Abstract
Cancers comprise a heterogeneous group of human diseases. Unifying characteristics include unchecked abilities of tumor cells to proliferate and spread anatomically, and the presence of clonal advantageous genetic changes. However, universal and highly specific tumor markers are unknown. Herein, we report widespread long interspersed element-1 (LINE-1) repeat expression in human cancers. We show that nearly half of all human cancers are immunoreactive for a LINE-1–encoded protein. LINE-1 protein expression is a common feature of many types of high-grade malignant cancers, is rarely detected in early stages of tumorigenesis, and is absent from normal somatic tissues. Studies have shown that LINE-1 contributes to genetic changes in cancers, with somatic LINE-1 insertions seen in selected types of human cancers, particularly colon cancer. We sought to correlate this observation with expression of the LINE-1–encoded protein, open reading frame 1 protein, and found that LINE-1 open reading frame 1 protein is a surprisingly broad, yet highly tumor-specific, antigen.
Global hypomethylation of genomic DNA is a well-established feature of human cancers1 and may be related to transcriptional derepression of noncoding and junk DNAs. Indeed, transcription from satellite repeats has recently been described in several common epithelial tumors compared with normal tissue types.2 Herein, we focused on a commonly occurring interspersed repeat in the human genome known to encode protein, the long interspersed element-1 (LINE-1) retrotransposon. Nearly one fifth of our genome is composed of LINE-1 sequences. LINE-1 is the only active autonomous mobile DNA in humans. This activity traces to a subset of approximately 100 genomic LINE-1 copies. These encode proteins that participate in the replicative propagation of LINE-1 and other interspersed repeats in the human germline.3 Recent studies have shown that LINE-1 also contributes to genetic changes in cancers, with somatic LINE-1 insertions seen in selected types of human cancers, particularly colon cancer.4,5 We sought to correlate this observation with expression of the LINE-1–encoded protein, open reading frame 1 protein (ORF1p), and found that the LINE-1 ORF1p is a surprisingly broad, yet highly tumor-specific, antigen.
Materials and Methods
Tissue Specimens
Archived pathological samples collected for diagnostic purposes were used for TMA assembly under Institutional Review Board–approved protocols at Johns Hopkins Hospital (Baltimore, MD). No identifying information was associated with samples, and no medical record review was conducted for the purposes of this study.
IHC Data
Tissues were procured as routine surgical specimens, embedded in paraffin, and fixed in formalin. We examined LINE-1 ORF1p antigenicity using immunohistochemistry (IHC). A rabbit polyclonal LINE-1 ORF1p antibody used was previously described.6 Additional antibody panel included antibodies against Ki-67 (catalog 790-2910; Ventana Medical Systems, Tucson, AZ; prediluted), TP53 (catalog 760-2542; Ventana Medical Systems; prediluted), and monoclonal LINE-1 ORF1p antibody (see Monoclonal Anti-Human ORF1p Antibody). Briefly, immunolabeling with polyclonal LINE-1 ORF antibody was performed using a standard overnight IHC protocol. Unstained sections (5 μm thick) of each TMA were baked for 25 minutes at 65°C. Slides were deparaffinized and hydrated before the antigen retrieval step. Heat-induced antigen retrieval was performed using EDTA buffer for 20 minutes. Slides were then washed with normalizing buffer (pH 7.2 Tris-buffered saline with Tween 20; Sigma-Aldrich, St. Louis, MO) and incubated overnight with primary antibody LINE-1 ORF1p (1:1000 dilution) in a 4°C humid chamber. Incubation time was set for 18 hours, after which slides were rinsed and washed with Tris-buffered saline with Tween 20 for 15 minutes. Reaction was developed with a biotin-free Bond-polymer detection system on a Bond-Leica autostainer (Leica Microsystems, Bannockburn, IL). 3′,3′-Diaminobenzidine chromogen-substrate was used for visualization of reaction. Slides were counterstained with hematoxylin, dehydrated, and coverslipped. LINE-1 ORF1p immunoreactivity was expressed using a nominal scale (ie, negative or positive). LINE-1 ORF1p intensity was expressed semiquantitatively using a four-level ordinal scale: 0, negative; 1, low; 2, intermediate; and 3, high. The level of LINE-1 ORF1p labeling was expressed using a cell count score, which was estimated as a percentage of LINE-1 ORF1p–positive lesional cells. The LINE-1 staining score (product score) was calculated by multiplying intensity and cell count scores.
