZ-F1 are induced in the proper sequence in ino80psg25 mutant animals following the late larval ecdysone pulse; however, ino80psg25 mutant animals exhibit significant delays in repression of all three of these genes. In contrast, the ecdysone biosynthetic gene spok, the ecdysone turnover gene Cyp18a1, and the ecdysone concentration-dependent target E74B are unaffected in ino80psg25 mutant animals. y-axis represents relative expression compared to the lowest point in control animals; x-axis represents developmental stage relative to puparium formation. Three independently-isolated whole animal samples were run for each timepoint and normalized to rp49. (B) Experimental paradigm to test efficiency of auto-inhibition by ectopic expression of the βFTZ-F1 and E74A proteins. Control and ino80psg25 animals were heat-shocked two hours prior to the endogenous peak of βFTZ-F1 (dark gray) or E74A (light gray) expression in each respective genotype, then allowed to recover for about 2 hours before isolation of total RNA. Timescale shows hours after puparium formation (APF). Arrows indicate time of heat-shock and isolation. (C) Auto-inhibition of βFTZ-F1 and E74A occurs inefficiently in ino80psg25 mutant animals. On left, expression of βFTZ-F1 protein from the hs-βFTZ-F1 transgene is sufficient to repress endogenous βFTZ-F1 transcription in control animals (white bars). The same treatment in ino80psg25 mutant animals results in incomplete repression of endogenous βFTZ-F1 transcription (blue bar). On right, similar results are obtained in a comparable experiment with ectopic expression of E74A. y-axis represents relative expression compared to the lowest point in each developmental profile; x-axis denotes heat-shock treatment. Asterisks denote significant differences between control and mutant samples (p<0.05).
In contrast, regression of these ecdysone-regulated genes does not occur in a timely manner following the late larval pulse of ecdysone in ino80psg25 mutant animals (Fig. 4A). In control animals, E74A transcription is shut off by 4 APF, while E74A expression is maintained until 6 APF in ino80psg25 mutant animals. Similarly, DHR3 and βFTZ-F1 expression persists about two hours longer in ino80psg25 compared to controls. Therefore, transcriptional responses to the late larval pulse of ecdysone are induced on time but regress two hours late in ino80psg25 mutant animals. These regression defects appear to affect only a subset of genes during prepupal development. For example, the ecdysone biosynthetic genes phantom (phm), spookier (spok), shadow (sad), and disembodied (dib) are properly induced prior to the late larval pulse of ecdysone, and are properly repressed at the end of the pulse in ino80psg25, although sad exhibits a subtle delay in repression (Fig. 4A, S4A). Additionally, Cyp18a1, a cytochrome P450 responsible for degradation of ecdysone, is also properly induced and repressed in ino80psg25 mutant animals (Fig. 4A).
In addition, ino80psg25 mutant animals also exhibit a delay in the onset of the responses to the prepupal ecdysone pulse. E74A expression is induced at high ecdysone titers and reflects the peak of the prepupal ecdysone pulse. In control animals, E74A is maximally induced by the prepupal ecdysone pulse at 10 APF, while E74A is not induced until 12 APF in ino80psg25 mutant animals (Fig. 4A), two hours later than controls. Additionally, E74B, which is induced at low ecdysone titers and repressed at high ecdysone titers, shows a normal expression profile in ino80psg25 mutant animals after the late larval pulse of ecdysone, but exhibits delayed induction during prepupal development (Fig. 4A), suggesting that ino80psg25 does not affect the titer of the prepupal ecdysone pulse, only when it is induced. Our data thus far suggests that delayed repression of ecdysone-induced transcription after the late larval ecdysone pulse results in delayed onset of the prepupal ecdysone pulse.
