Abstract
Mutations in the IDH1 and IDH2 genes encoding isocitrate dehydrogenases are frequently found in human glioblastomas1 and cytogenetically normal acute myeloid leukaemias (AML)2. These alterations are gain-of-function mutations in that they drive the synthesis of the ‘oncometabolite’ R-2-hydroxyglutarate (2HG)3. It remains unclear how IDH1 and IDH2 mutations modify myeloid cell development and promote leukaemogenesis. Here we report the characterization of conditional knock-in (KI) mice in which the most common IDH1 mutation, IDH1(R132H), is inserted into the endogenous murine Idh1 locus and is expressed in all haematopoietic cells (Vav-KI mice) or specifically in cells of the myeloid lineage (LysM-KI mice). These mutants show increased numbers of early haematopoietic progenitors and develop splenomegaly and anaemia with extramedullary haematopoiesis, suggesting a dysfunctional bone marrow niche. Furthermore, LysM-KI cells have hypermethylated histones and changes to DNA methylation similar to those observed in human IDH1- or IDH2-mutant AML. To our knowledge, our study is the first to describe the generation and characterization of conditional IDH1(R132H)-KI mice, and also the first report to demonstrate the induction of a leukaemic DNA methylation signature in a mouse model. Our report thus sheds light on the mechanistic links between IDH1 mutation and human AML.
IDH1/IDH2 mutations typically produce mutant enzymes with aberrant activity. Whereas wild-type (WT) Idh proteins metabolize isocitrate and NADP+ to yield α-ketoglutarate (αKG) and NADPH, mutant Idh proteins convert αKG into 2HG while consuming NADPH3.2HG competitively inhibits tet methylcytosine dioxygenases (Tet2), which regulate DNA methylation, as well as JmjC domain-containing histone demethylases4–6. Accordingly, human AML cells with IDH1/IDH2 mutation show global DNA hypermethylation5.
To create a murine model of the IDH1(R132H) mutation, we used the lox-stop-lox (LSL) system to generate a conditional Idh1 knock-in (KI) mouse (Idh1LSL/WT) (Supplementary Fig. 2). Mutant mice were crossed with LysMCre mice7 (Idh1LSL/WTLysMCre+/WT, LsyM-KI), because the LysM promoter is activated very early during myeloid development8 and human AML leukaemic stem cells show similarities with very early myeloid primed progenitors (LMPP)9.
LysM-KI mice were born at the expected Mendelian ratio, were viable and fertile, and had normal lifespans. However, serum 2HG levels were elevated approximately tenfold in both young (7–16 weeks old) and older (47–56 weeks old) LysM-KI mice (Supplementary Fig. 3a–c). Whereas peripheral blood cell counts of young LysM-KI mice were normal, older (42–46 weeks old) mutants developed anaemia (Supplementary Fig. 3d, e). In the OP9/OP9-DL1 in vitro differentiation system10, haematopoietic cell lineages developed normally from several IDH1(R132H)-KI embryonic stem cell clones (Supplementary Fig. 4). Macroscopic analysis of mice revealed mild splenic enlargement in young LysM-KI mice that progressed to overt splenomegaly in all older mutants. Histologically, splenic architecture became increasingly disorganized, with age-dependent expansion of spaces between lymphoid follicles and obvious extramedullary haematopoiesis (Fig. 1a, b).
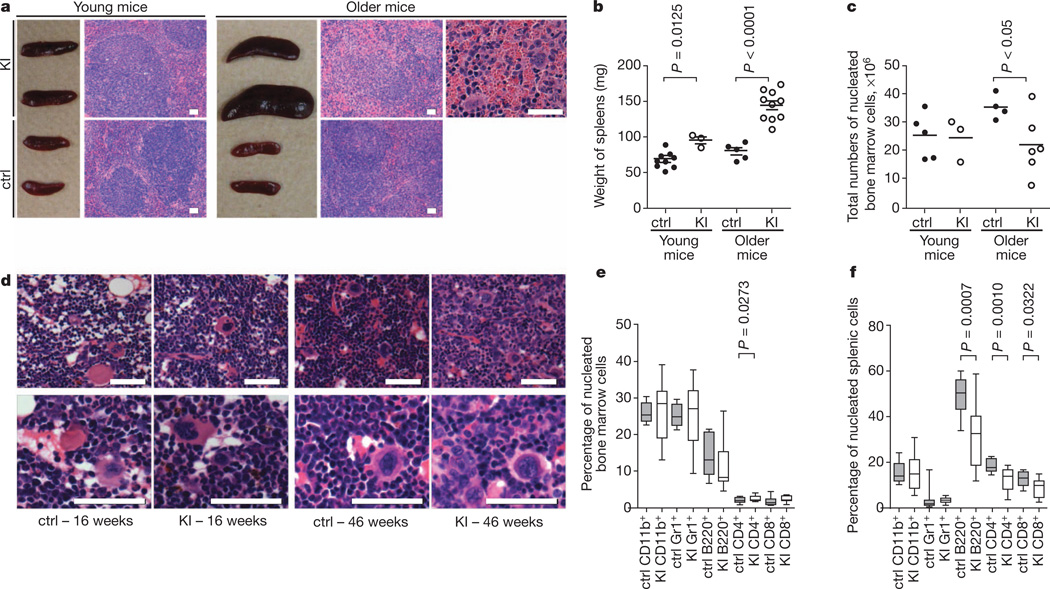
Figure 1. LysM-KI mice show age-dependent splenomegaly and decreased bone marrow cellularity.
a, Macroscopic appearance and haematoxylin-and-eosin-stained sections of spleens from young and older control (ctrl) and LysM-KI (KI) mice. b, c, Spleen weights (b) and total numbers of nucleated BM cells (c) from young and older control and LysM-KI mice (n = 3–5 per group). Horizontal line, mean. d, Representative haematoxylin-and-eosin-stained sections of BM from femurs of young and older control and LysM-KI mice (n = 4 per group). e, f, Quantifications of flow cytometric analyses of mature haematopoietic cell populations among total nucleated BM (e) and splenic cells (f) from older control and LysM-KI (n = 10–14 per group) mice. P values were determined using the unpaired t-test with Welch’s correction. Scale bars, 50 µm.