Monoclonal Anti-Human ORF1p Antibody
Fifteen peptides from the LINE-1 ORF1p consensus sequence (AAB60344.1, 338 amino acids) were selected as potential surface epitopes antibody library antigens by Abmart, Inc (Shanghai, China).
A synthetic gene encoding a multiple surface epitopes antibody library was expressed in Escherichia coli with proprietary immunogenicity enhancement factors. Each immunogen was used to immunize three BALB/c mice. The mouse with the best immune response was then used for antibody development. The monoclonal LINE-1 ORF1p antibody referenced herein targets the sequence corresponding to amino acids 35 to 44 of LINE-1 ORF1p (MENDFDELRE); this is in a region of relative divergence between human and mouse LINE-1 ORF1p proteins. The antibody purified by the manufacturer was validated in immunofluorescence, IHC, Western blot, and immunoprecipitation experiments against LINE-1 ORF1p expressed from plasmid constructs.
Statistical Analysis
Correlation of LINE-1 immunolabeling with TP53 genetic status was performed by using a binomial test. TP53 mutations were determined by bidirectional sequencing in pancreatic and ovarian carcinoma cases. TP53 status for lung carcinomas and secondary glioblastomas (GBMs) was estimated using a published IHC algorithm that closely approximates TP53 mutational status when compared with sequencing.7,8 For cases with both TP53 results available, 60 of 61 showed IHC and sequencing concordance. For comparison of LINE-1 staining scores, we used a two-sided Student’s t-test. LINE-1 immunoreactivity was scored by two of the authors (N.R. and K.H.B.).
Results
LINE-1 Retrotransposon ORF1p Protein Is Not Expressed in Normal Somatic Tissues
We used IHC to survey LINE-1 ORF1p protein expression in a collection of TMAs of human primary malignant tumors. We first used a rabbit polyclonal antibody previously shown to be specific by Western blot analysis.6 We validated staining conditions using a Tet-On inducible human LINE-1 retrotransposon expression vector. Uninduced cells showed no ORF1p immunoreactivity, whereas most induced cells showed high levels of immunolabeling in a speckled cytoplasmic pattern (Supplemental Figure S1). We next examined ORF1p immunolabeling in adult testis and embryonal carcinomas, which are reported to show LINE-1 protein expression.9,10 Consistent with these reports, we detected low LINE-1 immunoreactivity in a few human maturing spermatogonial cells and uniformly high levels of LINE-1 immunoreactivity in embryonal carcinomas (Supplemental Figure S2). We also developed a mouse monoclonal antibody against an epitope of LINE-1 ORF1p, which performed identically in these studies.
No LINE-1 protein expression was detected in mature nonneoplastic somatic tissues by IHC (Figure 1) or a series of Western blot analyses using tissue lysates (data not shown). Ki-67 (MKI67) was used to demonstrate proliferating cells in lymph node germinal centers and proliferative epithelial compartments of skin and gastrointestinal tract, which showed no corresponding LINE-1 ORF1p immunolabeling (Supplemental Figure S3).
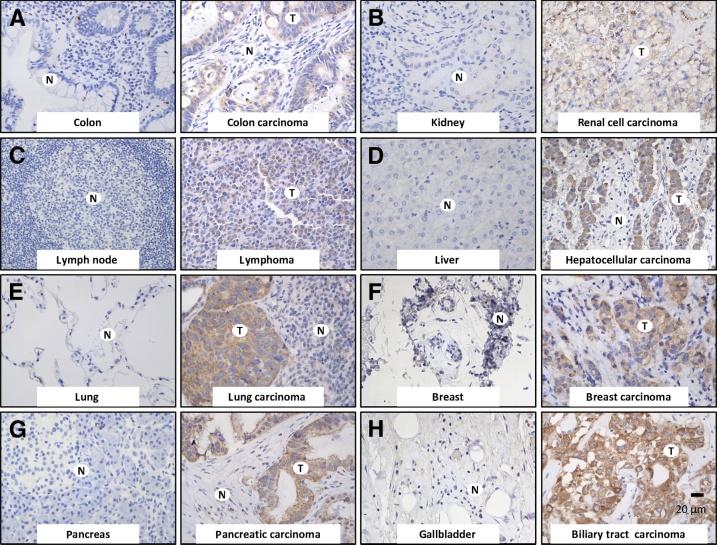
Figure 1.