Auto-inhibition of ecdysone-induced genes is inefficient in ino80psg25 mutant animals
To understand the nature of the regression defects, we examined the efficiency of auto-inhibitory mechanisms in ino80psg25 mutant animals. Although the hyper-induction and prolonged expression of E74A mRNA observed in ino80psg25 is also observed in mutants that disrupt E74A protein translation (Ihry et al., 2012), western blots with an E74A antibody show that both control and ino80psg25 mutant animals have robust expression of E74A protein (Fig. S4B). We then tested the possibility that ino80 is required to facilitate the auto-inhibitory activity of E74A protein. Precocious expression of E74A protein from a heterologous transgenic construct (hs-E74A) results in strong repression of the hormone-dependent transcription of endogenous E74A after the prepupal ecdysone pulse (Fig. 4B,C). In contrast, precocious expression of E74A protein in ino80psg25 mutant animals results in incomplete repression of ecdysone-dependent transcription of E74A. Similarly, precocious expression of βFTZ-F1 protein results in incomplete repression of endogenous βFTZ-F1 transcription in ino80psg25 mutant animals compared to control animals (Fig. 4B,C). Taken together, these results show that ino80 is required for the efficient auto-inhibitory activity of the E74A and βFTZ-F1 proteins, suggesting that the transcriptional regression defects during prepupal development are due to defects in auto-inhibition.
Loss of ino80 extends the duration of prepupal development
Our data thus far indicates that regression defects during ino80psg25 prepupal development result in delayed induction of the prepupal ecdysone pulse. Given that the prepupal ecdysone pulse triggers head eversion, we tested if the timing of head eversion was affected in ino80psg25 mutant animals. Control and ino80psg25 mutant animals were synchronized at puparium formation, then allowed to age until head eversion. Control animals begin to head evert at about 10.75 APF, and all animals complete head eversion by 12.5 APF, with about 50% of animals head everting by 11.5 APF (HE50∼11.5 APF, n=156)(black line, Fig. 5A). Control animals exhibit little variance in the duration of prepupal development, with nearly 90% of animals completing head eversion within a one-hour interval. The synchrony of this process is evident in the steep slope of the head eversion timing curve. ino80psg25 mutant animals do not begin head everting until about 12.5 APF, and all animals complete head eversion by 14.25 APF (HE50∼13.25 APF, n=152)(blue line, Fig. 5A), demonstrating that ino80psg25 animals head evert nearly two hours later than controls. A similar head eversion delay is observed in ino80ex64 (HE50∼13.5 APF, n=11). Despite a significant delay in the timing of head eversion, the ino80psg25 timing curve still exhibits a steep upward slope, suggesting that ino80psg25 does not disrupt the mechanisms that execute head eversion. The two hour delay in head eversion, coupled with the two hour delay in the onset of the prepupal ecdysone pulse shown above, suggest that the prolonged prepupal stage is likely a consequence of the regression defects in ino80psg25 mutant animals.
