Abstract
Autotransporters (ATs) represent a superfamily of proteins produced by a variety of pathogenic bacteria, which include the pathogenic groups of Escherichia coli (E. coli) associated with gastrointestinal and urinary tract infections. We present the first X-ray structure of the passenger domain from the Plasmid-encoded toxin (Pet) a 100 kDa protein at 2.3 Å resolution which is a cause of acute diarrhea in both developing and industrialized countries. Pet is a cytoskeleton-altering toxin that induces loss of actin stress fibers. While Pet (pdb code: 4OM9) shows only a sequence identity of 50 % compared to the closest related protein sequence, extracellular serine protease plasmid (EspP) the structural features of both proteins are conserved. A closer structural look reveals that Pet contains a β-pleaded sheet at the sequence region of residues 181-190, the corresponding structural domain in EspP consists of a coiled loop. Secondary, the Pet passenger domain features a more pronounced beta sheet between residues 135-143 compared to the structure of EspP.
Introduction
Autotransporters (ATs) represent a superfamily of proteins produced by a variety of pathogenic bacteria, which include the pathogenic groups of Escherichia coli (E. coli) associated with gastrointestinal and urinary tract infections. ATs compose of three functional domains; The signal peptide, β-domain and passenger domain. The Sec translocon recognizes the signal peptide and thus while being translocated, the protein is not completely folded. During translocation, the signal peptidase cleaves off the signal peptide, then, the AT devoid of the signal peptide, adopts a stable intermediate in the periplasm by interaction with periplasmic chaperones, such as Skp and SurA, as well as the outer membrane Omp85/Bam complex. The second domain is the β-domain that integrates the mature protein into the outer membrane, forming a β-barrel with a hydrophilic pore. After the beta-barrel has been inserted into the outer membrane the passenger domain is transported through the center of the barrel to the to the cell surface [1]. The barrel and passenger domain are corrected by a linker domain [1].
The passenger domain harbors a conserved serine protease motif (GDSGS) that contributes to different functions as well as adhesins, toxins, or digestive enzymes in order to facilitate invasion of the host cells during bacterial infection [2],[3],[4]. Serine protease autotransporters of the family Enterobacteriaceae (SPATEs) are a representative subfamily of the AT proteins. More than 20 SPATE's have been identified exclusively in pathogenic enteric bacterial species [5]. Members of SPATEs family include among others, EspP (extracellular serine protease plasmid (pO157)-encoded) [6] or PssA (protease secreted by STEC) [7] as initially designated, EpeA (EHEC plasmid-encoded autotransporter) from enterohaemorrhagic E. coli (EHEC) [8], Pic (protease involved in intestinal colonization) from enteroaggregative E. coli EAEC, uropathogenic E. coli (UPEC) and Shigella [9], Hbp (hemoglobin protease or hemoglobin binding protein) from enteropathogenic E. coli (EPEC) [10] and Sat (secreted autotransporter toxin) from UPEC and E. coli enteroaggregative (EAEC) [11]. The Plasmid-encoded toxin (Pet) is the prototypical SPATE member with different characteristic biological functions [12]. Pet was identified in the EAEC group as a cause of acute diarrhea in both developing and industrialized countries [13],[14]. Pet is a cytoskeleton-altering toxin that induces loss of actin stress fibers. The toxin cleaves epithelial α-fodrin (between M1198 and V1199) in vitro and in vivo, causing fodrin redistribution within the cells, to form intracellular aggregates [15]. Before disruption of the host-cell membrane Pet is associated with cytokeratin (CK8) and is internalized by the host cell where it undergoes retrograde trafficking. It utilizes the ER-associated degradation (ERAD) pathway to be released into the cytosol causing the cytotoxic cell damage [16],[17].
The crystallographic structures of the native passenger domains of the SPATEs Hbp from EPEC [18], IgAP from Haemophilus influenzae [19] and EspP from EHEC[20] have been described. The overall fold for passenger domains of most ATs is typical for both known and predicted structures where the β-helical subdomain or β-spine is believed to be conserved. [20],[21]. These folds allow the ATs to be assembled into the self-associating autotransporters (SAATs) to mediate cell-cell adhesion and facilitate biofilm formation [19]. The fold could also be responsible for oligomerization into rope-like structures, but more evidence is needed to support this hypothesis[22]. SPATE structures also include a globular subdomain or N-terminal serine protease domain with the canonical catalytic triad (His, Asp, Ser), the region responsible for effector function. Considering that most SPATEs are proven or putative virulence factors, understanding the details of their biogenesis and function would be particularly useful in designing new antimicrobial agents. In order to understand the structure-function relationship and the biological roles of the Pet passenger domain, it is essential to know the key structural features as described underneath.
