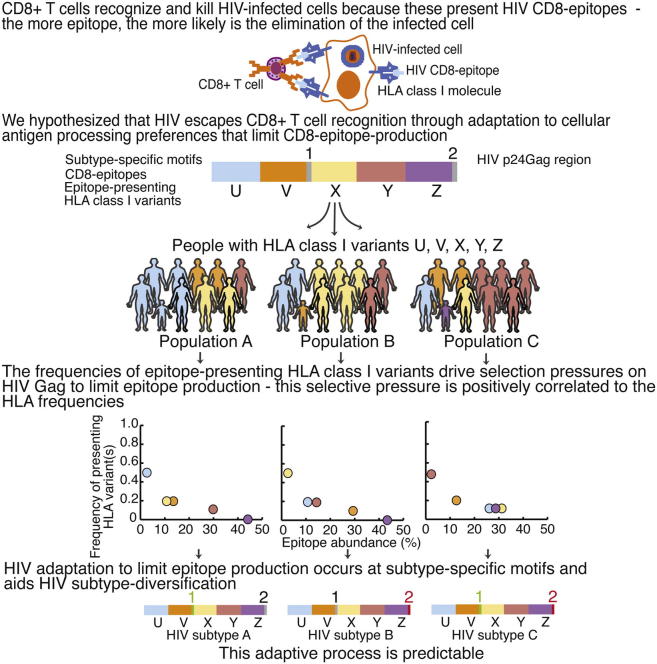
Summary
The recent HIV-1 vaccine failures highlight the need to better understand virus-host interactions. One key question is why CD8+ T cell responses to two HIV-Gag regions are uniquely associated with delayed disease progression only in patients expressing a few rare HLA class I variants when these regions encode epitopes presented by ∼30 more common HLA variants. By combining epitope processing and computational analyses of the two HIV subtypes responsible for ∼60% of worldwide infections, we identified a hitherto unrecognized adaptation to the antigen-processing machinery through substitutions at subtype-specific motifs. Multiple HLA variants presenting epitopes situated next to a given subtype-specific motif drive selection at this subtype-specific position, and epitope abundances correlate inversely with the HLA frequency distribution in affected populations. This adaptation reflects the sum of intrapatient adaptations, is predictable, facilitates viral subtype diversification, and increases global HIV diversity. Because low epitope abundance is associated with infrequent and weak T cell responses, this most likely results in both population-level immune evasion and inadequate responses in most people vaccinated with natural HIV-1 sequence constructs. Our results suggest that artificial sequence modifications at subtype-specific positions in vitro could refocus and reverse the poor immunogenicity of HIV proteins.
Graphical Abstract
Highlights
-
•
HLA class I variants drive selection pressures on Gag to limit epitope production
-
•
The strength of the selective pressure is positively correlated to HLA frequencies
-
•
HIV adaptation to limit epitope production occurs at subtype-specific HIV motifs
-
•
HIV adapts in a predictable way to HLA frequencies in newly infected populations
CD8+ T cell responses against HIV-1 effectively delay disease progression in a minority of patients with relatively rare HLA variants but are ineffective in most. Here, Tenzer et al. identify fundamental HIV-1 adaptation to the conserved human antigen-processing machinery that feeds epitopes to HLA. This adaptation occurs at subtype-specific motifs, facilitates subtype diversification, is predictable, and results in CD8 epitope abundances that correlate inversely with the HLA allele frequencies in affected populations. Thus, HIV vaccine immunogenicity might be increased by unnatural substitutions at subtype-specific motifs.
Introduction
Because a safe and effective HIV-1 vaccine remains an elusive goal, the basis for current vaccine strategies must be revisited. Although the immune system never successfully clears HIV in natural infection, the potential to contain the virus for decades through cytotoxic T cell (CTL) responses has been demonstrated in a minority of patients with rare human leukocyte antigen (HLA) variants, particularly HLA-B∗2705, -B∗5701, -B∗5703, and -B∗5801 (Goulder and Walker, 2012). The respective frequencies of these HLA variants are 0%–14%, 0%–11%, 0%–3%, and 0%–2% in whites and 0%–5%, 0%–6%, 0%–8%, and 0%–14% in Africans (Gonzalez-Galarza et al., 2011). These HLA molecules present key fragments (CD8 epitopes) of the HIV-1 capsid protein p24 Gag on the surface of infected cells to enable immune recognition and killing by CTL (Dahirel et al., 2011, Dinges et al., 2010, Streeck et al., 2007). However, despite progress (Elahi et al., 2011, Pereyra et al., 2010), the reason these particular CTL responses are strongly linked to delayed disease progression is not fully understood.
In addition to these protective CTL responses, there is an association between CTL responses against p24 Gag and lower mean viral loads in patients (Kiepiela et al., 2007). HLA-B rather than the less diverse HLA-A or HLA-C restricts most of these responses although the underlying mechanism is unclear (Goulder and Walker, 2012, Kiepiela et al., 2004). The current paradigm is therefore that relatively few HLA variants are associated with reductions in viral load and so impose a significant selective pressure on HIV (Matthews et al., 2008).
Antigen processing is one crucial step in the pathway responsible for HLA presentation of HIV epitopes to CTL. It is a multistep process, where viral proteins synthesized in infected cells are first degraded by cytosolic proteasomes. The proteasomes appear in constitutive and interferon (IFN)-γ-inducible immunoproteasomal forms; the latter is normally expressed in lymphoid and antigen presenting cells but can be induced in other cell types during HIV infection (Kloetzel, 2001, Tenzer et al., 2004, Toes et al., 2001). Proteasomal cleavage products (epitope precursors) might be further digested by cytosolic peptidases before the transporter-associated-with-antigen-processing (TAP) transfers them into the endoplasmatic reticulum (ER). Here, they undergo N-terminal trimming by the ER aminopeptidases (ERAP1,2) and are loaded onto any “restricting” HLA class I molecules with suitable anchors.
All CTL responses can select for mutations within or flanking CD8 epitopes that allow HIV to escape CTL recognition. These mutations may affect HLA binding, T cell receptor (TCR) contact sites, and/or antigen processing (Draenert et al., 2004, Goulder and Walker, 2012, Iversen et al., 2006, Tenzer et al., 2009, Zhang et al., 2012). Escape mutations might confer reduced viral fitness by coincidentally increasing epitope processing (Tenzer et al., 2009) and/or reducing viral replication or infectivity (Goulder and Walker, 2012). Such mutations typically revert following transmission to HLA-mismatched recipients (“reverting mutations”), whereas mutations with no fitness cost persist (“nonreverting mutations”) (Matthews et al., 2008).
The most effective CTL responses select for reverting escape mutations (Matthews et al., 2008), and the accumulation of these mutations at the viral population level due to factors such as compensatory mutations might therefore affect HIV control (Kawashima et al., 2009). Nonreverting escape mutations are not associated with viral load reductions but might accumulate and eventually replace the wild-type population. This evasion of immune responses is so advantageous to HIV that positive selection of escape mutations drives HIV evolution within hosts (Rambaut et al., 2004).
Following HIV transmission, ∼35% of Gag substitutions in the recipient is associated with HLA-mediated selection, and ∼70% of these are reversions (Brumme et al., 2008). However, the extent to which HIV evolves at the population level in response to immune pressures is a matter of debate because of other influences, such as the founder effect (the random initiation of epidemics in uninfected areas by a single or a few individuals’ HIV lineages), transmission bottlenecks that reduce genetic diversity, and variation in the rates of partner exchange involving sexual transmission. Bioinformatics studies have shown that many polymorphisms in HIV are associated with particular host HLA class I alleles (Bhattacharya et al., 2007); however, the impact of these changes on antiviral immune responses is often unclear, and evolution among hosts is dominated by genetic drift (Rambaut et al., 2004). Globally, HIV-1 forms geographically distinct clusters, known as subtypes, which are labeled alphabetically. These subtypes are believed to be caused by founder effects (Rambaut et al., 2004).
We and others have previously demonstrated that the amount of epitopes made following in vitro proteasomal digestions correlate with the magnitude and frequency of in vivo CTL responses (Lazaro et al., 2009, Schmidt et al., 2012, Tenzer et al., 2009), and studies have shown that epitope-specific CTL recognition increases with increasing epitope production and/or concentration (Almeida et al., 2007, Ranasinghe et al., 2011, Zhang et al., 2012). Because of these correlations, HIV sequence combinations that minimize or eliminate CD8 epitope production through adaptation to conserved antigen-processing preferences (Paulsson, 2004) would potentially be a very efficient immune escape mechanism if they were to occur. Here, we hypothesized that HIV adaptation to antigen-processing preferences might limit CD8 epitope production at the viral population level in a manner dependent on the HLA allele frequency within a host population, thereby reflecting the sum of all intrapatient adaptations and their transmissibility.