Figure 5. ino80 regulates the duration of development.
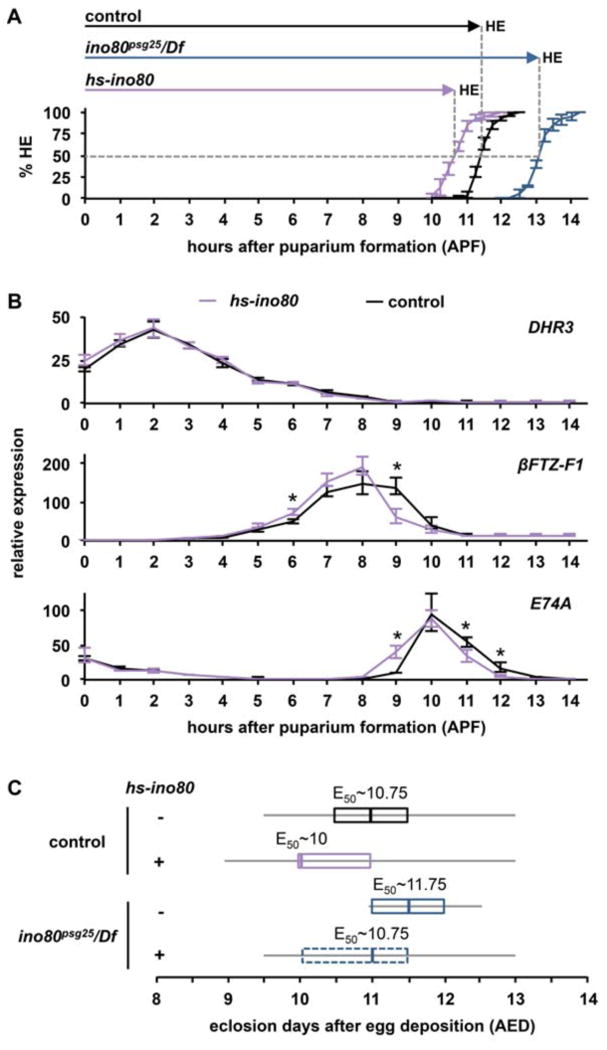
(A) Head eversion timing in control (black line), ino80psg25 (blue line), and hs-ino80 (purple line) animals. 50% of control animals head evert by 11.5 APF (HE50∼11.5, total n=156). ino80psg25 mutant animals have delayed head eversion, with 50% of the animals head everting by 13.25 APF (HE50∼13.25, total n=152). hs-ino80 accelerates head eversion, with 50% of animals head everting by 10.75 APF (HE50∼10.75, total n=151). y-axis plots the percentage of animals completing head eversion; x-axis plots time in hours relative to puparium formation (APF). Three independent samples of n∼50 animals were analyzed for each genotype; error bars represent plus or minus one standard deviation. (B) qPCR analysis of ecdysone-regulated transcription in control (black line) and hs-ino80 (purple line) whole animals staged relative to puparium formation. The DHR3 expression profile is nearly identical in control and hs-ino80 animals. In contrast, βFTZ-F1 regresses significantly faster in hs-ino80 animals compared to controls. E74A also displays a significant acceleration in onset and repression in hs-ino80. y-axis plots relative expression compared to the lowest point in controls; x-axis represents stages relative to puparium formation. Three independently-isolated whole animal samples were run for each timepoint and normalized to rp49. Asterisks denote significant differences between control and mutant samples (p<0.05). (C) Box plot of eclosion time in control and ino80psg25 mutant animals with or without the hs-ino80 transgene. Control animals (black box) begin to eclose at 9.5 days AED, and 50% eclose by 10.75 AED (n=624). Few ino80psg25 animals eclose (blue box), but those that do begin to eclose at 11 days AED, and 50% eclose by 11.75 AED (n=19). Increased expression of ino80 from the hs-ino80 transgene (purple box) accelerates eclosion timing, with animals beginning to eclose before 9 days AED (E50∼10 AED, n=906). Leaky expression of ino80 from the hs-ino80 transgene (dotted blue box) is sufficient to rescue the eclosion delay observed in ino80psg25 mutant animals, as hs-ino80/+; ino80psg25/Df animals begin to eclose at 9.5 days AED (E50∼10.75 AED, n=224). For each genotype, the gray horizontal lines (whiskers) represent the range for all animals analyzed, the boxes outline the middle two quartiles, and vertical lines within the boxes denote the median. x-axis represents time to eclosion in days after egg deposition (AED); y-axis shows each respective genotype plus (+) or minus (−) the hs-ino80 transgene. The time when 50% of animals eclose (E50) is listed for each respective genotype.