MATERIALS AND METHODS
Protein Expression and Purification
Expression and purification of the protein under study were performed following the procedure described by Villaseca et al. [23], with the following modifications. The Pet clone from E. coli strain HB101 (pCEFN2) [24] was used to overexpress the Pet protein in Luria broth (LB) medium. The supernatant of the cell culture was incubated with ammonium sulfate (1.7 M) for protein precipitation (Overnight at 37 C) and the pellet thus obtained was re-suspended in 50 mM Tris-HCL, 5 mM EDTA, pH 8.0, desalted and equilibrated using 30,000 Da molecular-weight-cutoff (MWCO) Amicon Ultra centrifugal filters (Millipore Co.). Subsequently, 50 ml of this solution was loaded onto a preequilibrated Q-sepharose anionic exchange column (Amersham Biosciences) equilibrated with 50 mM Tris-HCL, 5 mM EDTA, pH 8.0. A second ion exchange chromatography purification was performed by fast-protein liquid chromatography (FPLC) using Mono S HR 5/5 columns (High performance, Amersham Biosciences). The process of the purification was monitored by SDS-PAGE (8 %) and protein concentration was determined by Bradford protein assay or by O. D.280nm measurements [25].
N-terminal sequence determination
To confirm the identity of the purified protein the N-terminal sequence was determined using the automated Edman degradation on a gas phase protein sequencer (LF 3000; Beckman Instruments), which was equipped with an online Beckman System Gold high-performance liquid chromatography (HPLC) system. The HPLC equipment included a model 126 pump and a 168-diode array detector set at 268 and 293nm for detection of signal and reference, respectively. A Beckman Spherogel Micro PTH (2 by 150) column was used for the HPLC runs. The standard Beckman sequencing reagents were used for the analysis. Protein samples for sequencing were prepared by SDS-PAGE and electro blotting on polyvinylidene difluoride membranes (Millipore Co.), as described by Towbin et al. [26].
Crystallization
The Pet protein (13 mg/ml in 50 mM Tris-HCL, 5 mM EDTA, pH 6.5 and 0.3 M NaCl) was crystallized by the sitting-drop vapor diffusion method [27]. Crystallization was performed using 0.2 M proline, 0.1 M HEPES (pH 7.5), 10 % w/v PEG 3350 at 18 °C on intelli-plate 96-2 shallow well profile (Hampton Research Co), with a drop volume of 2 μL that composed 1 μL of protein and 1 μL of reservoir solution with100 μl of reservoir solution in the well.
Structure Determination and Refinement
The initial set of phases was determined by molecular replacement using EHEC EspP protease (pdb code: 3SZE) or E. coli Hbp (pdb code: 2WXR) as base for low sequence identity search models. The phase was assigned using the program phaser, version 2.5.1[28].
Results
Purification of Pet passenger domain
The Pet passenger domain was purified with the goal to crystallize the protein and determine the structure for analysis of the structure-function relationship of the Pet passenger domain. We purified the S260I mutant of the Pet passenger domain (969 residues, numbered 1-969) from E. coli HB101 transformed with plasmid pCEFN2. The purification process (Fig 1), was performed by ammonium sulphate precipitation, anion exchange chromatography (Q-Sepharose), where the protein elutes in the void volume, followed by cation exchange chromatography (Mono S HR). The identity of the isolated Pet protein was confirmed by N-amino terminal sequencing. The ANMDISKAWARDYLDLAQN sequence was obtained corresponding to the previously predicted pet gene product of the 042 EAEC strain [14].
Figure 1.
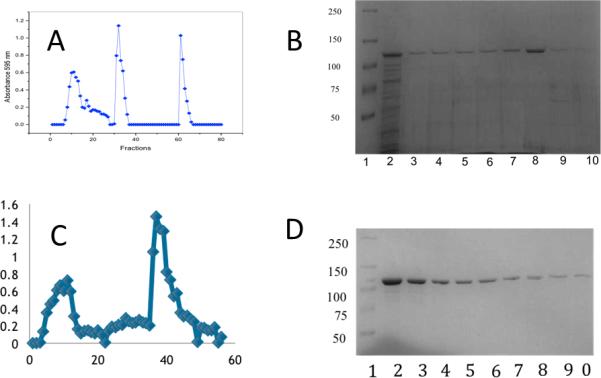
Results of anion exchange chromotography using Q-Sepharose are depicted in A. (B) SDS-PAGE of the fractions of the flow through containing Pet is shown (Well 1, molecular mass of protein standards in kDa, wells 2-10, fractions collected from the second peak in the chromatogram.) (C) the chromatographic profile for the pet purification of Pet by cation exchange chromatography (Mono S). (D) Corresponding SDS-PAGE of the second peak in C, well 1, molecular mass of protein standards (kDa), wells 2-10, fractions (second peak).