Results
HIV CD8 Epitope Abundance Correlates Inversely with Presenting HLA Frequencies in Infected Populations
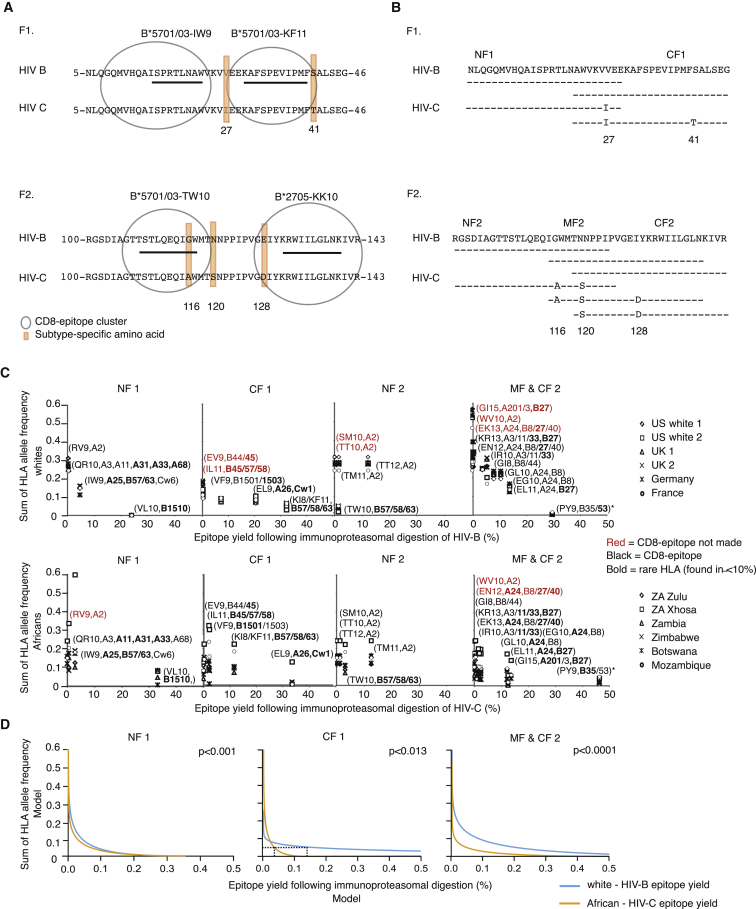
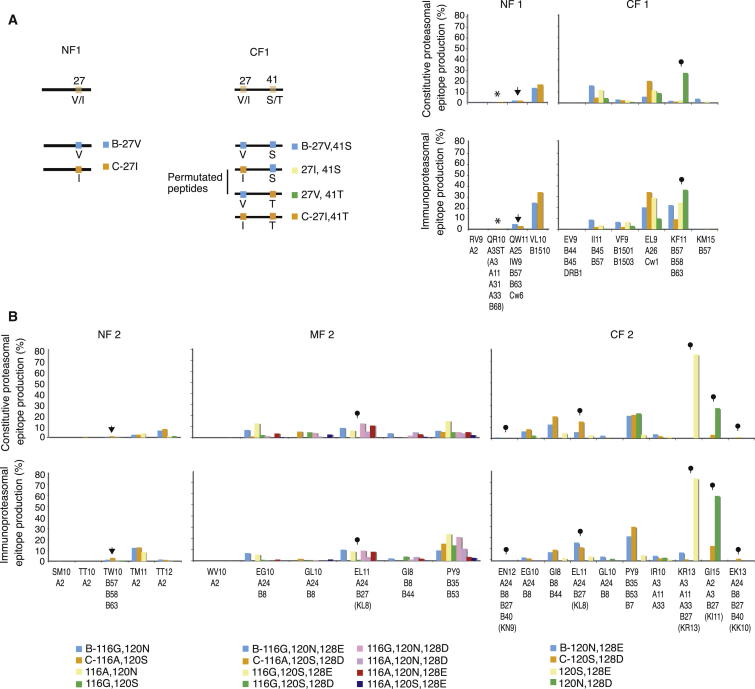
To investigate the regulation of CD8 epitope production at the viral population level, we studied the proteasomal processing of HIV subtypes B (HIV B) and C (HIV C); HIV B predominates in Western Europe and the US and accounts for approximately 11% of all infections worldwide, whereas HIV C prevails in sub-Saharan Africa and is responsible for approximately 50% of infections (Buonaguro et al., 2007). We focused on p24 Gag and concentrated on the two conserved regions that dominate the clinically effective HIV-specific CTL response in patients infected by either HIV subtype (fragments 1 [F1] and 2 [F2], Figure 1A; Goulder and Walker, 2012).
Figure 1.
Overview of the HIV p24 Gag Regions Analyzed
(A) Outline of p24 Gag fragment 1 (F1, amino acids 5–46, HIVHXB2 numbering) and fragment 2 (F2, amino acids 100–143) that highlights the overlapping CTL epitope clusters and subtype-specific positions (27, 41, 116, 120, and 128: orange boxes). The four epitopes presented by protective HLA class I molecules are underlined; all epitopes are listed in Table S1.
(B) Outline of the overlapping 25-mer peptides used as proteasomal substrates using the HIV B and HIV C consensus sequences as examples. F1 was divided into two parts (NF1 [N-terminal F1] and CF1 [C-terminal F1]), and F2 was divided into three parts (NF2, MF2 [Middle F2], and CF2).
(C) Scatterplots of the relationship between the allelic frequency of a restricting HLA variant (or the sum of HLA variants if the CD8 epitope and/or epitope precursor is presented by more than one HLA molecule or if it contains more than one epitope presented by different HLA variants) in six white and African populations (infected primarily with HIV B or HIV C, respectively) and “epitope yield” (i.e., the abundance of individual epitopes and/or epitope precursors represented as percentages of all peptide fragments) following 4 hr of immunoproteasomal digestion of either the consensus HIV B or HIV C sequence. KI8 and KF11 are grouped together as they are two forms of the same epitope. HLA restriction details are shown in Table S1. Red epitopes and their restricting HLA variants signify epitopes that are not produced but are made following the processing of either the other subtype or one of the sequence combinations (Table 1). Asterisk, the HLA allelic frequency for the PY epitope is set at zero in HIV-B and HIV-C infection because of intraepitope CTL-escape mutations (Matthews et al., 2012, Sabbaj et al., 2003). The figure represents one of three independent experiments.
(D) Multilevel analysis with the three-way interaction of fragment (NF1, CF1, or MF2&CF2), the presenting HLA allele frequency and ethnic group on epitope yield. NF2 could not be analyzed due to differential epitope processing and restriction by only two HLA class I variants.
The key p24 regions contain clusters of partially overlapping CD8 epitopes, including the only four CD8 epitopes that are strongly associated with delayed disease progression and 52 other epitopes that have no association with disease development and are rarely recognized (Table S1) (Dinges et al., 2010, Goulder and Walker, 2012, Streeck et al., 2007). Three of the beneficial epitopes are presented by HLA-B∗5701 and -B∗5703 (15IW923, 30KF1140, and 108TW10117), and one is presented by HLA-B∗2705 (131KK10140); KF11 and TW10 may also be presented by HLA-B∗5801 and -B∗63, and IW9 is also presented by HLA-A∗25, -B∗63, and -Cw∗06 (Figure 1A; Table S1). KF11 also encodes a shorter epitope form, 30KI837 (Goulder et al., 2000), presented by the same HLA variants. The nonprotective epitopes are presented by ∼30 HLA class I variants, and, similar to three of the protective epitopes, many can be presented promiscuously by more than one HLA variant (Frahm et al., 2007). These regions also contain five subtype-specific substitutions that are situated either between or within the epitope clusters (Figure 1A; [subtype B↔C] 27V↔I and 41S↔T in F1; 116G↔A, 120N↔S, and 128E↔D in F2).
To test the hypothesis that HIV adaptation to escape CTL responses may limit the production of viral epitopes in a manner dependent on the HLA class I allelic frequency within a host population, we first generated proteasomal digestion data. We designed overlapping HIV B and HIV C peptides (Figure 1B), digested these with purified human constitutive proteasomes and immunoproteasomes, and analyzed the resulting fragments by mass spectrometry as in Tenzer et al. (2009).
Whereas proteasomal digestion produces the carboxyl (C)-terminal end of most epitopes, several aminopeptidases in the cytosol and the ER can trim the amino (N)-terminal peptide extensions (van Endert, 2011). Therefore, we aligned the digestion fragments with reported CD8 epitopes and defined an epitope precursor as a peptide fragment that contained the correct C terminus of an epitope but could be extended by one or several residues at the N terminus. Although most known CD8 epitopes are defined as “optimal epitopes,” i.e., the artificially synthesized epitope-peptide form that elicits the greatest CTL responses in vitro, we found that most epitopes were processed as epitope precursors with N-terminal extensions (Table S1). For each epitope, the intensities of the epitope precursor(s) and the optimal epitope were combined for the calculation of the total epitope production of a given epitope (“epitope yield”). Occasionally, two or more epitopes, presented by different HLA class I variants, shared the same C-terminal end but differed with respect to the start of the N terminus; such epitope-precursor fragments would be analyzed on their own.
We next used scatterplots to illustrate the relationship between HIV B or HIV C epitope yields and the HLA allelic frequencies of epitope-presenting HLA variants in six white and six sub-Saharan populations predominantly infected with HIV B or HIV C, respectively (Figure 1C). If a given epitope-precursor fragment contained a promiscuously presented epitope and/or more than one epitope, we summed the frequencies of all HLA variants that could present the epitope(s) encoded by that epitope precursor. The abundance of epitope precursor fragments containing the protective epitopes IW9, KF11/KI8, or KK10 correlated with the summed frequencies of the presenting HLA variants, whereas the production of TW10 was less than expected considering that the HLA-driven selective pressures on TW10 and KF11 are similar (Figure 1C). However, TW10 differs in two important respects from KF11: first, most of TW10 is included in epitopes presented by the common HLA-A∗02 variant, and the combined selective pressures may further reduce TW10 processing, and second, the cytoplasmatic peptidase stability of TW10 (t1/2 = 31 min) is far greater than that of KF11 (t1/2 = 13 s) and most other epitopes (Lazaro et al., 2011). Because of this, the selective pressure to reduce the production of TW10 might be stronger than for KF11.
Subsequently, we took within-population correlation into account by fitting a multilevel linear model to examine the influence of the presenting HLA allelic frequencies, HIV region (NF1, CF1, MF2&CF2), and ethnic group (Africans, whites) on epitope yield (Table S2). This analysis demonstrated a significant three-way interaction among fragment, HLA frequency, and ethnic group on epitope yield. A strong relationship between epitope yield and HLA allelic frequency was found for NF1, CF1 and MF2&CF2 (Figure 1D), but, whereas similar variation in HLA frequency had similar effects on epitope yields in NF1 and MF2&CF2 regardless of ethnic group, a significant difference between ethnic groups was found with respect to CF1 (p < 0.0001). As shown in the graph (Figure 1D, CF1), identical HLA-mediated selection pressures in Africans and whites were typically associated with lower HIV C than HIV B epitope yields, especially at low HLA allelic frequencies (p < 0.0001). Specifically, at an HLA allelic frequency of 0.05 (equivalent to HLA variants found in 10% of the population), the epitope yields from HIV C were only approximately 25% of those of HIV B (Figure 1D, CF1, dashed line). The lower epitope yields might reflect a combination of a greater adaptive potential of this region in HIV C, differences between African and white HLA alleles, a bias in the database toward HIV B epitopes, and/or the longer history of the HIV epidemic in Africa. We next fitted the same model to epitope yields from the constitutive proteasomal digestions and found similar patterns (t test, p = 0.39–0.95) (Table S2).