We next evaluated the haematopoietic stem cell (HSC) and haematopoietic progenitor cell (HPC) compartments in LysM-KI mice. Bone marrow (BM) cellularity was normal in young LysM-KI mice but reduced in older mutants (Fig. 1c), where it correlated inversely with splenomegaly and the degree of anaemia. Histologically, the BM of young mutants appeared normal but the BM of older mutants showed fewer mature cells and more immature cells (Fig. 1d). Flow cytometric analyses of BM and spleen revealed altered numbers of mature cells (CD11b+, Gr1+, B220+, CD4+, CD8+) in LysM-KI mice (Fig. 1e, f).
More refined flow cytometric analyses confirmed that lineage-negative (Lin−) cells underwent an age-dependent expansion in LysM-KI BM (Fig. 2a, b and Supplementary Table 1). Moreover, LSK (Lin−Sca1+cKit+) cells were increased by approximately 1.8-fold in young LysM-KI mice and by approximately 5.4-fold in older mutants (Fig. 2a, b and Supplementary Table 1). The LSK population contains long-term HSCs (LT-HSCs; CD150+CD48−), short-term HSCs (ST-HSCs; CD150+CD48+), and lineage-restricted progenitors (LRPs; CD150−CD48+). We found that LysM-KI mice demonstrated an age-dependent increase in LRPs (Fig. 2b and Supplementary Table 1). There were no significant alterations to the LK population (Lin−Sca1−cKit+), which contains common myeloid progenitors (CMPs;CD34+CD16/32medLK), granulocyte-macrophage progenitors (GMPs; CD34+CD16/32highLK), and megakaryocyte-erythroid progenitors (MEPs; CD34−CD16/32lowLK), or to the common lymphoid progenitor population (CLPs; Lin−Il7Ra+Sca1medcKitmed) (Fig. 2b and Supplementary Table 1). In colony-forming cell (CFC) assays, BM cells from young or older LysM-KI mice showed statistically normal production of granulocyte (G), granulocyte-macrophage (GM), granulocyte-erythrocyte-macrophage-megakaryocyte (GEMM), and macrophage (M) colonies (Fig. 2c). Flow cytometric analysis of nucleated splenic cells from older LysM-KI mice revealed significantly elevated numbers of LSK and LRP cells, recapitulating the pattern observed in the bone marrow of these mice (Fig. 2a, d). CFC assays of nucleated splenic cells showed increased numbers of all four myeloid colony types in LysM-KI mice (Fig. 2e). These data confirm that extra-medullary haematopoiesis occurs in older LysM-KI mice.
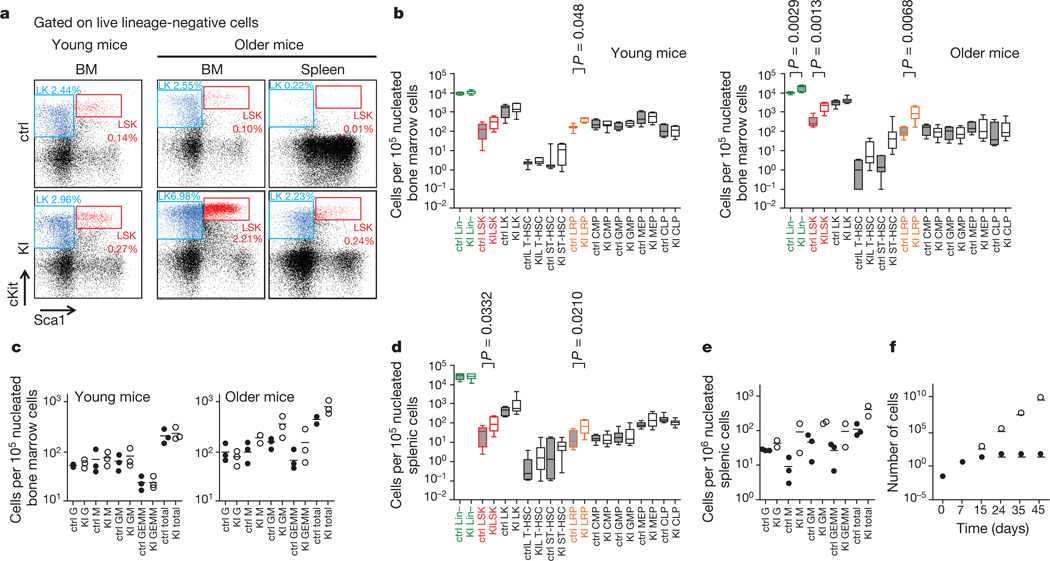
Figure 2. LysM-KI mice showage-dependent increases in lineage-restricted progenitors and extramedullary haematopoiesis.
a, Flow cytometric analysis of live lineage-negative cells for cKit and Sca1 in cells from the BM of young (left), and from the BM and spleen of older control and LysM-KI mice (right). LSK (red) and LK (blue) populations are highlighted. The percentage of a given population among total viable BM cells is indicated. b, Quantification of flow cytometric analyses for the indicated HSC and HPC populations in BM of young (n = 3–14 per group) and older (n = 4–8 per group) control and LysM-KI mice. Horizontal line, median; box, median±25%; end of bars, minimum and maximum. c, CFC assays of BM cells from young and older control and LysM-KI mice (n = 3 per group). Data points are the mean number of colonies of the indicated lineage for individual mice. Horizontal line, mean of values for all three mice in a group. d, Quantification of flow cytometric analyses for the indicated HSC and HPC populations in the spleens of older (n = 7–8 per group) control and LysM-KI mice, determined as for b. e, CFC assays of nucleated splenic cells from older control (n = 3) and LysM-KI (n = 2) mice determined as for c. f, Serial replating in methylcellulose of BM cells from control and LysM-KI mice (n = 2 per group; control, filled circles; LysM-KI, empty circles). Data are the mean total cell number for both mice in a group (in units of 106).