Increased levels of ino80 shorten the duration of prepupal development through acceleration of transcriptional regression
To demonstrate a causal relationship between INO80-dependent transcriptional repression mechanisms and the duration of prepupal development, we first tested whether increased expression of ino80 could accelerate transcriptional repression and/or developmental timing. Strikingly, increased expression of ino80 does both. We collected control and hs-ino80 animals staged relative to puparium formation and used qPCR for gene expression analysis, using a one-hour sampling rate during prepupal development in order to detect smaller shifts in gene expression. Leaky expression of ino80 from the heat-inducible transgenic construct (hs-ino80) results in about a 20-fold increase in ino80 mRNA levels at 25°C. In hs-ino80 animals, transcription of E74A and DHR3 in response to the late larval pulse appears to regress normally (Fig. 5B). However, βFTZ-F1 regresses faster in hs-ino80 animals, with significantly lower levels at 9 APF compared to controls. This accelerated regression coincides with precocious induction of E74A at the prepupal ecdysone pulse, one hour earlier than in controls. These results indicate that increased expression of ino80 is sufficient to accelerate regression of βFTZ-F1. I importantly, hs-ino80 animals head evert faster than control animals. Animals overexpressing ino80 have an HE50 of ∼10.75 APF (n=151, purple line, Fig. 5A), about 45 minutes faster than control animals. Similar results were obtained with an independent insertion of the hs-ino80 transgene (HE50 ∼10.75 APF, n=85). These effects on developmental timing were not limited to prepupal development. Control animals begin to eclose at 9.5 days after egg deposition (AED), and over 50% eclose by 11 days AED (E50∼10.75 AED, n=624) (black box, Fig. 5C). In contrast, increased expression of ino80 results in nearly a one day acceleration of overall development (E50∼10 AED, n=906) (purple box, Fig. 5C). Although only a few ino80psg25 animals eclose, those that do survive to adulthood eclose about one day later than controls (E50∼11.75 AED, n=19) (solid blue box, Fig. 5C), demonstrating that ino80psg25 mutants exhibit developmental delays throughout the entire life cycle. Importantly, this delay in ino80psg25 is fully rescued by leaky expression of ino80 from the hs-ino80 transgene (hs-ino80/+; ino80psg25/Df, E50∼10.75 AED, n=224) (dashed blue box, Fig. 5C).
Accelerated transcriptional repression of βFTZ-F1 is sufficient to shorten the duration of prepupal development
To confirm a causal relationship between transcriptional repression mechanisms and the duration of prepupal development, we focused on independently modifying the rate of regression of βFTZ-F1. To do this, we exploited the auto-inhibition assay described earlier (Fig. 4B). When βFTZ-F1 protein is expressed from a heterologous promoter prior to initiation of endogenous βFTZ-F1 transcription, the endogenous transcriptional response is ablated (Woodard et al., 1994). However, we show that when βFTZ-F1 protein is expressed immediately after the start of endogenous transcription, it does not block induction of βFTZ-F1; instead it appears to accelerate regression of βFTZ-F1 transcription. Animals were heat-shocked at 6 APF, at the start of endogenous βFTZ-F1 expression, then collected at one-hour intervals following the heat-shock. In this assay, the endogenous transcriptional response does occur, as βFTZ-F1 transcript levels are equal in both control and hs-βFTZ-F1-treated animals at 0.5 hrs after heat-shock (AHS) (Fig. 6A). However, in hs-βFTZ-F1-treated animals, endogenous βFTZ-F1 transcription regresses significantly faster, with transcription shut off by 3.5 AHS, while endogenous βFTZ-F1 transcription is still detectable at 4.5 AHS in control animals (cf. solid and dashed black lines, Fig. 6A). These results demonstrate that increased expression of βFTZ-F1 protein, delivered at the proper time, is sufficient to accelerate the rate of βFTZ-F1 transcriptional regression. Importantly, this faster regression results in a dramatically shorter prepupal stage (HE50 ∼10.75 APF, n=101), accelerating the timing of head eversion by almost 2 hours compared to comparably-treated controls (cf. solid and dashed black lines, Fig. 6B).
Figure 6. Precocious transcriptional repression of endogenous βFTZ-F1 is sufficient to accelerate developmental timing.