As a result of the HLA-mediated adaptation of HIV to proteasomal preferences, the CD8 epitopes presented by rare HLA variants are produced in greater quantities than those presented either by common variants or by a combination of rare and common variants and might therefore be more likely to prime (Faroudi et al., 2003) and/or be recognized by CTL (Purbhoo et al., 2004). Collectively, our data provide compelling evidence that these clinically important p24 HIV Gag regions adapt to proteasomal preferences at the population level in response to selection pressures that are imposed by all epitope-restricting HLA class I variants in an HLA frequency-dependent manner.
Processing Outcomes Are Supported by TAP Binding and ERAP Trimming Preferences
To examine how TAP transport and ERAP trimming preferences might affect the predicted epitope presentation and recognition at the cell surface, we tested the subset of epitope precursors with the protective IW9, KF11, KI, or TW10 motifs (Figure 2A). To induce a CTL response, the epitope-precursor fragments must first be transported by TAP into the ER, and, because TAP transport rate is determined by binding, not translocation (Gubler et al., 1998), we measured the normalized TAP affinities of the precursors (Figure 2B). TAP binding affinity was sequence but not length specific, given that TAP sometimes bound long epitope precursors (>16 amino acids) better than shorter peptides containing only parts of the longer sequence (e.g., AF19 versus VF14 and VF15, Figure 2B). Overall, most precursors were transported well, especially those produced in the greatest abundance (affinity <50).
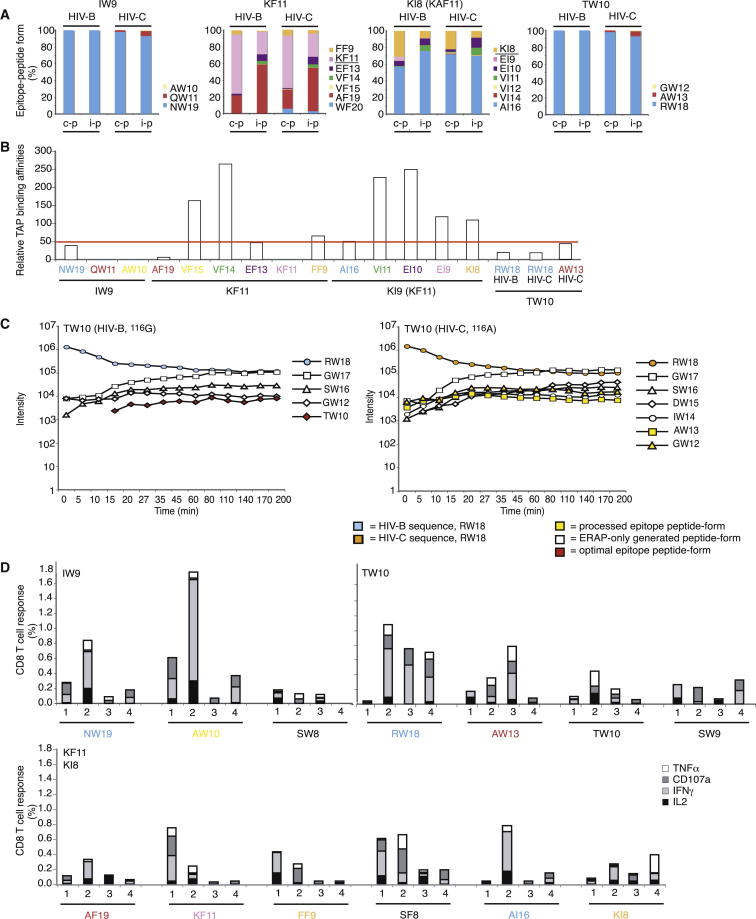
Figure 2.
TAP Binding, ERAP Trimming, and CD8+ T Cell Analyses
(A) Overview of the abundance of all IW9, KF11, KI8, and TW10 epitope forms produced after 4 hr constitutive (c-p) or immunoproteasomal (i-p) digestion.
(B) TAP binding affinity of the IW9, KF11, KI8, and TW10 epitope forms generated by proteasomal digestion. GW12 could not be tested because it was insoluble and WF20, VI14, and VI12 were not tested. The epitope forms are colored as in (A). The results represent one of at least two independent experiments.
(C) ERAP1,2 digestion of the TW10 epitope precursor RW18 from HIV B and HIV C. The result represents one of three independent experiments.
(D) PBMCs from four untreated HIV-1-infected patients with HLA-B∗5701 (Table S3) were stimulated with a subset of IW9, KF11, KI8, and TW10 HIV B epitope-precursor peptides because of limited sample availability. IL-2, IFN-γ, TNF-α, and CD107a responses were analyzed using flow cytometry. The percentages of responding CD8+ T cells are shown after background subtraction, and each peptide was tested two to three times. The epitope-peptide form is indicated along the x axis, and the optimal epitope is shown in the top-left corner of the graph. The epitope forms made by the proteasome are colored as in (A) and (B).
Following TAP transport, epitope precursors associate with HLA molecules, and the N-terminal part of the peptide may be trimmed by ERAP before or after binding (van Endert, 2011). We found that the longer epitope precursors were trimmed more slowly. Furthermore, internal subtype-specific amino acid differences could affect N-terminal trimming rates and thus affect the repertoire of trimmed epitope-containing fragments and the production of optimal epitopes (Figures 2C and S1). Specifically, the optimal TW10 epitope was observed only following ERAP trimming of the RW18 precursor from HIV B, not HIV C, because of a single amino acid variation at position 9 within the epitope (position 17 within the 18-mer precursor, Figure 2C), and the optimal KF11 epitope was undetectable due to rapid degradation (Figure S1).
However, functional analyses of CD8+ T cell responses in four untreated HIV-positive patients with HLA-B∗5701 (Table S3) demonstrated that the patients could recognize different length variants of the same epitope and that recognition often resulted in polyfunctional cytokine responses (Figure 2D). Taken together, our TAP, ERAP, and CTL-recognition data suggest that proteasomal digestion preferences often produce fragments that accommodate TAP binding preferences, that most epitope precursors are transported well into the ER, that a fraction is trimmed to shorter epitope precursors and optimal epitopes and that patients can recognize a variety of epitope-peptide forms. Thus, the observed differences in proteasomal epitope abundance appear to reflect differences in predicted epitope abundances on the cell surface.
Intrahost HIV Adaptation to Antigen Processing Decreases Epitope Production
Next, we investigated whether the inverse correlation between the HIV B and HIV C CD8 epitope yields and the frequency of the presenting HLA variants in the infected populations reflected the sum of individual adaptations within patients. To investigate intrahost viral adaptation, we combined bioinformatics analyses of HIV sequences from HLA-typed patients infected with either HIV B or HIV C with processing analyses of 22 peptides containing all of the possible permutations of the consensus sequences of the two subtypes (Figure 3). Each variant differed by only one amino acid from the next, and, whereas some substitution patterns were observed at a high frequency in vivo, other patterns were rare (Table 1).
Figure 3.
Epitope Production after Proteasomal Digestion of F1 and F2 Variants
(A) Overview of the NF1 and CF1 subtype-specific motifs (orange boxes), the HIV B and HIV C consensus sequences, and all combinations of the subtype-specific amino acids with a color-code key. The abundances of individual epitopes were represented as percentages of all peptide fragments following constitutive and immunoproteasomal digestion, and the epitopes are shown with the restricting HLA variants. RV9 and EV9 were produced in amounts too small to be easily visible in this figure. The results represent one of three independent experiments. F1 contains the protective epitopes IW9 (NF1) and KF11 (CF1) and overlapping epitopes (Figure 1A, Table S1). Asterisk, QR10; arrow, QW11 (IW9); pin, KF11.
(B) As in (A) except that all F2 peptide variants are shown in Table 1. SM10, TT10, EN12, and WV10 were produced in amounts too small to be easily visible in this figure. F2 contains the protective TW10 epitope (NF2) and several KK10 epitope forms (MF and CF2; in brackets) and overlapping epitopes (Table S1). Arrow, TW10; pins, KK10 epitope forms.
Table 1.
Frequencies of Subtype-Specific Amino Acid Combinations in HIV-B- and HIV-C-Infected Individuals and an Outline of the Overlapping 25-mer Peptides Used as Proteasomal Substrates
| Fragment | Subtype | Subtype-Specific Motif | Amino Acid Sequence | HIV B (%) | HIV C (%) |
|---|---|---|---|---|---|
| NF1 | B | 27V | NLQGQMVHQAISPRTLNAWVKVVEE | 23 | 3 |
| C | 27I | NLQGQMVHQAISPRTLNAWVKVIEE | 8 | 35 | |
| CF1 | B | 27V, 41S | AWVKVVEEKAFSPEVIPMFSALSEG | 44 | <1 |
| 27I, 41S | AWVKVIEEKAFSPEVIPMFSALSEG | 13 | 1 | ||
| 27V, 41T | AWVKVVEEKAFSPEVIPMFTALSEG | 11 | 6 | ||
| C | 27I, 41T | AWVKVIEEKAFSPEVIPMFTALSEG | 5 | 63 | |
| NF2 | B | 116G, 120N | RGSDIAGTTSTLQEQIGWMTNNPPI | 36 | <1 |
| 116A, 120N | RGSDIAGTTSTLQEQIAWMTNNPPI | 5 | 4 | ||
| 116G, 120S | RGSDIAGTTSTLQEQIGWMTSNPPI | 10 | 1 | ||
| C | 116A, 120S | RGSDIAGTTSTLQEQIAWMTSNPPI | 1 | 14 | |
| MF2 | B | 116G, 120N, 128E | IGWMTNNPPIPVGEIYKRWIILGLN | 29 | <1 |
| 116A, 120N, 128E | IAWMTNNPPIPVGEIYKRWIILGLN | 5 | 1 | ||
| 116G, 120S, 128E | IGWMTSNPPIPVGEIYKRWIILGLN | 8 | <1 | ||
| 116G, 120N, 128D | IGWMTNNPPIPVGDIYKRWIILGLN | <1 | <1 | ||
| 116A, 120S, 128E | IAWMTSNPPIPVGEIYKRWIILGLN | 1 | 3 | ||
| 116A, 120N, 128D | IAWMTNNPPIPVGDIYKRWIILGLN | <1 | 4 | ||
| 116G, 120S, 128D | IGWMTSNPPIPVGDIYKRWIILGLN | <1 | <1 | ||
| C | 116A, 120S, 128D | IAWMTSNPPIPVGDIYKRWIILGLN | <1 | 16 | |
| CF2 | B | 120N, 128E | TNNPPIPVGEIYKRWIILGLNKIVR | 36 | 3 |
| 120S, 128E | TSNPPIPVGEIYKRWIILGLNKIVR | 12 | 5 | ||
| 120N, 128D | TNNPPIPVGDIYKRWIILGLNKIVR | 2 | 6 | ||
| C | 120S, 128D | TSNPPIPVGDIYKRWIILGLNKIVR | <1 | 22 |
The first p24 fragment was divided into two parts (NF1 and CF1), and the second was divided into three parts (NF2, MF2, and CF2). The underlined amino acids are specific to HIV C.