We speculated that the LRP accumulation in LysM-KI BM (and spleen), despite the near-normal peripheral blood counts of these animals and their normal BM cell CFC activity, might be due to increased symmetric cell division and proliferation, and/or a partial block in differentiation. Serial plating experiments showed that, whereas control BM cells stopped proliferating after three rounds of plating (24 days), LysM-KI BM cells continued to grow at an exponential rate for six rounds of plating (45 days) (Fig. 2f).
To extend our findings, we crossed Idh1LSL/WT mice with VavCre mice11 to generate Vav-KI mutants expressing IDH1(R132H) in all haematopoietic cells, from LT-HSCs to terminally differentiated cells. Lin-negative, LSK, LRP and CLP cells were all significantly increased in BM of young Vav-KI mice (Supplementary Fig. 5a), as were splenic LSK, LK, LRP, MEP and CLP cells (Supplementary Fig. 5b). Like young LysM-KI mice, young Vav-KI mice had normal total BM cell numbers and showed no overt haematological changes (Supplementary Fig. 5c–e).
Next, we performed competitive BM repopulation assays in which equal numbers (105) of nucleated BM cells from donor mice (control or LysM-KI; CD45.2+) and from recipient mice (control; CD45.1+) were injected into lethally-irradiated recipients (CD45.1+). LysM-KI BM cells showed no defects in short-term or long-term repopulation capacity for at least 170 days after transplantation, and relative peripheral blood cell counts were not altered in recipients of LysM-KI BM (Supplementary Fig. 6a, b).
The homeostasis of HSCs and HPCs is influenced by reactive oxygen species (ROS)12,13. Because high ROS reduces HSC longevity, these cells strive to keep ROS low13. In contrast, higher levels of ROS in developing myeloid cells may be necessary for their differentiation13.ROS elevation in AML cells triggers their differentiation14. It has been assumed that IDH1/IDH2 mutations increase ROS3. However, when using CM-H2DCFDA (5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester; a membrane-permeable indicator of reactive oxygen species) to measure total ROS in granulocytes (CD11b+Gr1+), macrophages/monocytes (CD11b+Gr1low), LSK and LK cells in BM of young and older LysM-KI mice, no abnormalities were observed (Supplementary Fig. 7a, b). Liquid chromatography-mass spectrometry (LC-MS) analysis of 2HG as well as of NAD+, NADH, NADP+ and NADPH in CD11b+ cells isolated from BM and spleen of control and LysM-KI mice, and in BM-derived macrophages (BMDMs) revealed highly elevated 2HG in all LysM-KI samples, whereas NAD+, NADH, NADP+ and NADPH levels were not altered (Supplementary Fig. 7c–e). Taken together, these data indicate that the phenotype of LysM-KI mice is not due to alterations in ROS or NADPH levels, nor to 2HG-mediated complete abolition of myeloid differentiation.
The hypoxia-inducible transcription factors Hif1α and Hif2α are critical for HSC survival and maintenance15 but their involvement in HPC differentiation and beyond is unclear. IDH1 mutations have been shown to alter Hif1α stability6,16. We compared messenger RNA levels of several Hif1α target genes in sorted control and LysM-KI LSK cells but observed no differences (Supplementary Fig. 8). Thus, it is unlikely that Hif1α signalling is altered in LysM-KI LSK cells.
To analyse global mRNA expression, we performed gene expression microarray analyses of sorted LSK cells from young LysM-KI and control mice. Two-way hierarchical clustering of significantly altered genes (P value < 0.05, >1.5-fold change) revealed a clear separation of control and LysM-KI samples (Supplementary Fig. 9). Gene ontology analysis of the identified gene signature (Supplementary Table 3) revealed a significant enrichment of genes in several categories related to cellular growth and proliferation (Supplementary Table 4).
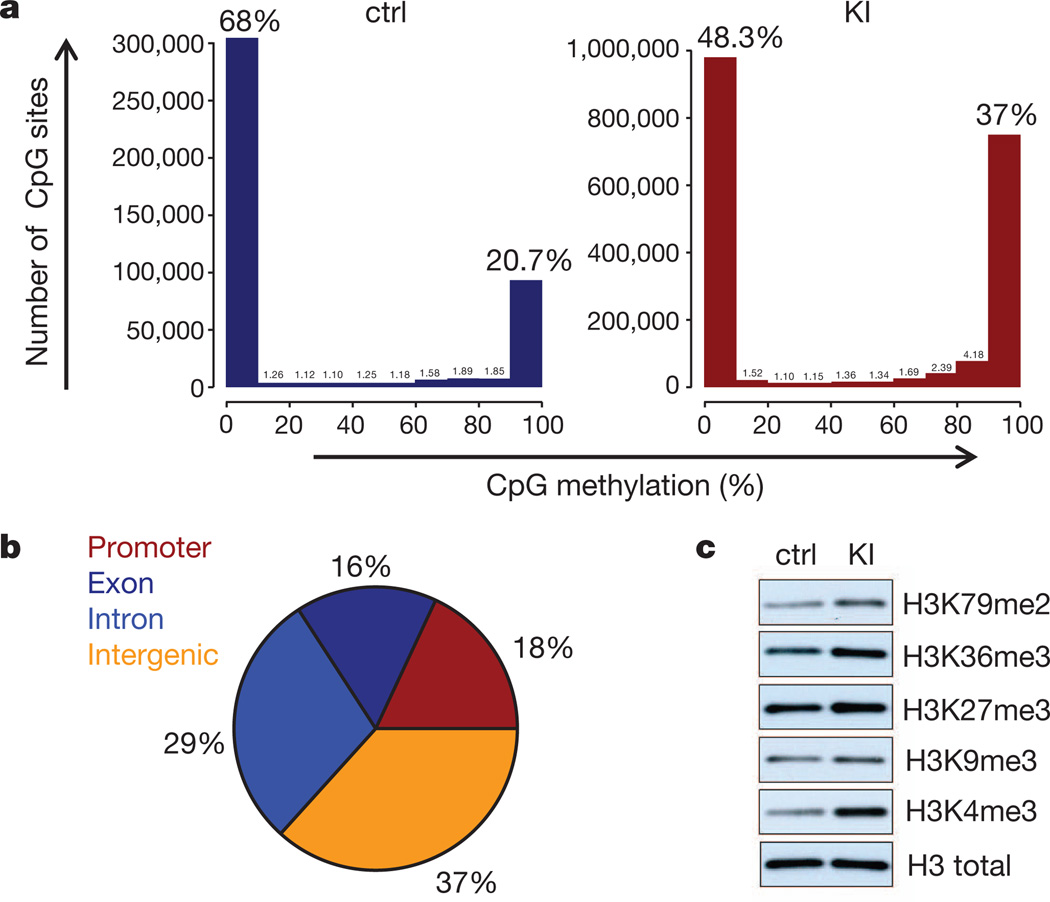
Human gliomas17 and AMLs with IDH1/IDH2 mutations have DNA hypermethylation that, in AML, is caused by 2HG-mediated inhibition of Tet2 (ref. 5). 2HG also inhibits histone demethylases, resulting in hypermethylation of histone lysine residues (especially in H3)4,6.To examine DNA methylation, we performed high-throughput sequencing of bisulphite-treated DNA from sorted LSK cells from young LysM-KI and control mice. DNA from LysM-KI LSK cells showed a significantly greater proportion of highly methylated CpG sites, with a marked increase in CpG sites showing greater than 80% methylation (Fig. 3a). A more detailed analysis of the differential distribution pattern of DNA methylation revealed wide-spread global changes, with all chromosomes equally affected (Supplementary Fig. 10a, b and Supplementary Table 5). A clear preference for promoters and intragenic regions was observed that paralleled the changes in DNA methylation observed in human IDH1/IDH2-mutant AMLs (Fig. 3b, Supplementary Fig. 10a, b and Supplementary Table 5)18. Comparison of the 1,559 hypermethylated murine genes in LysM-KI LSK cells to the 8195 hypermethylated human genes in IDH1/IDH2-mutant AMLs revealed an overlap of 784 genes (P = 1.63 × 10−36; Supplementary Table 6). Ingenuity pathway analysis (IPA) implicated several signalling pathways involved in haematopoietic cell proliferation and differentiation, leukaemogenesis and leukaemic stem cell maintenance (Supplementary Table 7 and Supplementary Fig. 11)19–22 including the WNT, NOTCH and TGF-β pathways, which are also targeted by aberrant methylation in human IDH1/IDH2-mutant AMLs18. Immunoblotting of lysates from control and LysM-KI BMDMs revealed increased methylation of multiple H3 lysine residues (Fig. 3c).