Crystallization and Data Collection
Crystals grew within 18 days of incubation at 18 °C. Single crystals were harvested from the sitting drop using a cryoloop, cryo protected by passing through a crystallization solution with 25 % glycerol (added to the mother liquor) and flash cooled in liquid nitrogen. X-ray diffraction data were collected at Advanced Photon Source (APS), Argonne (USA) at beam line 21-ID-F. X-ray diffraction data were collected on a Mar 225 CCD detector, with the exposure time of 1 sec per frame. The oscillation per frame was 1 degree; the beam line wavelength was 0.9787 Å (single wavelength beam line) and the complete data set consisted of 250 frames. The data have been integrated by XDS [29] and scaled by Scala from the ccp4i [30] package. The unit cell was assigned to the orthorhombic space group P212121 by the program pointless (ccp4i). The initial molecular replacement solution was improved by the rebuild program ARP/wARP [31] and the phenix suite [32] with complete rebuild. For last improvements on the refinement of the structure the program DEN [33] in the version from phenix and CNS 1.3 were used [34],[35]
X-ray model description
The structural model of the Pet passenger domain extends from the N-terminus residue 1 of the mature protein to residue 963(with in total 931 assigned residues), the C-terminal residue of the passenger domain as depicted in figure 2. One molecule of Pet passenger domain is in the asymmetric unit (ASU), the cell constants were determeined to α= 77.76Å β= 95.93Å and γ=164.87 Å in the orthorhombic space group P212121. The calculated solvent content is 58.4 % corresponding to a Matthews coefficient of 2.96 [36].
Figure 2.
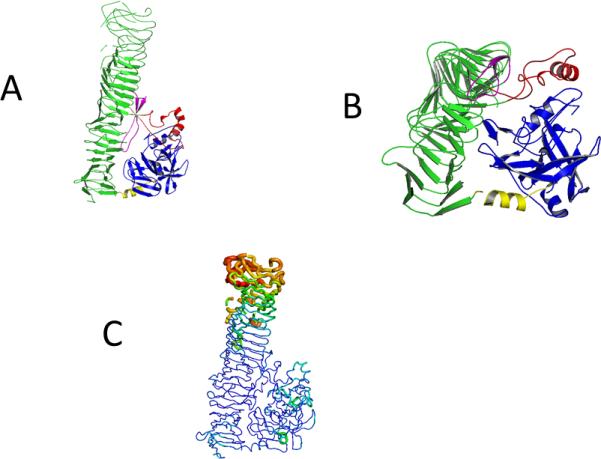
Model (A and B) of the Pet protein (pdb 4OM9) with the five subdomains, the serine protease domain (blue), connecting α-Helix (yellow), the β- Helix domain (green), intermitted by stabilizing α-Helix (red) and the spacer beta pleaded sheet (purple). In A the green subdomain is shown in maximal length. B shows a 90 ° backwards turn where the smaller subunits are featured.
C: B-putty representation of the structure to illustrate the relative B-factor distribution in the Pet passenger domain. Lower B-factors (blue) and higher B-factors (in red) are represented in different thickness and color of the c-alpha chain.
The Ramachandran plot has 87% of the residues in the most favorable regions and none in disallowed regions. The model is refined to R/Rfree= 0.22/0.263 with a maximal resolution of 2.3Å. A summary of the data collection and refinement statistics is given in Table I. The final model and structure factors have been deposited to the Protein Data Bank (pdb code: 4OM9).
Table 1.