We used subtype-permutated peptides because sequence variations in both p24 regions usually occur at the subtype-specific positions (Figure S2) and because changes typically occur to the preferred amino acid in the other subtype (Figure S3). In the minority of patients able to present the protective IW9, KF11, TW10, or KK10 epitopes, additional variation could sometimes be found at positions associated with well-described CTL-escape mutations (e.g., 14A→P [IW9], 110T→N [TW10], Table S4).
We compared variations in epitope-abundance for every epitope in F1 and F2 with the specific amino acid that was selected for at subtype-specific positions in HIV isolated from patients with HLA variants able to present each epitope (Table S4). The consensus HIV B (or HIV C) sequence was typically observed in carriers of common HLA variants (e.g., HLA A∗02, HLA A∗03, HLA B∗07, and HLA B∗44) and was the sequence combination associated with the minimal generation of presentable epitopes.
In contrast, in most HIV-B- or HIV-C-infected individuals with rare HLA variants, we found one or more substitutions at subtype-specific positions that disrupted the HIV consensus sequence. Because of these substitutions, the abundance of the epitopes that these HLA variants could potentially present was reduced relative to that found following the processing of the consensus sequences. These substitution dynamics could deviate within HLA suballeles; for example, in HIV-B-infected patients with HLA-B∗5701 or -B∗5703, which present the same three protective epitopes (IW9, KF11 and TW10), the selection at subtype-specific positions either concurred (1/4; 41S→T) or diverged (3/4; 27V→I, 116G→A, 120N→S; Table S4).
Divergent selection could result in quite large differences as, e.g., 120S was found in over 80% of HIV B sequences from patients with HLA-B∗5701, but in none with HLA-B∗5703 (Figures 4 and S4). As a result of these differences, the proteasomal production of IW9 and TW10 will likely be lower in patients with HLA B∗5701 than in those with HLA B∗5703, and the greater epitope abundance in the latter group may lead to stronger CTL responses and help to explain the higher frequency of associated flanking and intraepitope CTL-escape mutations in HIV from these patients (Table S4; 14A→P: HLA B∗5701, 2%; HLA B∗5703, 41%; 110T→N: HLA B∗5701, 19%; HLA B∗5703, 41%).
Figure 4.
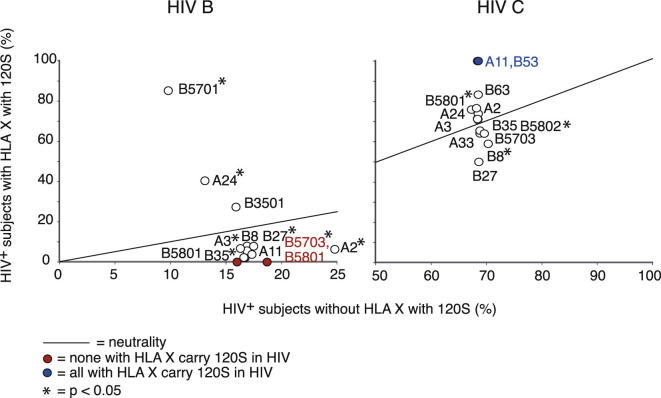
Intrahost Selection for the Subtype-Specific 120N→S Substitution
Analyses of intrahost selection for the subtype-specific HIV 120N (HIV B) → 120S (HIV C) substitution in HIV-B- and HIV-C-infected subjects with HLA variants restricting CD8 epitopes in F2 (all subtype-specific amino acid substitution analyses are presented in Figure S4 and Table S4). The circles represent the percentages of 120S in HIV from infected individuals who carry a specific HLA variant (y axis: 120S +, HLA X+) and the percentage of 120S in infected individuals without this HLA variant (x axis: 120S +, HLA X−). Therefore, the data illustrate the extent to which a specific HLA variant is associated with HIV evolution at position 120; specifically, circles above the line suggest positive selection, circles on the line suggest no selection, and circles below the line suggest a negative selection of 120S in patents with HLA X; Fisher’s exact test is used to estimate significance and asterisks indicate that the two-tailed p value is <0.05. Fewer than three HIV-B-infected patients carried HLA-B∗63, and an insufficient number of sequences were available from patients with HLA-B∗5801 and from HIV-C-infected patients with HLA-A∗11, -B∗53, -B∗63, and -B∗27 to test for significance. The frequencies of 120S in HIV B and HIV C from patients without HLA variants that could present epitopes from F2 were 6% and 72%, respectively.
Some epitope precursors contained overlapping epitopes that were restricted by more than one HLA variant. One example is the in vitro defined optimal KK10 epitope, which is presented by the protective and rare HLA B∗2705 molecule (whites, 0%–12%; absent in most African and Asian populations). This 10-mer epitope form is either processed in low amounts or not at all, whereas the shorter or longer “KK10” epitope forms (KL8, KN9, KI11, KR13), which are also presented by HLA B∗2705, are produced in high abundance, primarily as N-terminally extended epitope precursors (EL11 [KL8], EN12 [KN9], GI15 [KI11], KR13) and are associated with suboptimal CTL responses (Tenzer et al., 2009) (Figure 3B; Table S1). These epitope precursors contain overlapping epitopes that share the same C terminus and can be presented by other, more common HLA class I molecules and may encompass one embedded epitope, PY9, presented by HLA-B∗35 and -B∗53; the only epitope in this study that is surrounded by other epitopes (Figure 5A; Table S1). These epitope precursors were processed with different efficiencies from HIV B and HIV C, and their abundance was inversely correlated with the combined allelic frequencies of the presenting HLA variants (Figures 1C and 3B, MF and CF2, pins).
Figure 5.
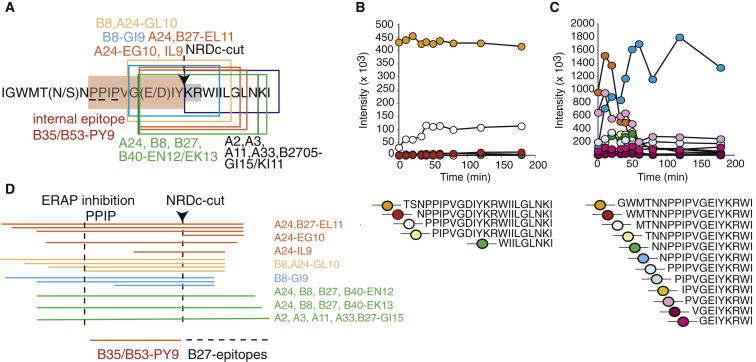
Selective Production of the Internal PY9 Epitope and KK10 Epitope Variants
(A) Outline of the overlapping epitope region comprising the B27-KK10 epitope; prolines 122, 123, and 125 are underlined, the internal B35/53-PY epitope is shaded red, and the cytoplasmic nardilysin (NRDc) recognition motif 131KR132 is gray (Kessler et al., 2011). Epitopes are boxed and HLA restriction elements are displayed in the same color as the box. Arrow, NRDc cut site.
(B) ERAP1,2 digestion of the HLA-A∗02, -A∗03, -A∗11, -A∗33, and -B∗2705-restricted GI15 precursor epitope. The result represents one of three independent experiments.
(C) ERAP1,2 digestion of the HLA-B∗08-restricted GI8 precursor epitope. The result represents one of three independent experiments.
(D) Outline of all proteasomal digestion fragments with different KK10 epitope forms produced following 4 hr of immunoproteasomal digestion of HIV B F2. The position of prolines 122, 123, and 125 and the NRDc cut site are indicated; production of the internal PY9 epitope and the KK10 epitope forms are delineated. HLA restriction elements are displayed next to the epitope precursors using the same color as in Figure 5A.
The cell-surface presentation of all epitopes encoded by these “KK10”-containing epitope precursors may be modified by other components of the antigen-processing machinery. More specifically, the production of many of the optimal epitopes is likely hampered because three proline (P) residues upstream of most of these epitopes inhibit ERAP trimming (Figure 5B, accumulation of KK10-containing trimming product with N-terminal PP; Figure 5C, similar accumulation of an HLA B∗8-restricted GI8-containing trimming product with N-terminal NPP) and because some precursors are cleaved by the cytosolic endopeptidase nardilysin (NRDc) (Kessler et al., 2011) (Figures 5A and 5D; Table S1).
NRDc digestion selectively destroys the epitopes presented by the common HLA-A∗24 and -B∗08 variants (Figure 5A), whereas it releases all of the KK10 epitope forms together with the internal PY9 epitope (Figure 5D). This PY9 production is unlikely to affect CTL responses in most patients because the HIV B and HIV C consensus sequences each contain CTL-escape mutations within PY9 that abolish the HLA-B∗35-restricted CTL response in HIV B infection, and the HLA-B∗53-restricted CTL response in HIV C (Matthews et al., 2012, Sabbaj et al., 2003). The presence of these escape mutations at the subtype level may reflect the benefit of a PY9-specific CTL response (Matthews et al., 2012) and the opposing frequencies of these HLA variants in whites and Central and Southern Africans (Gonzalez-Galarza et al., 2011).
Thus, overall intrahost selection can result in immune evasion by favoring HIV sequence combinations that inhibit proteasomal digestion and/or ERAP trimming and/or exploit NRDc to decrease both overall and specific CD8 epitope presentation and may also select for intraepitope CTL-escape mutations that prevent epitope recognition. When we combined the changes in specific epitope-precursor abundances and the favored HIV selection within patients who shared the presenting HLA variant, we found a strong, highly significant host-specific bias toward residues that specifically decrease the yield of epitopes that the host could potentially target (Table S4, p < 0.0001 [HIV B] and p = 0.0005 [HIV C], sign test). Potentially unrecognized CD8 epitope promiscuity will add noise to these analyses, but does not compromise them. This result provides strong evidence that all epitope-presenting HLA class I variants exert a selective pressure on these clinically important HIV Gag regions.