Figure 3. Altered methylation of DNA and histones in LysM-KI cells.
a, Sequencing data from bisulphite-treated DNA from sorted BM LSK cells of young control (n = 2) and LysM-KI (n = 3) mice. Numbers of fragments showing the indicated percentage of CpG methylation were calculated as detailed in Methods. Data shown are from one LysM-KI and one control mouse and are representative of all animals in a group. b, Graphic representation of the differentially methylated genomic regions with respect to their genomic location. c, Immunoblot analysis of methylation of the indicated H3 lysine residues in lysates of BMDMs derived from young control and LysM-KI mice. Results are representative of two independent experiments.
In conclusion, we show that the IDH1(R132H) mutation in all haematopoietic cells (Vav-KI), and in the myeloid lineage in particular (LysM-KI), results in the accumulation of LSK and LRP cells first in the bone marrow and later in the spleen. We believe that the accumulations of these cells are most likely due to 2HG-induced DNA and histone hypermethylation that affect LSK cell division and/or differentiation (Supplementary Fig. 1). Our finding that IDH1(R132H) does not completely block myeloid differentiation may explain why IDH1/IDH2 mutations are found in almost all FAB (French-American-British classification) AML subtypes23. Moreover, our mutants show extramedullary haematopoiesis, suggesting a dysfunctional bone marrow niche. Extramedullary haematopoiesis occurs in myelodysplastic syndromes, which frequently progress to AML and harbour IDH1/IDH2 mutations in 10–15% of cases24. Our study is, to our knowledge, the first to model epigenetic changes of human IDH1/IDH2-mutant AML in mice and thereby advances and will advance our understanding of the links between IDH1/IDH2 mutations and leukaemogenesis.
METHODS
Generation of LSL-Idh1R132H mice and breeding
The targeting vector containing both the loxP-flanked STOP (LSL) cassette and the IDH1(R132H) mutation is shown in Supplementary Fig. 2. The LSL cassette (Addgene) is composed of the puromycin resistance gene, a splicing acceptor sequence that stops transcription at the insertion point, and four repeats of the SV40 polyA sequence. Thus, the presence of LSL inhibits expression of the IDH1(R132H) protein, and Idh1LSL/WT mice are therefore heterozygous for the Idh1 wild-type allele. Cre-mediated excision of LSL allows expression of the IDH1(R132H) protein (Idh1LSL/WT Cre+).
The targeting vector was assembled by cloning the LSL cassette plus PCR-generated genomic fragments of the Idh1 gene representing short, middle and long arms of homology into the DT-A vector where the middle fragment contains murine Idh1 exon 4 bearing the R132H mutation, which was created by PCR-based mutagenesis. The linearized targeting vector was electroporated into E14K or Bruce4 embryonic stem cells. Homologous recombination was confirmed by Southern blotting using 5′-flanking, 3′-flanking, and puromycin-specific probes. Chimaeric mice were produced by microinjection of targeted embryonic stem cells into E3.5 blastocysts. Chimaeras derived from E14K or Bruce4 embryonic stem cells were bred to C57BL/6J or B6-Tyrc-2J/J mice, respectively. Germline transmission of the mutant Idh1 allele was confirmed by genomic Southern blotting. Both E14K-derived and Bruce4-derived strains were crossed to C57Bl/6J, and F2–F5 generations were used for analysis. The phenotypes of the resulting progeny were identical. For transplantation studies only offspring derived from the Bruce4-derived chimaeras was used. Pups were genotyped by PCR analysis using specific primer pairs to detect the wild-type and mutant Idh1 alleles. Sense and antisense primers for the KI, wild-type and LSL alleles were 5′-ACCAGCACCTCCCAACTTGTAT-3′, 5′-AGGTTAGCTCTTGCCGATCCGT-3′, and 5′-CAGCA GCCTCTGTTCCACATAC-3′, which yielded PCR products of the predicted sizes of 394 base pairs (bp), 307 bp, and 223 bp, respectively (Supplementary Fig. 2c). All animals were treated in accordance with the NIH Guide for Care and Use of Laboratory Animals as approved by the Ontario Cancer Institute Animal Care Committee (Toronto, Ontario, Canada).