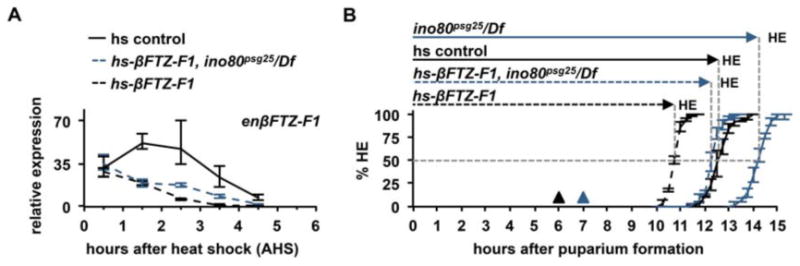
(A) Endogenous βFTZ-F1 regresses significantly faster after expression of βFTZ-F1 protein from a heterologous promoter (hs-βFTZ-F1). All genotypes have similar levels of induction of endogenous βFTZ-F1 at 0.5 hrs after heat shock (AHS). Ectopic βFTZ-F1 protein shuts off endogenous βFTZ-F1 transcription by 3.5 AHS in control animals (dashed black line), but it takes longer in ino80psg25 mutant animals (dashed blue line). y-axis plots relative expression compared the lowest point in the developmental profile; x-axis represents hours after heat-shock (AHS). Three independently-isolated whole animal samples were run for each timepoint and normalized to rp49. (B) Accelerated repression of βFTZ-F1 is sufficient to decrease the duration of prepupal development. 50% of heat-shocked control animals head evert by 12.75 APF (HE50∼12.5 APF, total n=104). However, additional βFTZ-F1 protein accelerates the timing of head eversion by 2 hours (dashed black line, HE50∼10.75 APF, total n=101). 50% of heat-shocked ino80psg25 mutants head evert by 14.25 APF (solid blue line, HE50∼14.25 APF, total n=97), but ectopic expression of βFTZ-F1 protein in the ino80psg25 mutant background accelerates the timing of head eversion by about 2 hours (dashed blue line, HE50∼12.25 APF, total n=99). x-axis represents hours after puparium formation (APF); y-axis represents the percentage of animals head everted. Three independent samples of n∼33 animals were analyzed for each genotype; error bars represent plus or minus one standard deviation. Triangles denote time of heat-shock in each genotype (black=control; blue=ino80psg25/Df).
Given that auto-inhibition of βFTZ-F1 appears to be inefficient in ino80psg25 (Fig. 4C), we next analyzed the effects of ectopic βFTZ-F1 expression in the ino80psg25 mutant background. Endogenous βFTZ-F1 expression starts about one hour later in ino80psg25 mutant animals compared to controls (Fig. S5A); therefore, as expected, heat-shock-induced expression of βFTZ-F1 at 6 APF abolishes the endogenous βFTZ-F1 transcriptional response (blue dashed line, Fig. S5B) and results in prepupal lethality. These effects of precocious expression of βFTZ-F1 on survival are consistent with similar experiments in control animals (Yamada et al., 2000). However, hs-βFTZ-F1 treatment in ino80psg25 mutant animals at 7 APF, when βFTZ-F1 transcripts are at a comparable level to controls at 6 APF (Fig. S5A), allows the endogenous βFTZ-F1 transcriptional response to occur (Fig. 6A). Addition of βFTZ-F1 protein accelerates regression of endogenous βFTZ-F1 in ino80psg25 mutant animals (blue dashed line, Fig. 6A). Strikingly, this regression occurs slightly faster than in control animals lacking the hs-βFTZ-F1 transgene (cf. blue dashed and black solid lines, Fig. 6A). As a result, head eversion in these hs-βFTZ-F1-treated ino80psg25 mutant animals occurs nearly two hours faster (HE50∼12.25 APF, n=99) than comparably treated ino80psg25 animals lacking the transgene (HE50∼14.25 APF, n=97) (cf. blue dashed and blue solid lines, Fig. 6B), effectively rescuing the timing of head eversion to that of control animals. Taken together, these results demonstrate that ino80-dependent repression of transcriptional responses, particularly those of βFTZ-F1, determines the duration of prepupal development.
Discussion
The duration of the Drosophila life cycle is determined by the timing of ecdysone pulses that regulate each developmental transition. Here, we demonstrate the importance of regression of ecdysone-induced transcriptional responses in the regulation of developmental timing. The chromatin remodeling protein ino80 is required for efficient repression of ecdysone-induced genes. ino80psg25 mutant animals fail to shut off ecdysone-induced transcription in a timely manner, resulting in delayed onset of head eversion and prolonged prepupal development. Furthermore, accelerating regression of the mid-prepupal competence factor βFTZ-F1 through overexpression of ino80 or enhancement of the auto-inhibitory activity of βFTZ-F1 protein is sufficient to accelerate the timing of head eversion and shorten the duration of prepupal development. Therefore, INO80 function is required for timely regression of target genes, revealing a critical role for transcriptional regression in the timing of prepupal development.