Statistics of the data sets collected at APS, beam line 21-ID-F. Numbers in parentheses refer to values for the highest resolution shells.
| Pet protein from E.coli | |
|---|---|
| Wavelength (Å) | 0.9787 |
| Resolution range (Å) | 41.46-2.3 |
| Space group | P212121 |
| Unit Cell length (Å) | 77.76 95.93 164.87 |
| Total Reflections | 1,330,454(166,646) |
| Unique Reflections | 55340(5472) |
| Multiplicity | 25.0(22.0) |
| Completeness (%) | 99.6(99.96) |
| Mean I/σ(I) | 69.28(4.52) |
| CC1/2* | 0.901(0.83) |
| R-work | 0.22(0.287) |
| R-free | 0.263(0.325) |
| Number of atoms | 7591 |
| Water molecules | 543 |
| Protein Residues | 931 |
| RMS* (bonds) Å | 0.008 |
| RMS* (angles) deg | 1.14 |
| Ramachandran* favored (%) | 87 |
| Ramachandran* Outliers (%) | 2.7 |
| Clashscore (MolProbity) | 13.1 |
| Average B-factor# Å2 | 59 |
CC1/2 is Pearson's coefficient calculated as described in Karplus & Diederichs[[39]]
Structure of the Pet passenger domain
The overall structure of Pet passenger domain is shown in Fig. 2A. The most striking feature is the right-handed ß-helical C-terminal domain (in green). Three large and two small subdomains, interrupting the ß-helical C-terminal domain were identified in the Pet passenger protein. The three large domains consist of the N-terminal serine protease domain (in blue), a connecting α-Helix (in yellow) and a characteristic β-helix domain that consists an helical arrangement of alternating β-sheet-loop-beta-sheets segments (in green). At two points the β-helix is interrupted, first by an alpha helical motif (in red) and a beta pleaded sheet (in pink). While Pet (pdb code: 4OM9) shows only a sequence identity 50 % compared to the closest related protein sequence (the EspP protein) in the pdb (pdb code: 3SZE), the structural features of both proteins are conserved. Most parts of the Pet passenger domain are in ordered structural folds and this indicates that the protein is in a stable conformation. The mutation of the protein, which eliminated the protease cleavage site, stabilized the protein, which was essential for the success of crystallization and structure determination. The relative high overall B-factor of 59 is due to the relatively flexible C-terminus of the Pet passenger domain this is illustrated in the B-putty representation in Figure 2 C, a similar observation was reported for the EspP protein [37].
The quality of the structure can be accessed from the representation of the connecting α-helix between the protease subdomain and the β-helical stalk (Figure 3 bottom). Superimposing the known EspP structure (3SZE) to the new Pet passenger domain structure results in a RMSD of 1.31 Å. While the overall fold of both proteins is similar, the overlay (Figure 3B) reveals a closer look (Figure 4) and identifies important differences between EspP and the Pet passenger domain in two regions; in the protease domain. While Pet contains a β-pleaded sheet at the sequence region of residues 181-190, the corresponding structural domain in EspP consists of a coiled loop. (Fig 4 insert A). Secondary, the Pet passenger domain features a more pronounced beta sheet between residues 135-143 compared to the structure of EspP (Fig 4 insert B).
Figure 3.
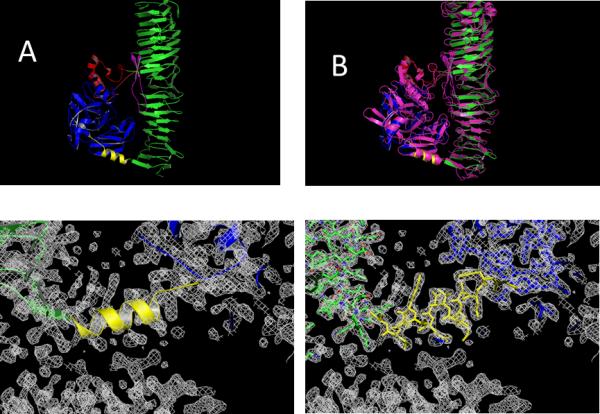
Comparison between the Pet structure, determined in this work (top left side A) with the closest related structure in the pdb (3SZE) of the EspP protein in purple (on the top right side B). The electron density map depicted on the bottom shows the connecting α-helix (yellow) between the serine protease (blue) and the β-Helical subdomain on the C-terminus (green). The model is represented in cartoon style. On the bottom right the same electron density is shown with the protein represented as a stick model. The contour level is at 1.5 σ for the 2Fo-Fc electron density maps.
Figure 4. Differences between Pet passenger domain and EspP in the protease region.