HLA-Driven Selection of HIV-C-like Subtype-Specific Amino Acids in HIV B over Time in the Caribbean
To investigate how HLA-driven selection might impact HIV B evolution over time, we studied longitudinal sequences from five ethnically diverse populations (Figure 6A). The Caribbean epidemic is particularly interesting because it is a unique example of the spread of HIV B in populations that are primarily of African ancestry. Specifically, 94/99 HIV B sequences (95%) originated from Caribbean countries where >90% of the population is black (Barbados, Haiti, and Jamaica). The region currently has the second-highest HIV prevalence rate worldwide, but the start of the epidemic within the region varies greatly; HIV B appears to have spread from Africa to Haiti in approximately 1970 (Gilbert et al., 2007), to Jamaica in approximately 1987 (Losina et al., 2008), to Barbados in about 1990 (http://www.unaids.org), and to Trinidad and Tobago between 1993 and 1996 (Gilbert et al., 2007).
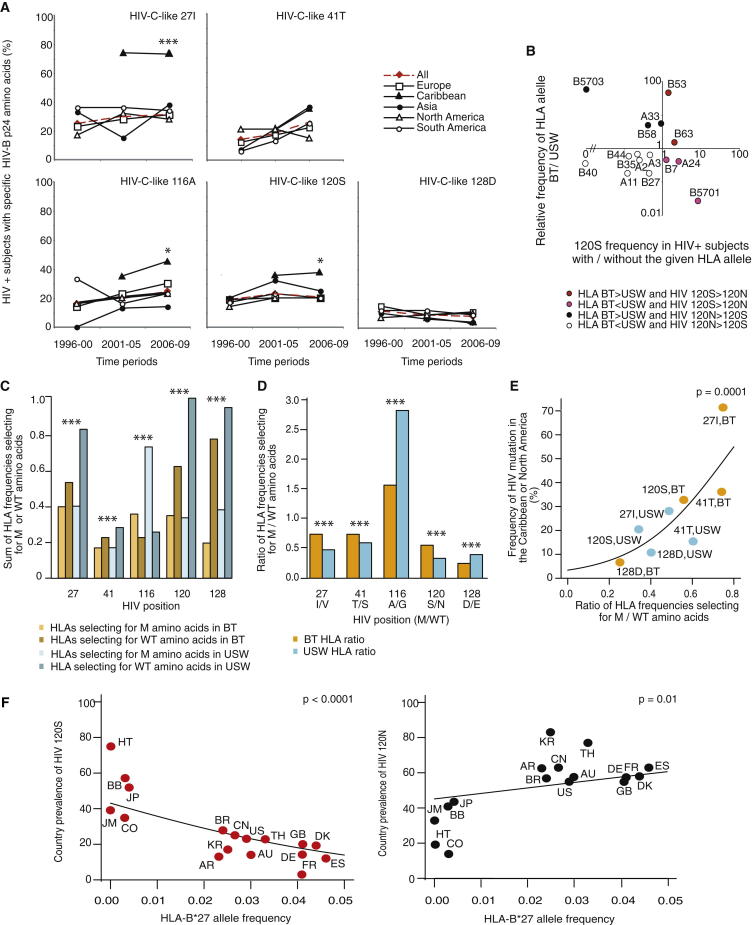
Figure 6.
HIV B Adaptation to the HLA Profiles of Host Populations
(A) One sequence from each HIV-B-infected patient was selected from the HIV database. The frequencies of HIV-C-like p24 Gag 27I, 41T, 116A, 120S, and 128D amino acids in HIV B were calculated across the data set at the indicated time periods in five geographic regions (region, n from time periods 1, 2, and 3), all minus Caribbean, 160, 575, and 556; Europe, 69, 254, and 375; Caribbean, 2, 68, and 31; Asia, 27, 96, and 97; North America, 47, 111, and 40; and South America, 17, 114, and 44. Fisher’s exact test was used to compare mutation frequencies at time period 3 between sequences from the Caribbean and all other cohorts combined (∗p < 0.05, ∗∗∗p < 0.0001).
(B) The frequency of an HLA variant in black Trinidadians (BT)/US whites (USW) versus the HIV B 120N→S substitution frequencies in subjects with or without that HLA variant is illustrated using log scales (all analyses, Figure S5 and Table S4).
(C) The summed allelic frequencies of HLA variants selecting for HIV B wild-type (“WT”) and HIV-C-like amino acids (“Mutant” or “M”), respectively, at each of the five subtype-specific positions for BT and USW; p values for the ethnic differences were calculated by fitting a binomial generalized linear model (GLM) (∗∗∗p < 0.0001) (Table S5).
(D) The ratios of the summed weighted HLA allelic frequencies in BT and USW that select for M/WT amino acids at each subtype-specific position (∗∗∗p < 0.0001) (Table S4; Table S5; Figure S5).
(E) A binomial GLM produced the graph of the frequency of p24 Gag 27I, 41T, 120S, and 128D in HIV B sequences from the Caribbean or North America versus the ratios of the summed HLA allelic frequencies in BT or USW that select for M/WT amino acids at each subtype-specific position. Position 116 was excluded because of competing selective pressures.
(F) HLA-B∗2705 frequencies versus the frequencies of HIV B p24 Gag 120N and 120S in sequences from each country during 2000–2009. The superposed curve was obtained by fitting a binomial GLM to the points. n = number of sequences, AR, Argentina (n = 78); AU, Australia (n = 21); BB, Barbados (n = 64); BR, Brazil (n = 68); CN, China (n = 75); CO, Columbia (n = 7); DE, Germany (n = 14); DK, Denmark (n = 23); ES, Spain (n = 8); FR, France (n = 7); GB, United Kingdom (n = 526); HT, Haiti (n = 16); JM, Jamaica (n = 14); JP, Japan (n = 27); KR, Korea (n = 18); TH, Thailand (n = 17); and US, USA (n = 122).
Despite the timing of the Caribbean epidemic, large numbers of HIV B p24 sequences only became available after 2000 (http://www.hiv.lanl.gov/content/index). Subtype-specific 27V→I and 41S→T (“HIV B consensus” or wild-type → “HIV-C-like” or mutant) substitutions accumulated rapidly in HIV B in the Caribbean cohort, reaching approximately 75% and 40%, respectively, after 5–10 years versus approximately 25%–35% (27I) and 10%–41% (41T), respectively, in the other geographic regions (Figure 6A). At time period 3, this accumulation was significantly greater than that found in all other cohorts combined for 27V→I (p < 0.0001) but not for 41S→T because of a comparable increase in the Asian cohort. Moreover, we observed parallel increases in HIV-C-like 116A (p = 0.05) and 120S frequencies (p = 0.05), whereas the HIV B wild-type 128E→D substitution appeared less frequently, although this difference did not reach significance. These observations collectively provide evidence of distinct HIV B adaptation in the Caribbean.
To investigate the underlying selective pressures, we examined the relationship between the frequencies of these HIV-C-like substitutions in HIV-B-infected patients with and without HLA variants that restrict epitopes in F1 and F2 (Table S4) and the relative frequencies of these HLA variants in black Caribbeans and whites (Figures 6B and S5) (Gonzalez-Galarza et al., 2011). Because only HLA information from black Trinidadians (BT) covered all of the necessary HLA variants, BT were used to represent all the black Caribbeans, and all whites were represented by data for US whites (USW). These analyses revealed which HLA variants select for HIV B wild-type amino acids and which select for HIV-C-like amino acids, and if a given HLA variant is more, or less, frequent in BT relative to USW; thus, Figures 6B and S5 illustrate the different, and similar, selective forces in the two ethnic groups. We frequently observed distinct HLA-associated selection pressures in each group. For example, some HLA variants associated with selection of 120S were underrepresented (HLA-A∗24, B∗07, and B∗5701) in BT when compared to USW (Figure 6B).
To examine the combined effect of the HLA-associated selective forces in each ethnic group, we summed the weighted allelic frequencies of HLA variants selecting for HIV B wild-type or HIV-C-like mutant amino acids at each of the five subtype-specific positions for both BT and USW (Figure 6C). We then examined whether the distribution of the sums was similar at each position within and between the two populations (i.e., the distribution of the light and dark orange [BT] and the light and dark blue [USW] bar heights). Although we observed a similar distribution between the sums of HLA alleles selecting for either mutant or wild-type amino acids at positions 27 and 41 for both BT and USW, we found a highly significant difference in the distribution pattern between the two ethnic groups (p < 0.0001) (Table S5). Similar strong ethnic differences were observed for the remaining four positions (p < 0.0001 for each) combined with distinct distribution patterns within the two populations when considering positions 116, 120, and 128 (p < 0.0001 for each).
For all positions, we next investigated whether the ratio of HLA variants selecting for mutant over wild-type amino acids was different between BT and USW (Figure 6D). We found highly significant differences in odds ratios between the two ethnic groups (Table S5), which provides evidence of distinct HLA-mediated selective pressure on HIV in each population.
Our results demonstrate that increases in the 27I, 41T, and 120S HIV-C-like amino acids in HIV-B-infected BT are associated with a higher prevalence ratio of HLA variants that select for these amino acids relative to those selecting for the wild-type amino acids 27V, 41S, and 120N compared with USW (Figure 6D). However, this finding did not reflect a higher relative abundance of HLA variants that select for HIV-C-like amino acids, but rather, a relative decrease of HLA variants that select for wild-type amino acids at these positions in BT relative to USW because the proportion of HLA variants selecting for wild-type amino acids in BT relative to USW was 0.65, 0.81, and 0.60 for positions 27, 41, and 120, respectively (Figure 6C; Table S5).
Similarly, the slight increase in the HIV B wild-type 128E among BT was associated with a decrease in the frequency of HLA variants selecting for the HIV-C-like 128D substitution, because the proportion for USW was 1.6 times larger than the proportion for BT (Table S5). The small increase in 116A over time is an anomaly because the proportion for USW was 1.8 times that of BT, but this increase is likely due to strong competing selective pressures affecting infectivity and natural killer cell recognition, which have distinct effects on HIV B evolution in the two ethnic groups (Fadda et al., 2011, Martinez-Picado et al., 2006).
When we examined how the USW and BT ratios of HLA variants that select for mutant over wild-type amino acids affected substitution patterns at subtype-specific positions, we found a highly significant positive correlation between these HLA ratios and the frequency of the HIV-C-like 27I, 41T, 120S, and 128D substitutions in HIV-B-infected individuals from North America and the Caribbean (Figure 6E, p = 0.0001). Position 116 was omitted from this analysis due to the competing selective pressures. Because the HLA ratio variation affected HIV B evolution at the viral population level in a similar manner at all four subtype-specific positions in both ethnic groups, this ratio is the key driver of HIV adaptation at these subtype-specific motifs.