The role of ino80 in efficient repression of ecdysone-regulated transcriptional responses is consistent with the previously-described biochemical functions of this chromatin remodeler. Although loss of INO80 changes expression levels of a significant fraction of the yeast genome (Jónsson et al., 2004; Shimada et al., 2008), the function of INO80 in transcription appears to be primarily in modulating the kinetics of gene expression (Barbaric et al., 2007). These effects on transcription are mediated by the ability of the INO80 complex to promote ATP-dependent nucleosome sliding (Shen et al., 2000). This sliding activity results in nucleosome eviction at some sites while increasing nucleosome density at others (Morrison and Shen, 2009), raising the question of whether the primary role of INO80 in transcription is in activation or repression of its target genes. Our data suggests that ino80 plays a more critical role in repression of target genes in Drosophila. Consistent with our data, studies in a Drosophila cell culture system indicate that INO80 tends to increase nucleosome density at target loci (Moshkin et al., 2012).
All of the genes that show ino80-dependent regression defects--E74A, DHR3, and βFTZ-F1--were originally identified as ecdysone-induced “puffs” on Drosophila polytene chromosomes (Ashburner et al., 1974). In other words, the expression of these genes occurs on such a large scale that they elicit visible “puffs” of open chromatin on the polytene chromosomes of the larval salivary glands. Given that INO80 also appears to regulate repair and resolution of complex DNA rearrangements (R. C. Conaway and J. W. Conaway, 2009; Morrison and Shen, 2009), it is possible that INO80 is required to restore normal chromatin structure to chromosomal puffs following ecdysone-induced transcription, thereby facilitating regression of these hormone-dependent responses. Genomic studies show that INO80 binds many, but not all, loci in the genome (Moshkin et al., 2012); however, it is unclear whether there is a direct correlation between INO80 binding and target gene expression levels. Moreover, given that INO80 does not directly bind DNA in vivo, how INO80 selects its target genes remains unclear. One possibility is that Ying-Yang 1 (YY1), or Pleiohomeotic (Pho) in Drosophila, a zinc finger transcription factor that is part of the INO80 complex in all metazoans (Cai et al., 2005; Jin et al., 2005; Klymenko et al., 2006), may play that role. In fact, in human cells, YY1 plays a role in localizing the INO80 complex to some target genes (Cai et al., 2007), raising the possibility that this transcription factor aids in recognition of INO80-target genes in Drosophila.
Our results indicate that βFTZ-F1 is a biologically-relevant target of ino80-dependent transcriptional repression. Under all experimental conditions tested, head eversion does not begin until βFTZ-F1 transcription is shut off, showing a direct correlation between the rate of regression of βFTZ-F1 and the onset of head eversion. Additionally, the timing of shut-off of βFTZ-F1 directly correlates with the onset of E74A, raising the possibility that regression of βFTZ-F1 serves as a temporal checkpoint for initiation of the prepupal ecdysone pulse. The mechanism by which βFTZ-F1 regulates the prepupal ecdysone pulse has yet to be determined, although ectopic expression of βFTZ-F1 protein is sufficient to amplify early-gene expression in response to ecdysone (Woodard et al., 1994). Given that βFTZ-F1 expression is critical at various stages of development, it is possible that the developmental delays observed in ino80 mutant animals are mediated by its effects on repression of genes, like βFTZ-F1, that play a critical role in temporal checkpoints during development.
Our ino80 data highlights a novel class of developmental timing phenotypes. Many developmental timing phenotypes disrupt the order of developmental programs. For example, heterochronic genes in C. elegans specify temporal cell fate decisions during development, thereby regulating the proper sequence of developmental events (Ambros, 2000; Thummel, 2001). Another class of timing phenotypes affects the ability to progress from one developmental program to the next. For example, an inability to remove ecdysone during prepupal development prevents the transition to pupal development (Rewitz et al., 2010). Our ino80 mutations highlight the subtle but essential role of the timely progression through development. Although the progression and order of developmental programs do not appear to be disrupted in ino80 mutant animals, most animals fail to complete development, highlighting the importance of proper control of the timing of events during development.
Supplementary Material
Highlights.