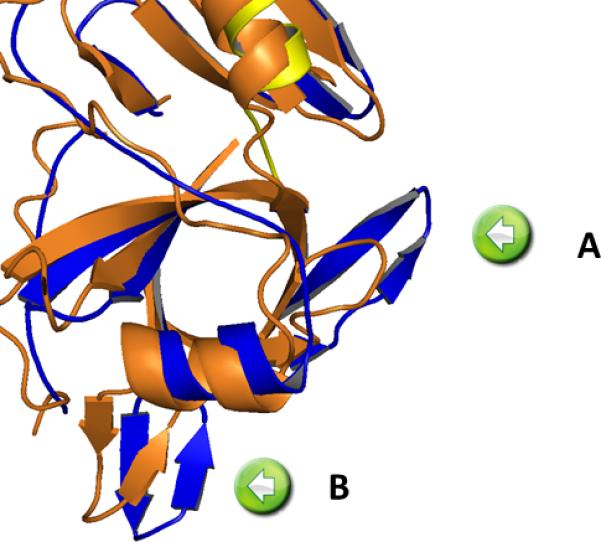
A coiled loop in EspP (insert A) and the more pronounced β-sheet between residues 135-143(insert B).
The Pet structure is represented in blue (serine protease) and yellow (helix) compared with the closest related protein in the pdb EspP (3SZE) depicted in orange.
Discussion
Despite great effort, the number of known 3D structures with atomic detail of Toxins from E. coli is still very limited. Periodic outbreaks of deadly variations of EHEC demonstrate the huge need to study toxins from the ubiquitous companions of human life[38]. Our newly solved structure of Pet reported here enlightens our understanding for these medically important toxins. Other than the low sequence identity (50%) between Pet passenger domain and EspP, the currently closest “relative” in the pdb, the overall structural homology between the two proteins is quite striking. We found significant differences, in the protease domain which may be important for the development of detoxifying agents. In particular, the pronounced beta sheets in the Pet passenger domain may be used to design drugs binding to this region that target this toxin for degradation or inhibit its toxicity. The relatively flexible β-helical part which forms the C-terminal domain of the Pet passenger domain is prominent and we may speculate that it could play a role in interaction of the pathogen with the host cell or may facilitate biofilm formation. The high flexibility may be an intrinsic feature of the β-helical domain, which could be discovered in the presented structure, because the structural features of this domain are not influenced by crystal packing. The monomeric packing of the 100kDa Pet passenger domain occurs without a reinforcing domain nearby forming crystal contact which could stabilize or alter the fold of this domain. This first crystal structure of Pet passenger domain opens the way for site directed mutagenesis in combination with structure/function studies, which could unravel the function of the individual Pet domains in pathogenesis and the development of new drugs against this notorious pathogenic bacterial strain of E.coli in the future.
X-Ray Crystal Structure of the passenger domain of Plasmid encoded toxin at 2.3 Å.
Structural differences between Pet passenger domain and EspP protein are described.
High flexibility of the C-terminal beta helix is structurally assigned.
Acknowledgments
The use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, is acknowledged. This work was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
The use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). ATL is supported by Consejo Nacional de Ciencia y Tecnología grant CB2011-164838; Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México (PAPIIT-DGAPA) grant IN201213. Special thanks should be given to Shatabdi Roy Chowdhury for proofreading of the manuscript. José Domingo Meza Aguilar appreciates the support from Doctorado en Ciencias Biomédicas, Facultad de Medicina UNAM, Consejo Nacional de Ciencia y Tecnología and the opportunity to work on this project during several research visits at Arizona State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. MMBR. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dutta PR, Cappello R, Navarro-García F, Nataro JP. Functional comparison of serine protease autotransporters of enterobacteriaceae. Infect. Immun. 2002;70:7105–7113. doi: 10.1128/IAI.70.12.7105-7113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henderson IR, Nataro JP. Virulence functions of autotransporter proteins. Infect. Immun. 2001;69:1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dautin N. Serine protease autotransporters of enterobacteriaceae (SPATEs): biogenesis and function. Toxins. 2010;2:1179–1206. doi: 10.3390/toxins2061179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsang C, Malik H, Nassman D, Huang A, Tariq F, Oelschlaeger P, Stathopoulos C. Intramolecular interactions between the protease and structural domains are important for the functions of serine protease autotransporters. Infect. Immun. 2010;78:3335–3345. doi: 10.1128/IAI.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 1997;24:767–778. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 7.Djafari S, Ebel F, Deibel C, Krämer S, Hudel M, Chakraborty T. Characterization of an exported protease from Shiga toxin-producing Escherichia coli. Mol. Microbiol. 1997;25:771–784. doi: 10.1046/j.1365-2958.1997.5141874.x. [DOI] [PubMed] [Google Scholar]