This correlation directly links the distinct HIV B evolutionary patterns to differences in the HLA haplotype distribution in the two populations because the HLA data are not linked to the acquisition of the HIV sequences. Because of this relationship, our data provide compelling evidence that the specific amino acid at a given subtype-specific position in HIV B Gag affects the processing of all CD8 epitopes in nearby epitope clusters and that viral evolution is influenced by the frequencies of all HLA variants that can present CD8 epitopes from these clusters. Together, our data strongly suggest that the combined effect of these HLA-driven selective pressures on viral evolution is predictable and that viral founder effects are overlaid by rapid adaptation to local HLA population frequencies.
Collectively, our data strongly suggest that the HIV B Gag-consensus sequence is selected mainly by white HLA allelic frequencies and that the ancestral African HLA distribution in the Caribbean has shaped recent HIV B evolution in this location in a manner similar to the manner in which the HLA profile of infected sub-Saharan populations may have affected HIV C evolution.
HLA-Driven Selection of Amino Acids in Subtype-Specific Positions in HIV B across Populations
We next studied the influence of HLA frequency differences on HIV B evolution across populations worldwide. The 120N (HIV B) ↔ 120S (HIV C like) interchange is particularly informative because 120S simultaneously decreases B57-TW10 processing and increases B27-KK10 production and because both epitopes are associated with clinically beneficial CTL responses (Table S4). We correlated HLA-B∗2705 frequencies with the corresponding HIV 120S and 120N frequencies and found a strong inverse correlation between the HLA B∗2705 and 120S HIV frequencies (p < 0.0001), which was mirrored by a positive correlation between HLA-B∗2705 and 120N (p = 0.01) (Figure 6E). In contrast, a trend toward a positive correlation was observed for the HLA B∗5701 and 120S frequencies despite the small size and restricted range of the data set, resulting from the absence of this HLA variant in most populations (n = 8, p = 0.18). The rarity of HLA B∗5701 made it difficult to estimate any impact on 120N frequencies because 120N is positively selected in most HIV-B-infected individuals (p = 0.24). Together, the correlation between HLA frequencies across populations and the corresponding selection for evasive substitutions in the circulating virus argue strongly for the immune selection of HIV and viral adaptation to antigen-processing preferences.
Discussion
Here, we present an extensive study of HIV evolution and the effect of HIV subtype-specific substitutions on proteasomal processing of more than 50 epitopes presented by approximately 30 HLA class I variants in two key clinically relevant HIV B and HIV C Gag regions. We have identified a hitherto unrecognized adaptation of HIV Gag to the antigen-processing machinery at the viral population level. This adaptation occurs through substitutions at subtype-specific motifs, and multiple HLA variants presenting nearby epitopes all drive selection at the same subtype-specific position.
We provide evidence of a three-way interaction among HIV region, HLA frequency, and ethnic group and demonstrate that the proteasomal processing of all CD8 epitope precursors inversely correlates with the HLA allelic frequency of the presenting HLA variant(s) in the infected populations. Because of this adaptive process, epitopes presented by common HLA variants are generated in low amounts, whereas epitopes presented by rare HLA variants are made in abundance. Taken together with the reported weak and infrequent CTL responses associated with common HLA variants, and the strong and frequent CTL responses associated with rare HLA variants in these Gag regions (Goulder and Walker, 2012, Streeck et al., 2007), our results support previous findings linking epitope-abundance to the magnitude and frequency of epitope-specific CTL responses (Lazaro et al., 2009, Ranasinghe et al., 2011, Schmidt et al., 2012, Tenzer et al., 2009). Our data provide clear evidence that all epitope-presenting HLA class I variants exert selective pressures on subtype-specific positions in these key HIV Gag regions, resulting in population-level immune evasion in both HIV B and HIV C infection.
We demonstrate that this population-level immune evasion reflects the sum of intrapatient adaptations to reduce or obliterate the production of potentially presentable CD8 epitopes. Specifically, we show that the prevailing amino acid in either the HIV B or HIV C consensus sequence is associated with the least production of the epitopes presented by the most common HLA variants in both the HIV-B- and HIV-C-infected populations. In contrast, in patients with relatively rare HLA variants, HIV was often found to have nonconsensus amino acids at subtype-specific positions that were associated with a reduction in processing of the epitope presented by these rare HLA variants.
Thus, the HIV consensus sequences might be perceived as the best HIV sequence for immune evasion in the majority of infected individuals in whom additional individual HIV adaptations to escape the antiviral immune response take place. These individual adaptations may include intraepitope CTL escape mutations that affect, e.g., HLA binding or TCR recognition and/or further adaptations to limit antigen processing (Goulder and Walker, 2012). Notably, these individual adaptations typically differ from the HIV B or HIV C consensus sequence, which suggests that most of these substitutions impose fitness costs on HIV that select for reversion upon transmission to an HLA-mismatched recipient. Therefore, this process is very different from the adaptive process we describe here, which helps to create and/or modify HIV subtypes through adaptations to proteasomal preferences at subtype-specific motifs. This HLA-frequency-driven process is so advantageous to viral survival and transmission that it largely shapes the HIV B and HIV C Gag consensus sequence.
This process of adaptation to all HLA-A, -B, and -C variants may also help to explain the generally better control of HIV replication by HLA-B-restricted CTL responses (Kiepiela et al., 2004). HLA-B has more alleles (2,068) than either HLA-A (1,518) or HLA-C (1,016) (Gonzalez-Galarza et al., 2011); the HLA-B locus has the highest occurrence of heterozygosity because of a more even distribution of allele frequencies (Cao et al., 2001); and HLA-B variants bind an even greater variety of peptides (Marsh et al., 2000). As a result, the selective pressures on each peptide:HLA B combination may be lower, and this will lead to slower adaptation. Hence, we hypothesize that HLA-B-restricted epitopes are likely to be produced in higher amounts than those restricted by HLA-A and HLA-C, and this higher epitope abundance is likely to result in a more frequent priming of CD8+ T cells and more efficient elimination of infected cells (Faroudi et al., 2003). Thus, it is likely due to the generally reduced selective pressure by HLA-B to limit or obliterate CD8 epitope production that HLA-B-restricted CTL responses dominate the CTL response against HIV.
Our study of HIV evolution over time in five ethnically diverse populations perhaps most clearly demonstrates the broader importance of the correlation between HLA and subtype-specific polymorphisms in HIV Gag. We observed that HIV B in the black Caribbean population evolved in a manner resembling that of HIV C in Central and Southern Africa and that this pattern was associated with a relative decrease of HLA variants selecting for HIV B consensus amino acids when compared with whites in the US. Moreover, we found evidence of a worldwide adaptation of HIV B to the HLA-B∗2705 allelic frequency in infected populations, providing evidence of adaptation both between and within ethnic groups. Thus, HIV Gag subtypes have diversified in part because of their adaptation to the local HLA allelic frequency distributions, which contrasts with the current paradigm that HIV subtype diversification is mainly due to random founder effects (Rambaut et al., 2004). Although founder effects likely also contribute, our data provide strong evidence that founder effects are followed by the rapid adaptation to local HLA population frequencies in a predictable manner. This rapid adaptation may not have been observed previously because of insufficient sequence data from most countries during the start of the epidemic and a lack of data relating to antigen processing of HIV.
Our data should be interpreted cautiously when considering other HIV proteins because the extent of HIV adaptation to antigen processing will likely depend on the strength of the selective pressure exerted by CTL responses and because discordant associations exist between viral load and CTL responses toward different HIV proteins (Kiepiela et al., 2007). Vulnerable regions within other HIV proteins may also demonstrate variable adaptation to the same HLA selective pressure, in line with the different HIV B and HIV C CF1 epitope yields. The Gag regions we analyzed in this study are the only regions in the HIV proteome where CTL responses are strongly associated with delayed disease progression (Goulder and Walker, 2012), and these regions are therefore potentially important to target through vaccination. Our data provide compelling evidence that differences in epitope abundance are part of the reason these two key p24 regions only elicit strong and frequent CTL responses in the minority of patients with a few rare HLA-B variants.
Although the high evolutionary rate of HIV makes it possible to study the ongoing coevolution of HIV and the host immune response, any pathogen that is largely controlled by CTL responses might similarly adapt to the human antigen-processing machinery to limit epitope production. Differences in HLA frequencies between populations are thought to reflect differences in infectious challenges; however, it is likely that the distinct HLA profiles in different populations also impact the evolution of pathogens other than HIV. Possible candidates may include pathogens largely controlled by the CTL response that have long coexisted with humans and have diversified into subgroups in distinct geographic regions such as hepatitis B virus and hepatitis C virus.
Because HIV-1 selectively adapts to the HLA distribution of the infected population, vaccines based on locally prevalent HIV sequences would be predicted to result in weaker and less frequent CD8+ T cell responses than vaccines containing a mosaic of subtypes or ancestral or consensus HIV sequences inferred from patient-derived virus. However, all these naturally selected HIV sequences will likely result in weak CD8+ T cell responses in the majority of vaccinated HIV-uninfected individuals, regardless of how they are chosen and combined, due to similarities among HLA frequencies in different ethnic groups (Cao et al., 2001). Therefore, even the creation of mosaic HIV inserts based on natural HIV sequences is unlikely to substantially increase the processing of epitopes presented by most of the common HLA variants.
We demonstrate that viral evolution in vivo can alter the processing of all epitopes in overlapping epitope clusters through substitutions at subtype-specific motifs, which suggests that artificial sequence modifications at these positions in vitro could refocus and reverse the poor immunogenicity of HIV proteins. This vaccine concept would exploit the fact that CD8+ T cells have two distinct antigen-load-dependent activation thresholds: one for clonal expansion and one for target cell elimination (Faroudi et al., 2003). Processing of a modified insert might generate epitopes in high abundance that will elicit strong CTL responses capable of eliminating naturally infected cells that display as little as three peptide:HLA complexes on their surface (Purbhoo et al., 2004). Thus, vaccine efficacy might be improved by unnatural substitutions that increase the processing of subdominant CD8 epitopes presented by common HLA variants in vulnerable parts of the virus.