LysMCre mice (B6.129P2-Lyz2tm1(cre)Ifo/J; catalogue no. 004781), VavCre mice (B6.Cg-Tg(Vav1-cre)A2Kio/J; catalogue no. 008610) and CD45.1 mice (B6.SJL-Ptprca Pepcb/BoyJ; catalogue no. 002014) were purchased from the Jackson Laboratory. LysMCre mice and VavCre mice were backcrossed into the C57Bl/6J background for at least 10 generations. LysM-KI mice were generated by breeding Idh1LSL/WT mice with LysMCre+/WT mice as well as by crossing LysM-KI mice with wild-type C57BL/6 mice. Vav-KI mice were generated by breeding Idh1LSL/WT mice with VavCre+/WT mice as well as by crossing Vav-KI mice with wild-type C57BL/6 mice.
For all experiments, LysM-KI mice were heterozygous for both the IdhLSL allele and the LysMCre allele (Idh1LSL/WTLysMCre+/WT), and Vav-KI mice were heterozygous for both the IdhLSL allele and the VavCre allele (Idh1LSL/WTVavCre+/WT). Control mice were wild-type animals or animals heterozygous for either the Idh1LSL allele or the LysMCre allele, if LysM-KI mice were used, or the VavCre allele, if Vav-KI mice were used (Idh1WT/WTLysMCreWT/WT, Idh1WT/WTLysMCre+/WT, Idh1LSL/WTVavCreWT/WT, or Idh1WT/WTVavCre+/WT). All animal procedures were approved by the Animal Care and Use Committee of the University Health Network (Toronto, Ontario, Canada).
Generation of IDH1(R132H)-KI embryonic stem cell clones
KI embryonic stem cells which had been generated while making the KI mouse were expanded and transiently transfected with pCAGGS-nlsCre using Lipofectamine2000 (Invitrogen). After transfection, cells were trypsinized and plated on gelatin-coated cell culture plates. Single colonies were picked and plated in duplicates into 96-well plates to test for puromycin sensitivity as the Cre-mediated recombination of the LSL cassette results in loss of the puromycin-resistance cassette. Puromycin-sensitive clones were expanded, tested by PCR-genotyping, and then used for in vitro differentiation experiments.
OP9/OP9-DL1 embryonic stem cell in vitro differentiation system
Experiments including co-culture and differentiation of embryonic stem cells in the presence of OP9 or OP9-DL1 cells were carried out as described elsewhere9.
Histology
For histological analyses, mouse tissues were fixed in 10% buffered formalin. After fixation, femurs were decalcified in formic acid. Fixed tissues were paraffin-embedded, sectioned, and stained with haematoxylin and eosin according to standard laboratory protocols.
Flow cytometric analyses
Flow cytometric analyses were performed according to standard protocols. In brief, mice were euthanized, single-cell suspensions from bone marrow (BM) were generated by flushing out the bone marrow from tibia and femur in Iscove’s modified Dulbecco’s medium supplemented with 2% heat-inactivated fetal bovine serum (HI-FBS) (IMDM2). Single-cell suspensions of spleens were generated by mashing the spleen in IMDM2. Cell suspensions were passed through 70 µm cell strainers, centrifuged and resuspended in ammonium-based red cell lysis buffer (Sigma). Nucleated cells were washed in IMDM2, centrifuged and resuspended in flow cytometric buffer (PBS without Mg2+ or Ca2+ (PBS−/−), 2% HI-FBS, 5mMEDTA, pH8.0). Viable cell numbers were determined by cell counting after trypan blue staining.
For flow cytometric analyses, cells were incubated in Fc block (BD Biosciences) and then immunostained with fluorophore-linked antibodies (BD Biosciences, eBioscience). For immunostaining of lineage-positive cells, a cocktail containing biotin-labelled primary antibodies against CD5, B220, CD11b, 7-4, Gr-1 and Ter-119 (Miltenyi Biotec) was used, followed by staining with streptavidin-linked fluorophore-labelled secondary antibodies.
To detect total reactive oxygen species (ROS), immunostained cells were incubated with the live cell dye CM-H2DCFDA (100 µM) at 37 °C for 15 min followed by two washes in flow cytometric buffer. flow cytometric data were acquired on a BD CantoII and analysed using FlowJo (TreeStar). For flow cytometric analyses of differentiated cells, 104–105 events were acquired. For HSC and HPC analyses, up to 2.5 × 106 events were acquired. Statistical analyses of flow cytometric data were performed using the GraphPad Prism software.
LSK cell sorting
Single-cell suspensions from BM were generated as described above from young mice (8 to 12 weeks of age) and cells were resuspended in flow cytometric buffer and counted. Lineage-negative cells were isolated using the Miltenyi lineage cell depletion kit. Cells were counted and immunostained as described above with anti-lineage-biotin/streptavidin-APC-Cy7, anti-cKit-PE-Cy7 and anti-Sca-1-APC antibodies. Cells were then sorted on a BD FACSAria cell sorter, collected in IMDM2, centrifuged and pellets shock-frozen on dry ice and stored at −80 °C until further processing.
Real-time RT–PCR analysis
RNA was purified from sorted LSK cells using TRIzol (Invitrogen), resuspended in water and quantified on a NanoDrop spectrophotometer before transcription into cDNA using the iScript complementary DNA synthesis kit (Bio-Rad). Primers used for RT–PCR are listed in Supplementary Table 2. Real-time RT–PCR analyses were performed using Power SybrGreen on a 7900HT Fast-Real Time PCR system (Applied Biosystems). Ct values were normalized to the housekeeping gene Hprt (ΔCt) and then to the average of the Δ Ct values of all samples (ΔΔCt). Relative mRNA expression levels were calculated (2−ΔΔCt) and normalized to the average of the relative control mRNA expression levels.