- 8.Leyton DL, Sloan J, Hill RE, Doughty S, Hartland EL. Transfer region of pO113 from enterohemorrhagic Escherichia coli: similarity with R64 and identification of a novel plasmid-encoded autotransporter. EpeA, Infect. Immun. 2003;71:6307–6319. doi: 10.1128/IAI.71.11.6307-6319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 1999;67:5587–5596. doi: 10.1128/iai.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto BR, van Dooren SJ, Nuijens JH, Luirink J, Oudega B. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J. Exp. Med. 1998;188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guyer DM, Henderson IR, Nataro JP, Mobley HL. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 2000;38:53–66. doi: 10.1046/j.1365-2958.2000.02110.x. [DOI] [PubMed] [Google Scholar]
- 12.Huang DB, Nataro JP, DuPont HL, Kamat PP, Mhatre AD, Okhuysen PC, Chiang T. Enteroaggregative Escherichia coli is a cause of acute diarrheal illness: a meta-analysis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2006;43:556–563. doi: 10.1086/505869. [DOI] [PubMed] [Google Scholar]
- 13.Nataro JP. Enteroaggregative Escherichia coli pathogenesis. Curr. Opin. Gastroenterol. 2005;21:4–8. [PubMed] [Google Scholar]
- 14.Eslava C, Navarro-García F, Czeczulin JR, Henderson IR, Cravioto A, Nataro JP. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 1998;66:3155–3163. doi: 10.1128/iai.66.7.3155-3163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrada-Garcia T, Navarro-Garcia F. Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol. Med. Microbiol. 2012;66:281–298. doi: 10.1111/j.1574-695X.2012.01008.x. [DOI] [PubMed] [Google Scholar]
- 16.Clements A, Young JC, Constantinou N, Frankel G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes. 2012;3:71–87. doi: 10.4161/gmic.19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nava-Acosta R, Navarro-Garcia F. Cytokeratin 8 is an epithelial cell receptor for Pet, a cytotoxic serine protease autotransporter of Enterobacteriaceae. mBio. 2013;4:e00838–00813. doi: 10.1128/mBio.00838-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto BR, Sijbrandi R, Luirink J, Oudega B, Heddle JG, Mizutani K, Park S-Y, Tame JRH. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J. Biol. Chem. 2005;280:17339–17345. doi: 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- 19.Meng G, Spahich N, Kenjale R, Waksman G, St Geme JW., 3rd Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. EMBO J. 2011;30:3864–3874. doi: 10.1038/emboj.2011.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Charles IG, Fairweather NF, Isaacs NW. Structure of Bordetella pertussis virulence factor P69 pertactin. Nature. 1996;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- 21.Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xicohtencatl-Cortes J, Saldaña Z, Deng W, Castañeda E, Freer E, Tarr PI, Finlay BB, Puente JL, Girón JA. Bacterial macroscopic rope-like fibers with cytopathic and adhesive properties. J. Biol. Chem. 2010;285:32336–32342. doi: 10.1074/jbc.M110.162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villaseca JM, Navarro-García F, Mendoza-Hernández G, Nataro JP, Cravioto A, Eslava C. Pet toxin from enteroaggregative Escherichia coli produces cellular damage associated with fodrin disruption. Infect. Immun. 2000;68:5920–5927. doi: 10.1128/iai.68.10.5920-5927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro-García F, Sears C, Eslava C, Cravioto A, Nataro JP. Cytoskeletal effects induced by pet, the serine protease enterotoxin of enteroaggregative Escherichia coli. Infect. Immun. 1999;67:2184–2192. doi: 10.1128/iai.67.5.2184-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chayen NE, Saridakis E. Protein crystallization: from purified protein to diffraction-quality crystal. Nat. Methods. 2008;5:147–153. doi: 10.1038/nmeth.f.203. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions, Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W. XDS, Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP 4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Fromme P, Fromme R, Levitt M, Schroeder GF. Improving the Accuracy of Macromolecular Structure Refinement at 7 angstrom Resolution. Structure. 2012;20:957–966. doi: 10.1016/j.str.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 37.Khan S, Mian HS, Sandercock LE, Chirgadze NY, Pai EF. Crystal structure of the passenger domain of the Escherichia coli autotransporter EspP. J. Mol. Biol. 2011;413:985–1000. doi: 10.1016/j.jmb.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldwater PN, Bettelheim KA. Treatment of enterohemorrhagic Escherichia coli (EHEC) infection and hemolytic uremic syndrome (HUS) BMC Med. 2012;10:12. doi: 10.1186/1741-7015-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karplus PA, Diederichs K. Linking Crystallographic Model and Data Quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]