LINE-1–encoded protein ORF1p is undetectable in mature human somatic tissues and commonly expressed in a wide range of human cancers. Representative photomicrographs are shown in pairs, with normal tissues on the left; tissues are counterstained (blue). Photomicrographs on the right show LINE-1 ORF1p immunoreactivity (brown) detected with a rabbit polyclonal ORF1p antibody in various human neoplasms, including colon carcinoma (A), renal cell carcinoma (B), lymphoma (C), hepatocellular carcinoma (D), lung carcinoma (E), breast carcinoma (F), pancreatic carcinoma (G), and biliary tract carcinoma (H). Original magnification, ×160 (A–H). Scale bar = 20 μm. N, normal tissue; T, tumor.
LINE-1 Retrotransposon ORF1p Protein Is a Hallmark of Many Human Cancers
We next examined LINE-1 immunolabeling across a panel of different human neoplasms (n = 1027 cases) in a collection of TMAs assembled from anatomical pathology cases from the Sidney Kimmel Comprehensive Cancer Center at the Johns Hopkins Hospital. Almost half (47%) of these were LINE-1 immunoreactive (n = 482) (Supplemental Table S1). LINE-1 ORF1p immunolabeling was restricted to cellular cytoplasm in most cases, frequently with a fine punctate or speckled pattern. Immunolabeling was seen in neoplastic cells exclusively; no intervening benign components of tumors were immunoreactive (Figure 1). LINE-1 immunolabeling frequently highlighted the presence of isolated individual neoplastic cells or small aggregates of cancer cells. Individual cases showed little intratumoral variability in LINE-1 expression, but immunoreactivity varied between cases (Supplemental Figure S4). Identical staining patterns were seen using the polyclonal and monoclonal antibodies, and there was a strong, linear relationship in staining intensities comparing the two reagents [correlation coefficient (r) = 0.746 (n = 36)] (Supplemental Figure S5). Metastatic disease was not associable with increased LINE-1 immunolabeling in the one tumor type in which this was studied. LINE-1 immunoreactivity in matched primary and metastatic pancreatic cancer specimens did not differ (n = 27; primary pancreatic cancer LINE-1 score = 1.9; matched metastasis LINE-1 score = 1.9; P = 0.86604).
We subdivided neoplasms on the basis of their primary site of origin. Figure 2A illustrates the percentage of LINE-1–immunolabeling carcinomas within each group (n = 608 cases). Neoplasms most consistently LINE-1 positive include invasive breast carcinomas (66 of 68 LINE-1–positive cases; 97% LINE-1 positive), high-grade ovarian carcinomas (29 of 31 LINE-1–positive cases; 93.5% LINE-1 positive), and pancreatic ductal adenocarcinomas (56 of 63 LINE-1–positive cases; 89% LINE-1 positive). Carcinomas originating in the endometrium, biliary tract, esophagus, bladder, head and neck, lung, and colon were frequently LINE-1 immunoreactive (mean, 61% LINE-1 positive). Conversely, carcinomas that originated in the kidney, liver, cervix, and prostate infrequently express LINE-1 ORF1p (mean, 24% positive). The intensity of immunoreactivity was highest in cancer types with relatively high overall proportions of LINE-1–positive cases. More LINE-1 immunolabeling was seen in histologically high-grade sarcomas, lymphomas, pancreatic carcinomas, and secondary GBMs (Figure 2, B–E). The correlation between LINE-1 expression and histological hallmarks of aggressive neoplasms suggests that LINE-1 expression may be an acquired feature not seen frequently in early preneoplastic lesions or low-grade tumors, but rather restricted to high-grade lesions at more advanced phases of tumorigenesis.
Figure 2.
LINE-1 ORF1p is present in a wide range of human neoplasms. A: LINE-1 expression as a percentage of LINE-1–immunoreactive cases in different primary tumor types. LINE-1 immunolabeling in low- versus high-grade mesenchymal tumors (B), lymphomas (C), pancreatic ductal neoplasms (D), and astrocytic neoplasms (E), expressed as a percentage of cases. The number above each column in panels B–E designates the number of cases examined for LINE-1 immunolabeling.