Experimental Procedures
Patients and HLA Typing
Chronically HIV-1-infected patients were recruited from the Department of Infectious Diseases, Rigshospitalet, Denmark (Table S3). All patients gave informed consent and were studied according to the regulations of the Danish Board of Medical Ethics for human experiments. HLA genotyping was done using multiplex PCR (Dynal Biotech).
Peptide Synthesis and Purification
Peptides were synthesized on an MK-IV peptide synthesizer (Schafer-N) and purified to >98% purity using a JupiterProteo C12 column (Phenomenex) as in Tenzer et al. (2009).
Purification of Proteasomes and In Vitro Digestions
Constitutive proteasomes and immunoproteasomes (20S) were purified from LCL721.174 and LCL721 EBV-transformed human B cell lines as in Tenzer et al. (2004). We performed 2, 4, and 6 hr digestions with purified constitutive proteasomes and immune proteasomes (substrate:enzyme ratio:1,000:1); all peptide digests in one experimental set were performed on the same day. We used the 4 hr digestion results here to allow detection of all cleavage products while minimizing effects of proteasomal re-entry of already digested peptide as less than 40% of the initial substrate was degraded at this time in all experiments.
Analysis of Peptide Digests by Mass Spectrometry
We performed capillary liquid chromatography of all peptide digests using a Waters NanoAcquity UPLC system and mass spectrometry analysis as in Tenzer et al. (2009) and the Supplemental Experimental Procedures. Each sample was analyzed in triplicate. We used the ProteinLynx Global Server version 2.2 for the processing, identification and quantification of the LC-MSE data. The mass error tolerance values were typically <5 ppm. We used mass spectrometric fragment intensity as a surrogate marker for quantity because we previously found a highly significant correlation between these two parameters (Tenzer et al., 2009).
TAP-Peptide Binding Assays
The TAP-peptide binding of epitope precursors and optimal epitopes was determined as in Tenzer et al. (2009) and the Supplemental Experimental Procedures.
In Vitro Peptide Digestions with ERAP Enzymes
Each peptide was incubated with human ERAP1/2 complexes as in Tenzer et al. (2009) and the Supplemental Experimental Procedures. Aliquots were removed for analysis at the indicated time points.
Intracytoplasmic Cytokine Staining
About 150,000 peripheral blood mononuclear cells (PBMCs) per well were stimulated in a 96-well plate with peptides (2.5 ng/ml), 1 μg/ml anti-CD28, anti-CD49d, and anti-CD107a FITC (BioLegend) for 6 hr. After 30 min, 5 μg/ml Brefeldin A and 1× Monensin (BioLegend) were added. The positive control contained 2.5 ng/ml PMA and 25 ng/ml ionomycin, and the negative cells and CD28/CD49d. Cells were washed and stained with LIVE/DEAD Fixable Violet stain (Invitrogen), fixed, and permeabilized with BD Fix/Perm solution (BD) and stained with a mix of fluorochrome-conjugated antibodies: CD3 EDC, CD8 PerCP (BD), interleukin (IL)-2 PE, IFN-γ, Pe-Cy7, and tumor necrosis factor (TNF)-α APC (BioLegend). Unstained and singly stained cells were used as controls to calculate compensation. The cell fluorescence intensities were acquired on a Cyan ADP Cytometer (Beckman Coulter) and analyzed with Summit software (Dako).
Sequence Analyses
All sequences were obtained from the HIV database (http://www.hiv.lanl.gov/content/index). We grouped HIV B and HIV C sequences from 1980 to 2009 according to (1) 5 year time period and (2) 5 year period and geographic region. Group 3 consisted of all HIV B sequences from 2000 to 2009 grouped according to geographic region, and group 4 contained all sequences from HLA-typed HIV-B- or HIV-C-infected persons divided according to HLA group; this group was further divided into groups of patients with or without specific HLA variants (Table S4). We used the sequence tool in the HIV database to randomly select only one sequence from each patient or transmission cluster in groups 1–3 to avoid skewing. We excluded sequences with translational problems (i.e., frameshift or stop codons).
Shannon Entropy analyses (http://www.hiv.lanl.gov/content/index) were done on sequence set 1. Sequence changes in geographic regions over time were examined using set 2. HIV adaptation in specific countries was estimated using set 3. Differences in amino acid frequencies in HIV Gag from patients with or without specific HLA alleles were estimated using set 4. We counted and calculated the amino acid frequencies at each HIV-1 Gag p24 position; each query HIV-1 gag sequence was pairwise aligned to the HXB2 reference strain and mutations were defined as changes relative to this sequence. The frequency of mutations in the p24 gene was then calculated across each of the data sets 2–4. Because we analyzed all HIV sequences in the database, our intraepitope mutation frequencies (Table S4) might differ from those reported by smaller cohorts. The HIV-1 consensus sequences were obtained from the HIV database.
Epitope Designation and HLA Analyses
HLA allelic frequencies were obtained from the Allele Frequency Database (Gonzalez-Galarza et al., 2011) and (Assane et al., 2010, Novitsky et al., 2001). When more than one data set was available from a country, we used weighted mean values unless otherwise specified. For the analyses in Figure 1, we used HLA frequencies from the UK (UK1 = Leeds [n = 5,024], UK2 = Sheffield [n = 4,755]), the US (US1 = Los Angeles, Seattle, and Detroit [n = 1,070], US2 = Bethesda [n = 307]), Germany (n = 11,407), France (n = 130), South Africa (Zulu, n = 100; Xhosa, n = 50), Zambia (n = 256), Zimbabwe (n = 108), Botswana (n = 161), and Mozambique (n = 250). We labeled peptide fragments as CD8 epitopes based on the HIV immunology database (http://www.hiv.lanl.gov/content/immunology). Because studies disagree as to whether HLA-Cw∗03 can bind the VL10 epitope (Zappacosta et al., 1997, Zarling and Lee, 1998), we did not include a selection pressure from HLA-Cw∗03 in our analyses.
Statistical Analyses
We used multilevel modeling to examine the effects of HLA frequency, HIV region, and ethnic group on epitope yield. Further analyses were done using binomial generalized linear models (McCullough and Nelder, 1989) using the canonical logit link function, Fisher’s exact test, t tests, and a nonparametric sign test as described in the text and in the Supplemental Experimental Procedures. These analyses were done using GraphPad Prism 5 (GraphPad), the R statistical package (version 3.0.0 [2013-04-03]), Microsoft Excel 2007 (Microsoft), and Stat Trek (Stattrek.com) software. A value of p < 0.05 was considered significant. Graphs were made using GraphPad Prism 5, R, and Microsoft Excel 2007.
Author Contributions
A.K.N.I. conceived, designed, and organized the overall study. S.T., H.S., P.v.E., and A.K.N.I. planned and supervised experiments. S.T., H.C., P.P., A.B., N.A., and M.W. performed experiments. R.G., V.B.S., T.d.O., and A.K.N.I. analyzed sequences, S.T., H.C., P.P., A.B., H.S., L.F., D.L., P.v.E., and A.K.N.I. analyzed other data. P.v.E., J.B., J.G., A.K.N.I., and H.S. provided reagents. A.K.N.I. wrote the manuscript. S.T., H.S., D.L., P.v.E., and J.B. contributed intellectual input, and all authors commented on the manuscript. H.C. and P.P. contributed equally to the work and V.B.S. and M.W. contributed equally to the work.
Acknowledgments
We thank the patients for donating samples, M. Pagel, P. Klenerman, and N. Willcox for helpful discussions, B. Baadegaard and L.P. Jensen for patient management, and D. Hass, N. Philipp, and J. Forsch for technical assistance. H.C. was a Mary Goodger Postdoctoral Fellow. Funding was obtained from the German Research Foundation (SFB490/E6, Z3), the European Community’s 7th Framework Program (FP7/HEALTH-2007-1.1-4; no. 222773, acronym PEPCHIPOMICS), the Forschungszentrum Immunologie of the Johannes-Gutenberg University Mainz (H.S. and S.T.), the Wellcome Trust (082384/Z/07/Z; T.d.O.), the Thyssen Foundation (P.v.E.), the UK Medical Research Council (A.K.N.I., P.P., and V.B.S.), and the Medical Research Fund, Oxford University (A.K.N.I.).
Published: April 10, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Supplemental Experimental Procedures, five figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.03.031.