Colony-forming cell (CFC) assays
Single-cell suspensions from BM or spleen from control and LysM-KI mice were generated as described above. Cells were then plated at a density of 104 ml−1 for BM and 105 ml−1 for spleen in triplicates in complete methylcellulose media containing stem cell factor, Il-3, Il-6 and erythropoietin (Stem Cell Technologies, M3434). Colonies were counted after 7 days at 37 °C and 5% CO2. For serial plating cells were collected from methylcellulose media, washed once in IMDM2, counted and replated in complete methylcellulose media at a density of 104 ml−1.
Bone marrow competitive repopulation experiments
For competitive BM transfer, donor BM cells from control or LysM-KI mice (CD45.2+), and competitor BM cells from WT mice (CD45.1+), were isolated as described above and then washed in PBS−/− twice. Cell numbers were adjusted to 106 ml−1. Lethally-irradiated (10.5 Gy) recipient mice (CD45.1+) were injected with 105 donor BM cells plus 105 competitor BM cells (2 × 105 total). For flow cytometric analyses of peripheral blood, mice were bled from the tail vein and immunostaining was carried out as described above. Immunostained blood samples were fixed in FoxP3 fixation buffer (eBioscience) to lyse red blood cells, washed in flow cytometric buffer, and analysed by flow cytometry.
Generation of bone marrow-derived macrophages (BMDMs)
BMDMs were generated from single-cell BM suspensions of young control or LysM-KI mice using a standard in vitro M-CSF-based differentiation protocol. In brief, 5 × 106 cells were plated on a 10-cm tissue culture plate in 5 ml of RPMI1640 media supplemented with 10% HI-FBS and 50 ng ml−1 M-CSF. On days 2, 3, and 4 an additional 1 ml of the complete media was added to every tissue culture plate. Cells were collected on day 5 of the in vitro differentiation procedure and washed in PBS containing Mg2+ and Ca2+ (PBS+/+) before being used in experiments.
Immunoblot analysis
For immunoblot analysis of histone-3 methylation, equal numbers of BMDMs were generated as described above and lysed in 1 × cell lysis buffer (20mMTris-HCl pH7.5, 150mMNaCl, 1mMEDTApH8.0, 1mMEGTA pH8.0, 1% Triton X-100) supplemented with EDTA-free protease inhibitor cocktail (Roche). SDS (1%) was added to lysates and incubation was continued for another 30 min. Lysates were centrifuged, loading buffer was added to supernatants, and samples were subjected to electrophoresis on 12% Bis-Tris gels using MES running buffer (Invitrogen). Separated proteins were transferred onto nitrocellulose membranes (Invitrogen). Proteins were then detected using standard laboratory procedures. Primary antibodies used in this study recognized total histone-3 (Abcam ab10799), histone-3 K4me3 (Millipore 07-473), histone-3 K9me3 (Abcam ab8898), histone-3 K27me3 (Millipore 07-449), histone-3 K36me3 (Abcam ab9050), or histone-3 K79me2 (Cell Signaling Technology 9757).
Mass spectrometry
CD11b+ cells were isolated from single-cell suspensions of BM or spleen cells using a CD11b-magnetic bead purification kit (Miltenyi Biotec). BMDMs were generated by in vitro differentiation as described above. For serum, blood was collected from mice and separated by centrifugation. Metabolites were extracted from an equivalent number of CD11b+ cells or BMDMs using dry-ice-temperature 80% methanol or equal volumes of serum diluted to 80% methanol using dry-ice-temperature 100% methanol. Cells and serum were centrifuged at 14,000 r.p.m. (20,800g) for 10 min at 4 °C. Supernatants were dried down under nitrogen gas, resuspended in 50% methanol, and centrifuged at 13,000g for 5 min at 4 °C. The supernatant was transferred to HPLC sample vials and each extract was analysed by reverse-phase liquid chromatography (LC) with tributylamine as an ion pairing reagent. The LC was coupled to a triple-quadrupole mass spectrometer running in negative mode (Thermo Quantum Ultra). Specific chromatography conditions and mass spectrometry parameters were as described25. Peak heights of specific metabolites were measured by metabolite-specific multiple reaction monitoring (MRM) scans as previously reported26, with the exception of hydroxyglutarate, which was monitored using an MRM scan consisting of a 147 to 129 m/z transition and a collision energy of 13 eV. The mzrock mass spectrometry tool kit (http://code.google.com/p/mzrock/) was used for data analysis and visualization. To minimize the impact of inter-day instrument variation, extracted chromatogram peak heights for a given metabolite were normalized by the maximum sample value for that specific metabolite measured in the same experiment. These relative values were then scaled by 10,000.
DNA methylation analysis
High molecular weight genomic DNA (gDNA) was isolated from sorted LSK cells from two control and three LysM-KI mice (7 to 15 weeks of age) using the Puregene kit (Qiagen). Genomic DNA from two IDH1/IDH2-mutant AML samples was obtained from previously reported, de-identified patient samples5. Two normal CD34+ bone marrow control samples were purchased from AllCells. Institutional review board approval was obtained at Weill Cornell Medical Center and this study was performed in accordance with the Helsinki protocols. DNA was isolated from each primary sample using the Puregene kit (Qiagen). The gDNA was then characterized using a modified reduced representation bisulphite sequencing (RRBS)27 approach called enhanced reduced representation bisulphite sequencing (ERRBS)18.