LINE-1 Retrotransposon ORF1p Expression Occurs More Commonly in TP53-Deficient Human Cancers
To test the hypothesis that acquired mutations in tumor-suppressor genes may correlate with LINE-1 expression, we implemented a regression analysis comparing the percentage of LINE-1–positive cases we observed with the mean reported TP53 mutation rates for different primary tumor types. This suggested a positive correlation between LINE-1 immunolabeling and TP53 deficiency (P = 0.02) (Figure 3A and Supplemental Table S2).11–16 We examined this relationship further using specific lung, pancreatic, and ovarian carcinoma cases in which TP53 loss of function was determined by sequencing and supported by IHC labeling of overexpression of the mutated protein (Figure 3, B and C). In lung carcinoma, LINE-1 expression was associated with TP53 deficiency (n = 85, P = 0.035, binomial test). This was also true in pancreatic and ovarian carcinomas in which many cases expressed LINE-1 and were TP53 deficient (pancreas, n = 36, P < 10−6; ovary, n = 25, P < 10−5). Notably, LINE-1–immunoreactive lung and pancreatic cancers with TP53 deficiency also had higher LINE-1 staining intensity scores compared with LINE-1–positive cases with wild-type TP53 (n = 85, P < 0.05, two-tailed Student’s t-test, and n = 36, P < 0.05, respectively) (Supplemental Figure S6, A and B).
Figure 3.
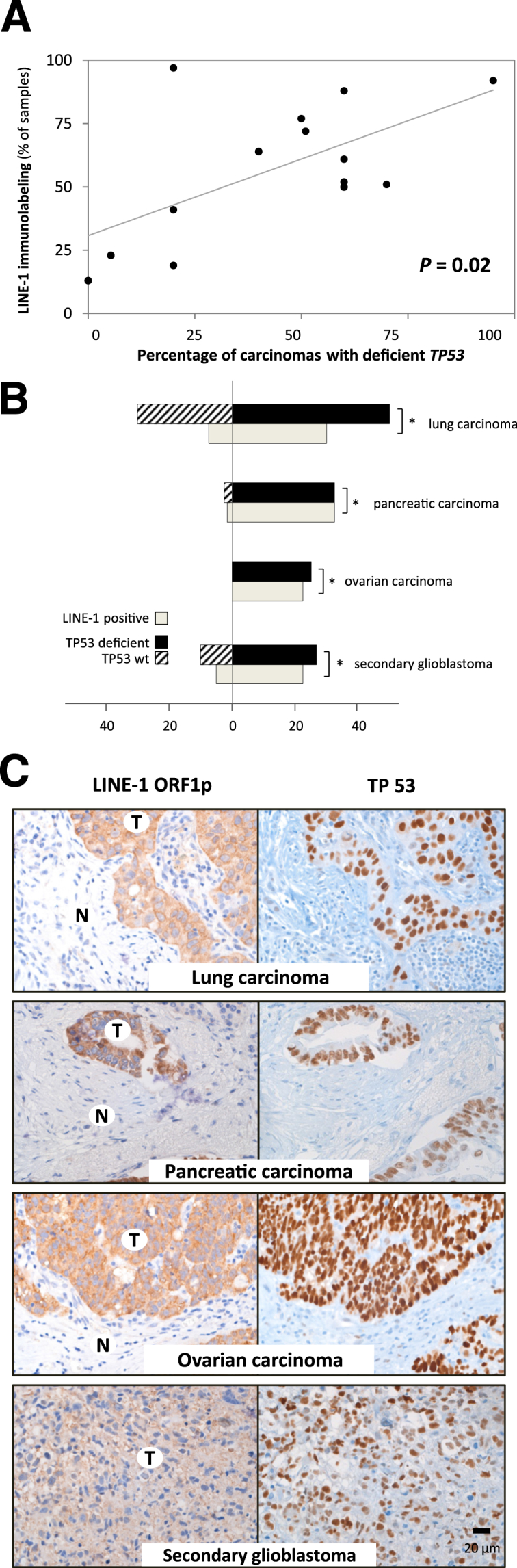
LINE-1 expression correlates with TP53 deficiency. A: Comparison of published TP53 mutation rates versus LINE-1 immunolabeling in different carcinomas. B and C: A diagram (B) and representative photomicrographs (B) depicting LINE-1 immunolabeling and TP53 deficiency in lung carcinomas, pancreatic carcinomas, ovarian carcinomas, and secondary GBMs. Immunoreactivity for TP53 is an indicator of overexpression of a mutated protein. The number of cases is denoted on the x axis of the bar graph. ∗P < 0.05, based on the binomial test. Original magnification, ×160 (A–C). Scale bar = 20 μm. N, normal tissue; T, tumor.