Supplemental Information
References
- Almeida J.R., Price D.A., Papagno L., Arkoub Z.A., Sauce D., Bornstein E., Asher T.E., Samri A., Schnuriger A., Theodorou I. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assane A.A., Fabricio-Silva G.M., Cardoso-Oliveira J., Mabunda N.E., Sousa A.M., Jani I.V., Ferreira O.C., Jr., Porto L.C. Human leukocyte antigen-A, -B, and -DRB1 allele and haplotype frequencies in the Mozambican population: a blood donor-based population study. Hum. Immunol. 2010;71:1027–1032. doi: 10.1016/j.humimm.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Bhattacharya T., Daniels M., Heckerman D., Foley B., Frahm N., Kadie C., Carlson J., Yusim K., McMahon B., Gaschen B. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- Brumme Z.L., Brumme C.J., Carlson J., Streeck H., John M., Eichbaum Q., Block B.L., Baker B., Kadie C., Markowitz M. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 2008;82:9216–9227. doi: 10.1128/JVI.01041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonaguro L., Tornesello M.L., Buonaguro F.M. Human immunodeficiency virus type 1 subtype distribution in the worldwide epidemic: pathogenetic and therapeutic implications. J. Virol. 2007;81:10209–10219. doi: 10.1128/JVI.00872-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K., Hollenbach J., Shi X., Shi W., Chopek M., Fernández-Viña M.A. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 2001;62:1009–1030. doi: 10.1016/s0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- Dahirel V., Shekhar K., Pereyra F., Miura T., Artyomov M., Talsania S., Allen T.M., Altfeld M., Carrington M., Irvine D.J. Coordinate linkage of HIV evolution reveals regions of immunological vulnerability. Proc. Natl. Acad. Sci. USA. 2011;108:11530–11535. doi: 10.1073/pnas.1105315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinges W.L., Richardt J., Friedrich D., Jalbert E., Liu Y., Stevens C.E., Maenza J., Collier A.C., Geraghty D.E., Smith J. Virus-specific CD8+ T-cell responses better define HIV disease progression than HLA genotype. J. Virol. 2010;84:4461–4468. doi: 10.1128/JVI.02438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draenert R., Le Gall S., Pfafferott K.J., Leslie A.J., Chetty P., Brander C., Holmes E.C., Chang S.C., Feeney M.E., Addo M.M. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S., Dinges W.L., Lejarcegui N., Laing K.J., Collier A.C., Koelle D.M., McElrath M.J., Horton H. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat. Med. 2011;17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda L., O’Connor G.M., Kumar S., Piechocka-Trocha A., Gardiner C.M., Carrington M., McVicar D.W., Altfeld M. Common HIV-1 peptide variants mediate differential binding of KIR3DL1 to HLA-Bw4 molecules. J. Virol. 2011;85:5970–5974. doi: 10.1128/JVI.00412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faroudi M., Utzny C., Salio M., Cerundolo V., Guiraud M., Müller S., Valitutti S. Lytic versus stimulatory synapse in cytotoxic T lymphocyte/target cell interaction: manifestation of a dual activation threshold. Proc. Natl. Acad. Sci. USA. 2003;100:14145–14150. doi: 10.1073/pnas.2334336100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm N., Yusim K., Suscovich T.J., Adams S., Sidney J., Hraber P., Hewitt H.S., Linde C.H., Kavanagh D.G., Woodberry T. Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur. J. Immunol. 2007;37:2419–2433. doi: 10.1002/eji.200737365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M.T., Rambaut A., Wlasiuk G., Spira T.J., Pitchenik A.E., Worobey M. The emergence of HIV/AIDS in the Americas and beyond. Proc. Natl. Acad. Sci. USA. 2007;104:18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Galarza F.F., Christmas S., Middleton D., Jones A.R. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39(Database issue):D913–D919. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Walker B.D. HIV and HLA class I: an evolving relationship. Immunity. 2012;37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Tang Y., Pelton S.I., Walker B.D. HLA-B57-restricted cytotoxic T-lymphocyte activity in a single infected subject toward two optimal epitopes, one of which is entirely contained within the other. J. Virol. 2000;74:5291–5299. doi: 10.1128/jvi.74.11.5291-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler B., Daniel S., Armandola E.A., Hammer J., Caillat-Zucman S., van Endert P.M. Substrate selection by transporters associated with antigen processing occurs during peptide binding to TAP. Mol. Immunol. 1998;35:427–433. doi: 10.1016/s0161-5890(98)00059-5. [DOI] [PubMed] [Google Scholar]
- Iversen A.K., Stewart-Jones G., Learn G.H., Christie N., Sylvester-Hviid C., Armitage A.E., Kaul R., Beattie T., Lee J.K., Li Y. Conflicting selective forces affect T cell receptor contacts in an immunodominant human immunodeficiency virus epitope. Nat. Immunol. 2006;7:179–189. doi: 10.1038/ni1298. [DOI] [PubMed] [Google Scholar]
- Kawashima Y., Pfafferott K., Frater J., Matthews P., Payne R., Addo M., Gatanaga H., Fujiwara M., Hachiya A., Koizumi H. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler J.H., Khan S., Seifert U., Le Gall S., Chow K.M., Paschen A., Bres-Vloemans S.A., de Ru A., van Montfoort N., Franken K.L. Antigen processing by nardilysin and thimet oligopeptidase generates cytotoxic T cell epitopes. Nat. Immunol. 2011;12:45–53. doi: 10.1038/ni.1974. [DOI] [PubMed] [Google Scholar]
- Kiepiela P., Leslie A.J., Honeyborne I., Ramduth D., Thobakgale C., Chetty S., Rathnavalu P., Moore C., Pfafferott K.J., Hilton L. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- Kiepiela P., Ngumbela K., Thobakgale C., Ramduth D., Honeyborne I., Moodley E., Reddy S., de Pierres C., Mncube Z., Mkhwanazi N. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- Kloetzel P.M. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2001;2:179–187. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- Lazaro E., Godfrey S.B., Stamegna P., Ogbechie T., Kerrigan C., Zhang M., Walker B.D., Le Gall S. Differential HIV epitope processing in monocytes and CD4 T cells affects cytotoxic T lymphocyte recognition. J. Infect. Dis. 2009;200:236–243. doi: 10.1086/599837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro E., Kadie C., Stamegna P., Zhang S.C., Gourdain P., Lai N.Y., Zhang M., Martinez S.A., Heckerman D., Le Gall S. Variable HIV peptide stability in human cytosol is critical to epitope presentation and immune escape. J. Clin. Invest. 2011;121:2480–2492. doi: 10.1172/JCI44932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losina E., Figueroa P., Duncan J., Divi N., Wolf L.L., Hirschhorn L.R., Robertson M., Harvey K., Whorms S., Freedberg K.A., Gebre Y. HIV morbidity and mortality in Jamaica: analysis of national surveillance data, 1993--2005. Int. J. Infect. Dis. 2008;12:132–138. doi: 10.1016/j.ijid.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S.G.E. Elsevier; London: 2000. The HLA FactsBook. [Google Scholar]
- Martinez-Picado J., Prado J.G., Fry E.E., Pfafferott K., Leslie A., Chetty S., Thobakgale C., Honeyborne I., Crawford H., Matthews P. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews P.C., Prendergast A., Leslie A., Crawford H., Payne R., Rousseau C., Rolland M., Honeyborne I., Carlson J., Kadie C. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 2008;82:8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews P.C., Koyanagi M., Kløverpris H.N., Harndahl M., Stryhn A., Akahoshi T., Gatanaga H., Oka S., Juarez Molina C., Valenzuela Ponce H. Differential clade-specific HLA-B∗3501 association with HIV-1 disease outcome is linked to immunogenicity of a single Gag epitope. J. Virol. 2012;86:12643–12654. doi: 10.1128/JVI.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough P., Nelder J.A. Chapman & Hall; London: 1989. Generalized Linear Models. [Google Scholar]
- Novitsky V., Flores-Villanueva P.O., Chigwedere P., Gaolekwe S., Bussman H., Sebetso G., Marlink R., Yunis E.J., Essex M. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum. Immunol. 2001;62:146–156. doi: 10.1016/s0198-8859(00)00236-6. [DOI] [PubMed] [Google Scholar]
- Paulsson K.M. Evolutionary and functional perspectives of the major histocompatibility complex class I antigen-processing machinery. Cell. Mol. Life Sci. 2004;61:2446–2460. doi: 10.1007/s00018-004-4113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra F., Jia X., McLaren P.J., Telenti A., de Bakker P.I., Walker B.D., Ripke S., Brumme C.J., Pulit S.L., Carrington M., International HIV Controllers Study The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purbhoo M.A., Irvine D.J., Huppa J.B., Davis M.M. T cell killing does not require the formation of a stable mature immunological synapse. Nat. Immunol. 2004;5:524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Posada D., Crandall K.A., Holmes E.C. The causes and consequences of HIV evolution. Nat. Rev. Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- Ranasinghe S.R., Kramer H.B., Wright C., Kessler B.M., di Gleria K., Zhang Y., Gillespie G.M., Blais M.E., Culshaw A., Pichulik T. The antiviral efficacy of HIV-specific CD8+ T-cells to a conserved epitope is heavily dependent on the infecting HIV-1 isolate. PLoS Pathog. 2011;7:e1001341. doi: 10.1371/journal.ppat.1001341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbaj S., Bansal A., Ritter G.D., Perkins C., Edwards B.H., Gough E., Tang J., Szinger J.J., Korber B., Wilson C.M. Cross-reactive CD8+ T cell epitopes identified in US adolescent minorities. J. Acquir. Immune Defic. Syndr. 2003;33:426–438. doi: 10.1097/00126334-200308010-00003. [DOI] [PubMed] [Google Scholar]
- Schmidt J., Iversen A.K., Tenzer S., Gostick E., Price D.A., Lohmann V., Distler U., Bowness P., Schild H., Blum H.E. Rapid antigen processing and presentation of a protective and immunodominant HLA-B∗27-restricted hepatitis C virus-specific CD8+ T-cell epitope. PLoS Pathog. 2012;8:e1003042. doi: 10.1371/journal.ppat.1003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck H., Lichterfeld M., Alter G., Meier A., Teigen N., Yassine-Diab B., Sidhu H.K., Little S., Kelleher A., Routy J.P. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 2007;81:7725–7731. doi: 10.1128/JVI.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenzer S., Stoltze L., Schönfisch B., Dengjel J., Müller M., Stevanović S., Rammensee H.G., Schild H. Quantitative analysis of prion-protein degradation by constitutive and immuno-20S proteasomes indicates differences correlated with disease susceptibility. J. Immunol. 2004;172:1083–1091. doi: 10.4049/jimmunol.172.2.1083. [DOI] [PubMed] [Google Scholar]
- Tenzer S., Wee E., Burgevin A., Stewart-Jones G., Friis L., Lamberth K., Chang C.H., Harndahl M., Weimershaus M., Gerstoft J. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat. Immunol. 2009;10:636–646. doi: 10.1038/ni.1728. [DOI] [PubMed] [Google Scholar]
- Toes R.E., Nussbaum A.K., Degermann S., Schirle M., Emmerich N.P., Kraft M., Laplace C., Zwinderman A., Dick T.P., Müller J. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med. 2001;194:1–12. doi: 10.1084/jem.194.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Endert P. Post-proteasomal and proteasome-independent generation of MHC class I ligands. Cell. Mol. Life Sci. 2011;68:1553–1567. doi: 10.1007/s00018-011-0662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappacosta F., Borrego F., Brooks A.G., Parker K.C., Coligan J.E. Peptides isolated from HLA-Cw∗0304 confer different degrees of protection from natural killer cell-mediated lysis. Proc. Natl. Acad. Sci. USA. 1997;94:6313–6318. doi: 10.1073/pnas.94.12.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarling A.L., Lee D.R. Conversion of a human immunodeficiency virus cytotoxic T lymphocyte epitope into a high affinity HLA-Cw3 ligand. Hum. Immunol. 1998;59:472–482. doi: 10.1016/s0198-8859(98)00053-6. [DOI] [PubMed] [Google Scholar]
- Zhang S.C., Martin E., Shimada M., Godfrey S.B., Fricke J., Locastro S., Lai N.Y., Liebesny P., Carlson J.M., Brumme C.J. Aminopeptidase substrate preference affects HIV epitope presentation and predicts immune escape patterns in HIV-infected individuals. J. Immunol. 2012;188:5924–5934. doi: 10.4049/jimmunol.1200219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.