We identified and characterized the first metazoan INO80 mutant
INO80 is essential for viability, and mutant animals die during metamorphosis
INO80 is required for efficient regression of hormone-dependent transcription
Regression of βFTZ-F1 is a checkpoint for the timing of head eversion
Failure to efficiently shut-off transcription causes developmental timing defects
Acknowledgments
We would like to thank the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, and C. Thummel for fly stocks; the Developmental Studies Hybridoma Bank and C. Thummel for antibodies; the Berkeley Drosophila Genome Project for reagents and Genetic Services, Inc. for transgenic generation; Yunsik Kang and Nick Wleklinski for technical assistance. This work was supported by an NSF Graduate Research Fellowship Program grant (DGE-1256259) to S.D.N. and an NIH/NIGMS grant (R01 GM095944) to A.B..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambros V. Control of developmental timing in Caenorhabditis elegans. Current Opinion in Genetics & Development. 2000;10:428–433. doi: 10.1016/s0959-437x(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. doi: 10.1016/s0091-679x(08)60932-2. [DOI] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B. QuantPrime--a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008;9:465. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Chihara C, Meltzer P, Richards G. Temporal control of puffing activity in polytene chromosomes. Cold Spring Harb. Symp Quant Biol. 1974;38:655–662. doi: 10.1101/sqb.1974.038.01.070. [DOI] [PubMed] [Google Scholar]
- Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. Journal of embryology and experimental morphology. 1981;66:57–80. [PubMed] [Google Scholar]
- Barbaric S, Luckenbach T, Schmid A, Blaschke D, Hörz W, Korber P. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J Biol Chem. 2007;282:27610–27621. doi: 10.1074/jbc.M700623200. [DOI] [PubMed] [Google Scholar]
- Bhatia S, Pawar H, Dasari V, Mishra RK, Chandrashekaran S, Brahmachari V. Chromatin remodeling protein INO80 has a role in regulation of homeotic gene expression in Drosophila. Genes to Cells. 2010 doi: 10.1111/j.1365-2443.2010.01416.x. [DOI] [PubMed] [Google Scholar]
- Broadus J, McCabe JR, Endrizzi B, Thummel CS, Woodard CT. The Drosophila beta FTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Molecular Cell. 1999;3:143–149. doi: 10.1016/s1097-2765(00)80305-6. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, Conaway JW. YY1 functions with INO80 to activate transcription. Nature Structural & Molecular Biology. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- Caldwell PE, Walkiewicz M, Stern M. Ras activity in the Drosophila prothoracic gland regulates body size and developmental rate via ecdysone release. Curr Biol. 2005;15:1785–1795. doi: 10.1016/j.cub.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Chiang PW, Kurnit DM. Study of dosage compensation in Drosophila. Genetics. 2003;165:1167–1181. doi: 10.1093/genetics/165.3.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Deng H, Kerppola TK. Regulation of Drosophila metamorphosis by xenobiotic response regulators. PLoS Genet. 2013;9:e1003263. doi: 10.1371/journal.pgen.1003263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 2010;17:534–539. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch O, Benvenuto G, Bowler C, Molinier J, Hohn B. The INO80 protein controls homologous recombination in Arabidopsis thaliana. Molecular Cell. 2004;16:479–485. doi: 10.1016/j.molcel.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Horner MA, Chen T, Thummel CS. Ecdysteroid regulation and DNA binding properties of Drosophila nuclear hormone receptor superfamily members. Dev Biol. 1995;168:490–502. doi: 10.1006/dbio.1995.1097. [DOI] [PubMed] [Google Scholar]
- Huet F, Ruiz C, Richards G. Sequential gene activation by ecdysone in Drosophila melanogaster: the hierarchical equivalence of early and early late genes. Development. 1995;121:1195–1204. doi: 10.1242/dev.121.4.1195. [DOI] [PubMed] [Google Scholar]
- Ihry RJ, Sapiro AL, Bashirullah A. Translational Control by the DEAD Box RNA Helicase belle Regulates Ecdysone-Triggered Transcriptional Cascades. PLoS Genet. 2012;8:e1003085. doi: 10.1371/journal.pgen.1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Baehrecke EH, Thummel CS. Steroid regulated programmed cell death during Drosophila metamorphosis. Development. 1997;124:4673–4683. doi: 10.1242/dev.124.22.4673. [DOI] [PubMed] [Google Scholar]