Generation of libraries
In brief 2.5–10 ng gDNA were digested with MspI. Digested DNA was isolated using a standard phenol-chloroform extraction followed by ethanol precipitation and resuspended in 10mMTris pH8.0. End-repair of digested DNA was performed using T4 DNA polymerase, Klenow DNA polymerase, T4 polynucleotide kinase, dNTPs in T4 DNA ligase buffer (New England Biolabs). After incubation the products were purified using QIAquick PCR purification columns (Qiagen). Adenylation was performed using Klenow fragment 3′ to 5′ exo minus (New England Biolabs) and dATP in Klenow buffer. Products were purified using MinElute PCR purification columns (Qiagen). Adenylated DNA fragments were ligated with pre-annealed 5-methylcytosine-containing Illumina adapters using T4 DNA ligase (New England Biolabs). Products were isolated using MinElute columns (Qiagen). Library fragments of 150–250 bp and 250–400 bp were gel-isolated using the QIAquick Gel Extraction kit (Qiagen). Bisulphite treatment was performed using the EZ DNA Methylation Kit (Zymo Research) with the incubation after the addition of CT conversion reagent being conducted in a thermocycler (Eppendorf MasterCycler). Purified products were subjected to PCR amplification using the FastStart High Fidelity PCR System (Roche). PCR products were isolated using AMPure XP beads. All amplified libraries underwent quality control using a Qubit 1.0 fluorometer and a QuantiT dsDNA HS Assay Kit for quantification (Invitrogen) as well as Bioanalyzer visualization (Agilent 2100 Bioanalyzer). The amplified libraries were sequenced on an Illumina Genome Analyzer II or HiSeq2000 per manufacturer’s recommended protocol for 50 bp single-end read runs. Image capture, analysis and base calling were performed using Illumina’s CASAVA 1.7.
Primary data analysis
For alignment of bisulphite treated reads and methylation calls, reads were filtered from the adaptor sequences using CUTADAPT software (http://code.google.com/p/cutadapt/). Adaptor sequence contamination usually occurs towards 3′ ends of some reads. The adaptor matching part of the read was removed if it aligned with the adaptor sequence at least 6 bp and had at most a 0.2 mismatch error rate. Reads were aligned to whole genome using the bismark alignment28 with a maximum of 2 mismatches in a directional manner and only uniquely aligning reads were retained. To call methylation score for a base position, we required that read bases aligning to that position have at least 20 phred quality score and the base position should have at least 10 × coverage. Only CpG dinucleotides that satisfied these coverage and quality criteria were retained for subsequent analysis. Percentage of bisulphite converted Cs (representing unmethylated Cs) and non-converted Cs (representing methylated Cs) were recorded for each C position in a CpG context.
Downstream data analysis
CpG islands, RefSeq genes and repeat sequences for the MM9 genome were downloaded from the UCSC genome browser29. CpG shores were defined as 1,000-bp flanking regions on upstream and downstream of a given CpG island. If a 1,000 bp shore overlapped with another island, then the shore was clipped so that its last base falls before the start of the overlapping CpG island. Similarly, if shores were overlapping they were merged into a single shore. In addition, the genome was partitioned into intergenic, intronic, exonic and promoter regions. Promoter regions were defined as the 2-kilobases window centred around the transcription start sites (TSS) of RefSeq genes. We classified CpG dinucleotides as promoter, intronic, exonic or intergenic based on their overlap with these predefined regions. In addition, we classified CpG dinucleotides as CpG island or shore overlapping.
Calculating differential methylation
Methylation values for genomic regions (intergenic, intronic, exonic and promoters, CpG islands and island shores) between different samples were compared by taking the mean methylation percentage of CpG dinucleotides overlapping those regions. To calculate the correlation between different samples and generate the appropriate scatter plots we required that in any given region at least 3 CpG dinucleotides were covered by reads in both control and KI samples. Testing for differential methylation was performed at both the single-base and predefined region levels. For the base-level comparison, coverage on base positions for each sample was required whereas for the region-level comparison we required at least 3 covered bases on all samples. The number of methylated and unmethylated Cs aligning to each base/region were counted and compared across samples. To determine significant differential methylation between two groups of samples we applied logistic regression and the likelihood ratio test was used. Observed P values were adjusted with the q value method30.
Pathway analysis of differentially methylated regions
Differentially methylated cytosines were annotated to the nearest RefSeq gene and pathway analysis performed using the Ingenuity Pathway Analysis software (Ingenuity Systems) and the Ingenuity knowledge database as reference.
mRNA expression profiling
RNA was extracted from LSK cell pellets from young mice using TRIzol (Invitrogen) followed by on-column DNase digest and purification using the RNeasy Micro Kit (Qiagen).RNA concentration and quality for all samples was assessed using the RNA 6000 Pico Kit (Agilent). Five nanograms of RNA from each of the samples as well as one Universal Mouse Reference RNA (Stratagene/Agilent) were amplified using the WT-Ovation Pico RNA amplification System Version 1.0 to generate cDNA (Nugen). cDNA samples were biotin labelled according to the Nugen Illumina protocol and hybridized to Mouse WG-6 V2.0 BeadChips which were hybridized, washed and stained according to the manufacturer’s protocol (Illumina) and scanned on the iScan (Illumina). The data files were quantified in GenomeStudio Version 2010.2 (Illumina).
Array processing and data analysis
Microarray data were quality-checked for technical outliers before analysis using the LUMI package31 and quality control metrics in the R/Bioconductor statistical software package (v2.14.1). All samples were of sufficient quality to proceed. Data were imported into the Genespring (v11.5.1, Agilent) software framework for statistical testing and visualization and normalized using a quantile normalization function followed by median centring and logging (log base 2). Of the 45,281 probes, 9,437 did not meet a minimum threshold of expression across both sample groups as defined by being higher than the 20th percentile of measured expression in at least 80 percent of the samples in either the wild-type or KI groups. To assess statistical differences between control and KI sample groups a straightforward t-test (P < 0.05) followed by a fold-change cut-off of 1.5 was used (Supplementary Table 3). False discovery rate (FDR)-corrected32 t-statistics as well as FDR-corrected moderated t-statistics33 were assessed but did not yield any significant results, probably as a result of the increased variability between samples due to the RNA amplification and the small number of replicates in each group. In total, 240 probes were found to be upregulated in KI samples along with 108 downregulated ones. Corrected hypergeometric test statistic following ref. 34 (FDR q < 0.03) was used to find genes in each list that were enriched for gene ontology categories (Supplementary Table 4). To visualize the differences that were found between sample groups, we performed a two-way hierarchical clustering using Pearson-centred distance metrics under average-linkage tree building rules (Supplementary Fig. 8).