Glial tumors provide an illustrative example of the relationship between LINE-1 protein expression, histological grade, and TP53 deficiency. We examined 244 astrocytic neoplasms, which were overall infrequently LINE-1 positive (mean, 20%) (Supplemental Table S1). Low-grade pilocytic astrocytomas were rarely LINE-1 positive [1 (1.5%) of 67]. Intermediate-grade brain cancers, diffuse astrocytomas, and clinically aggressive anaplastic astrocytomas expressed LINE-1 protein infrequently [3 (8%) of 39 LINE-1–positive cases]. High-grade GBMs expressed LINE-1 more commonly (46 (33%) of 138 LINE-1–positive cases), albeit less frequently, than high-grade carcinomas. We then separated GBMs into primary (de novo) cases and secondary cases; the latter are thought to arise from progression of antecedent low- and intermediate-grade astrocytomas and, more frequently, harbor TP53 mutations.17 We saw many LINE-1–positive secondary GBMs [29 (74%) of 39 cases]. In secondary GBMs, we observed an association between LINE-1 immunoreactivity and TP53-deficient genetic background (n = 39, P = 0.015) (Figure 3B and Supplemental Figure S6C).
Discussion
Our observation that LINE-1 expression is a hallmark of histologically aggressive cancers extends prior studies of LINE-1 in cancer. Several groups have described hypomethylation of genomic LINE-1 sequences in a variety of tumor types, including breast, colon, hepatocellular, prostate, and neuroendocrine tumors.2,18–21
LINE-1 may contribute to genetic lesions in tumorigenesis through isolated activity of its encoded endonuclease or by promoting new LINE-1 insertions that affect gene expression. Evidence for the latter has been described by targeted sequencing in colorectal cancers,4,5 lung cancers,22 and hepatocellular carcinomas23 and by analysis of whole-genome sequences of colorectal, ovarian, and pancreatic cancers.4 Direct comparisons of ORF1p expression levels and somatic retrotransposition rates are needed; presumably, new transposition events require a combination of aberrant LINE-1 expression and other host conditions permissive for transposition.
Although nuclear localization of a portion of LINE-1–encoded protein is required for transposition, we primarily observe a punctate cytoplasmic pattern of LINE-1 ORF1p immunolabeling in most cases. This largely agrees with published observations, from both in vitro expression of LINE-1 constructs used for retrotransposition assays24 and human tumors. However, two groups have described a small fraction, approximately 5%, of human breast carcinomas displaying nuclear LINE-1 ORF1p immunolabeling.6,25 More important, the same groups associated nuclear immunolabeling with poor clinical outcomes. It will be of considerable interest to enhance our sensitivity for detecting nuclear LINE-1 ORF1p and investigate whether this corresponds to somatic LINE-1 retrotransposition in tumors.
LINE-1 ORF1p may have clinical utility as a neoplastic marker. It is broadly expressed in a wide variety of high-grade human neoplasms, including highly lethal malignancies in which early detection remains a clinical challenge. How many genomic templates contribute to immunoreactivity for ORF1p and whether interpatient heterogeneity is related to differences in the inherited complement of active genomic LINE-1 sequences remain unknown. More important, we report minimal to no intratumoral heterogeneity and similar LINE-1 ORF1p immunolabeling in matched primary and metastatic specimens. These findings are in agreement with a prior study by Matsunoki et al,26 who reported little intrapatient heterogeneity in genomic LINE-1 methylation in colorectal carcinomas and metastatic samples. This property may make LINE-1 ORF1p expression useful in detecting residual disease and in evaluating small specimens or those limited by rare lesional cells. It is also possible that LINE-1 can be targeted in therapeutic interventions, developed either to prevent roles LINE-1 plays in clonal evolution of transformed cells or to potentiate LINE-1 expression to promote cell lethal genotoxicity.