- Jiang C, Lamblin AF, Steller H, Thummel CS. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Molecular Cell. 2000;5:445–455. doi: 10.1016/s1097-2765(00)80439-6. [DOI] [PubMed] [Google Scholar]
- Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutiérrez JL, Coleman MK, Workman JL, Mushegian A, Washburn MP, Conaway RC, Conaway JW. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- Jónsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Molecular Cell. 2004;16:465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Köcher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam G, Hall BL, Bender M, Thummel CS. DHR3 is required for the prepupal-pupal transition and differentiation of adult structures during Drosophila metamorphosis. Dev Biol. 1999;212:204–216. doi: 10.1006/dbio.1999.9343. [DOI] [PubMed] [Google Scholar]
- Lam GT, Jiang C, Thummel CS. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development. 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- Morrison A, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009 doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Chalkley GE, Kan TW, Reddy BA, Ozgur Z, van Ijcken WFJ, Dekkers DHW, Demmers JA, Travers AA, Verrijzer CP. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol Cell Biol. 2012;32:675–688. doi: 10.1128/MCB.06365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz KF, Yamanaka N, O'connor MB. Steroid hormone inactivation is required during the juvenile-adult transition in Drosophila. Dev Cell. 2010;19:895–902. doi: 10.1016/j.devcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford L. Hormones and Drosophila development. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila Melanogaster. 1993. pp. 899–939. The development of Drosophila melanogaster. [Google Scholar]
- Robertson CW. The metamorphosis of Drosophila melanogaster, including an accurately timed account of the principal morphological changes. Journal of morphology. 1936;59:351–399. [Google Scholar]
- Sapiro AL, Ihry RJ, Buhr DL, Konieczko KM, Ives SM, Engstrom AK, Wleklinski NP, Kopish KJ, Bashirullah A. Rapid Recombination Mapping for High-Throughput Genetic Screens in Drosophila. G3 (Bethesda) 2013;3:2313–2319. doi: 10.1534/g3.113.008615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Shen X, Ranallo R, Choi E, Wu C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Molecular Cell. 2003;12:147–155. doi: 10.1016/s1097-2765(03)00264-8. [DOI] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Thummel CS. Flies on steroids--Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 1996;12:306–310. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Urness LD, Thummel CS. Molecular interactions within the ecdysone regulatory hierarchy: DNA binding properties of the Drosophila ecdysone-inducible E74A protein. Cell. 1990;63:47–61. doi: 10.1016/0092-8674(90)90287-o. [DOI] [PubMed] [Google Scholar]
- Wang L, Evans J, Andrews HK, Beckstead RB, Thummel CS, Bashirullah A. A genetic screen identifies new regulators of steroid-triggered programmed cell death in Drosophila. Genetics. 2008;180:269–281. doi: 10.1534/genetics.108.092478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Peterson CL. The INO80 family of chromatin-remodeling enzymes: regulators of histone variant dynamics. Cold Spring Harb. Symp Quant Biol. 2010;75:35–42. doi: 10.1101/sqb.2010.75.063. [DOI] [PubMed] [Google Scholar]
- White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- White KP, Hurban P, Watanabe T, Hogness DS. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- Woodard CT, Baehrecke EH, Thummel CS. A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell. 1994;79:607–615. doi: 10.1016/0092-8674(94)90546-0. [DOI] [PubMed] [Google Scholar]
- Yamada M, Murata T, Hirose S, Lavorgna G, Suzuki E, Ueda H. Temporally restricted expression of transcription factor betaFTZ-F1: significance for embryogenesis, molting and metamorphosis in Drosophila melanogaster. Development. 2000;127:5083–5092. doi: 10.1242/dev.127.23.5083. [DOI] [PubMed] [Google Scholar]
- Yin VP, Thummel CS, Bashirullah A. Down-regulation of inhibitor of apoptosis levels provides competence for steroid-triggered cell death. J Cell Biol. 2007;178:85–92. doi: 10.1083/jcb.200703206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