Supplementary Material
Acknowledgements
We thank the Animal Research Colony (ARC) at the Ontario Cancer Institute for mouse care; I. Ng, A. Shahinian, J. Sylvester and S. McCracken for administrative and organizational expertise; M. Bailey and J. Tsao for technical assistance; F. Tong and R. Nayyar for assistance with flow cytometric analysis and sorting; the Weill Cornell Medical College (WCMC) Epigenomics Core Facility for technical help and expertise; G. Melino, D. Green, M. Minden, H. Chang and P. Lang for helpful discussions; J. Thomsen for figure layout and M. Saunders for scientific editing. C.B.K. and D.B. were supported in part by a Feodor-Lynen Postdoctoral Research Fellowship from the Alexander-von-Humboldt-Foundation, Germany. D.B. and A.B. were supported in part by a Fellowship from the German Research Foundation (DFG). J.C.M. is supported by a National Institute of Health grant (NIH R01AI081773) and is a Damon Runyon-Rachleff Innovation Awardee supported by the Damon Runyon Cancer Research Foundation (DRR-09-10). P.S.O. holds a Canada Research Chair in Autoimmunity and Tumor Immunity. M.E.F. is supported by the Leukemia & Lymphoma Society Special Fellow Award and a Doris Duke Clinical Scientist Development Award. A.M. is supported by an LLS SCOR grant (7132-08), a Burroughs Wellcome Clinical Translational Scientist Award and a Starr Cancer Consortium grant (I4-A442). J.-C.Z.-P. is supported by a Canada Research Chair in Developmental Immunology. This work was supported by grants from the Canadian Institutes of Health Research (CIHR) and the Ontario Ministry of Health and Long Term Care to T.W.M., and a program grant from the Terry Fox Foundation to P.S.O., J.-C.Z.-P. and T.W.M. Please note that the views expressed do not necessarily reflect those of the OMOHLTC.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions C.B.K., M.S. and T.W.M. initiated the project. M.S. designed the conditional IDH1(R132H)-KI mouse and generated it, as well as the IDH1(R132H)-KI embryonic stem cell clones with the help of S.S., S.M.S., A.W., J.H. and A.Y.-T.; C.B.K. designed and organized experiments involving the hematopoietic system in consultation with T.W.M., M.E.F., A.M., J.-C.Z.-P., G.R., P.S.O., C.V. and D.L.B.; C.B.K. performed the flow cytometry analyses, bone marrow transfers, cell sorting, RNA extraction and real-time PCR analyses, immunoblotting, colony formation assays and serial plating experiments. J.C.M. performed the LC-MS analyses. E.F.L., D.B., A.B. and I.S.H. helped with flow cytometric analyses and BM transfers. R.H. performed the OP9/OP9-DL1 embryonic stem cell in vitro differentiation experiments. W.Y.L. helped with flow cytometric analyses and cell culture experiments. M.E.F. and A.M. performed the DNA methylation experiments and analysed the data. C.V. performed the mRNA expression microarray experiments and analysed data. C.B.K. and T.W.M. wrote the manuscript with the help of A.B. and D.B. All authors discussed the results extensively and agree with the conclusions presented in the manuscript.
The microarray and sequencing data have been deposited in the Gene Expression Omnibus of the National Center for Biotechnical Information with the accession numbers GSE38589 and GSE38687.
The authors declare no competing financial interests.
References
- 1.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clausen BE, et al. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 8.Ye M, et al. Hematopoietic stem cells expressing the myeloid lysozymegene retain long-term, multilineage repopulation potential. Immunity. 2003;19:689–699. doi: 10.1016/s1074-7613(03)00299-1. [DOI] [PubMed] [Google Scholar]
- 9.Goardon N, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011;19:138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Holmes R, Zúñiga-Pflücker J-C. The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harb. Protoc. 2009 doi: 10.1101/pdb.prot5156. http://dx.doi.org/10.1101/pdb.prot5156. [DOI] [PubMed] [Google Scholar]
- 11.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 12.Ito K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nature Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 13.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Callens C, et al. Targeting iron homeostasis induces cellular differentiation and synergizes with differentiating agents in acute myeloid leukemia. J. Exp. Med. 2010;207:731–750. doi: 10.1084/jem.20091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eliasson P, Jönsson JI. The hematopoietic stem cell niche: low in oxygen but a nice place to be. J. Cell. Physiol. 2010;222:17–22. doi: 10.1002/jcp.21908. [DOI] [PubMed] [Google Scholar]
- 16.Zhao S, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akalin A, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. doi: 10.1371/journal.pgen.1002781. (in the press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell C, et al. Signal control of hematopoietic stem cell fate: Wnt, Notch, and Hedgehog as the usual suspects. Curr. Opin. Hematol. 2008;15:319–325. doi: 10.1097/MOH.0b013e328303b9df. [DOI] [PubMed] [Google Scholar]
- 20.Heidel FH, et al. Self-renewal related signaling in myeloid leukemia stem cells. Int. J. Hematol. 2011;94:109–117. doi: 10.1007/s12185-011-0901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klinakis A, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Söderberg SS, et al. Complex and context dependent regulation of hematopoiesis by TGF-β superfamily signaling. Ann. NY Acad. Sci. 2009;1176:55–69. doi: 10.1111/j.1749-6632.2009.04569.x. [DOI] [PubMed] [Google Scholar]
- 23.Patel KP, et al. Acute myeloid leukemia with IDH1 or IDH2 mutation: frequency and clinicopathologic features. Am. J. Clin. Pathol. 2011;135:35–45. doi: 10.1309/AJCPD7NR2RMNQDVF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patnaik MM, et al. Differential prognostic effect of IDH1 versus IDH2mutations in myelodysplastic syndromes: a Mayo Clinic Study of 277 patients. Leukemia. 2012;26:101–105. doi: 10.1038/leu.2011.298. [DOI] [PubMed] [Google Scholar]
- 25.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nature Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajad SU, et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storey JD, Tibshirani R. Statistical significance for genome wide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du P, et al. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B. 1995;57:289–300. [Google Scholar]
- 33.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 34.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.