Our data indicate a strong, but imperfect, correlation between TP53 mutation and LINE-1 expression. Congruous observations have been made recently in colorectal carcinomas, which show a tendency for LINE-1 DNA hypomethylation in TP53-deficient tumors.27 Thus, although the LINE-1 promoter can be bound and transcriptionally activated by p53 in vitro,28 it appears that this is dispensable for LINE-1 activity in TP53-mutated tumors and that other factors are primary determinants in the relationship between LINE-1 expression and the p53 pathway. One model is that loss of TP53 promotes the viability and proliferative potential of LINE-1–expressing cells, perhaps by escape from p53-mediated responses to LINE-1–induced DNA damage. In support of this, studies enforcing ectopic expression of LINE-1 have described double-stranded DNA breaks, cytotoxicity, and apoptosis in TP53 wild-type cells.24,29,30 However, although TP53 loss may promote LINE-1 transposition, LINE-1 can be expressed and transpose with TP53 intact.24,31,32 Interestingly, TP53 control of interspersed repeat expression may extend beyond LINE-1. Leonova et al33 have recently shown in mouse that DNA hypomethylation causes massive accumulation of short interspersed element transcription and double-stranded RNA accumulation specifically in TP53-deficient cells.
In conclusion, we estimate that as many as half of human cancers express ORF1p. Within many tumor classification schemes, we associate LINE-1 expression with higher histological grades and deficiency of the TP53 tumor suppressor.
Acknowledgments
We thank Drs. Jim Eshleman and Corrado Spadafora for critical review of the manuscript and the Johns Hopkins Biostatistics Center for providing expert consults.
Footnotes
Supported by the Johns Hopkins University School of Medicine Department of Pathology; NIH awards K08CA134746 (K.H.B.), R01CA161210 (J.D.B.), and R01CA163705 (K.H.B.); the Burroughs Wellcome Fund Career Awards for Medical Scientists (K.H.B.); Sol Goldman Pancreatic Cancer Research Fund award (N.R.); and the Howard Hughes Medical Institute Medical Research Fellows Program (J.Z.). Resources for TMAs used in this study include a Joseph C. Eggleston Fund award in Surgical Pathology (A.S.M.) and the Brady Urological Research Institute Prostate Specimen Repository award, which is supported by the National Cancer Institute/NIH Prostate SPOREP50CA58236 Pathology Core and the Department of Defense Funded Prostate Biospecimen Repository Network site at Johns Hopkins.
Disclosures: None declared.
Contributor Information
Nemanja Rodić, Email: nrodic1@jhmi.edu.
Kathleen H. Burns, Email: kburns@jhmi.edu.
Supplemental Data
The IHC (A) and Western blot (B) validation of exogenous LINE-1 ORF1p detection, using polyclonal anti-ORF1p antibody. Tet-On HeLa cells were stably transfected doxycycline (Dox)–inducible LINE-1 (LINE-1 RP) retrotransposon expression vector and either treated or not treated with 1 μg/mL Dox. Photomicrographs in A are shown in pairs, with the H&E-stained sections and IHC sections showing LINE-1-ORF1p immunoreactivity. ORF1p expression is seen as a brown signal; the counterstain is blue. Identical results were obtained using a mouse monoclonal anti–LINE-1 ORF1p antibody (data not shown). Original magnifications: ×160 (A and B). Scale bars: 20 μm (A and B).
The IHC validation of endogenous LINE-1 ORF1p immunolabeling. A representative LINE-1–ORF1p–positive embryonal carcinoma sample (A and B) and histologically normal adult testis, where LINE-1 ORF1p highlights infrequent spermatogonial cells (C). Original magnifications: ×160 (A and C); ×260 (B). Scale bar = 20 μm.
LINE-1 ORF1p is not expressed in proliferating benign human cells. Undetectable LINE-1 immunolabeling in proliferating compartments of a reactive lymph node (A), skin (C), and colon (E). Infrequent colonic crypt base cells exhibited a non-specific reactivity qualitatively consistent with neuroendocrine vacuoles is shown in E. Ki-67 cell cycle antigen is expressed in proliferating cells, specifically reactive lymphocytes (B), epidermal basal cells (D), and colonic mucosa cells lining the crypts (F).
Scoring system for intensity of LINE-1 IHC staining. Photomicrographs show semiquantitative intensity of LINE-1 immunoreactivity in representative TMA cores. Original magnification, ×20. Scale bar = 100 μm.
Representative photomicrographs showing side-by-side comparison of two pancreatic cancer cases immunolabeled with polyclonal and monoclonal LINE-1 ORF1p antibodies. A representative LINE-1–ORF1p low to negative pancreatic carcinoma case immunolabeled with polyclonal (A and B) and monoclonal (C and D) LINE-1 ORF1p antibodies. A representative LINE-1 ORF1p high positive pancreatic carcinoma case immunolabeled with polyclonal (E and F) and monoclonal (G and H) antibodies. Original magnifications: ×64 (A, C, E, and G); ×260 (B, D, F, and H).
LINE-1 immunolabeling intensity in TP53 wild-type (wt) and TP53 mutated tumors, related to Figure 3. Increased LINE-1 immunolabeling intensity was associated with deficient TP53 in lung carcinomas (A), pancreatic carcinomas (B), and secondary GBMs (C). ∗P < 0.05, based on a two-sided Student’s t-test.
References
- 1.Feinberg A.P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 2.Ting D.T., Lipson D., Paul S., Brannigan B.W., Akhavanfard S., Coffman E.J., Contino G., Deshpande V., Iafrate A.J., Letovsky S., Rivera M.N., Bardeesy N., Maheswaran S., Haber D.A. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–596. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns K.H., Boeke J.D. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee E., Iskow R., Yang L., Gokcumen O., Hanseley P., Luquette L.J., Lohr J.G., Harris C.C., Ding L., Wilson R.K., Wheeler D.A., Gibbs R.A., Kucherlapati R., Lee C., Kharchenko P.V., Park P.J., Network TCGAR Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solyom S., Ewing A.D., Rahrmann E.P., Doucet T., Nelson H.H., Burns M.B., Harris R.S., Sigmon D.F., Casella A., Erlanger B., Wheelan S., Upton K.R., Shukla R., Faulkner G.J., Largaespada D.A., Kazazian H.H., Jr. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22:2328–2338. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris C.R., Normart R., Yang Q., Stevenson E., Haffty B.G., Ganesan S., Cordon-Cardo C., Levine A.J., Tang L.H. Association of nuclear localization of a long interspersed nuclear element-1 protein in breast tumors with poor prognostic outcomes. Genes Cancer. 2010;1:115–124. doi: 10.1177/1947601909360812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu B., Yachida S., Morgan R., Zhong Y., Montgomery E.A., Iacobuzio-Donahue C.A. Clinicopathologic and genetic characterization of traditional serrated adenomas of the colon. Am J Clin Pathol. 2012;138:356–366. doi: 10.1309/AJCPVT7LC4CRPZSK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vang R., Visvanathan K., Gross A., Maambo E., Gupta M., Kuhn E., Li R.F., Ronnett B.M., Seidman J.D., Yemelyanova A., Shih Ie M., Shaw P.A., Soslow R.A., Kurman R.J. Validation of an algorithm for the diagnosis of serous tubal intraepithelial carcinoma. Int J Gynecol Pathol. 2012;31:243–253. doi: 10.1097/PGP.0b013e31823b8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ergün S., Buschmann C., Heukeshoven J., Dammann K., Schnieders F., Lauke H., Chalajour F., Kilic N., Strätling W.H., Schumann G.G. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J Biol Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 10.Bratthauer G.L., Fanning T.G. Active LINE-1 retrotransposons in human testicular cancer. Oncogene. 1992;7:507–510. [PubMed] [Google Scholar]
- 11.Bell D., Berchuck A., Birrer M., Chien J., Cramer D., Dao F., Dhir R., DiSaia P., Gabra H., Glenn P., Godwin A., Gross J., Hartmann L., Huang M., Huntsman D., Iacocca M., Imielinski M., Kalloger S., Karlan B., Levine D., Mills G., Morrison C., Mutch D., Olvera N., Orsulic S., Park K., Petrelli N., Rabeno B., Rader J., Sikic B., Smith-McCune K., Sood A., Bowtell D., Penny R., Testa J., Chang K., Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hruban R, Iacobuzio-Donahue C: The pancreas. Robbins and Cotran Pathologic Basis of Disease, Professional Edition, ch 19. Philadelphia, Elsevier Inc, 2010, pp 901.
- 13.Elleson L, Pirog E: The female genital tract. Robbins and Cotran Pathologic Basis of Disease, Professional Edition, ch 22. Philadelphia, Elsevier Inc, 2010, pp 1032.
- 14.Khan S.A., Thomas H.C., Toledano M.B., Cox I.J., Taylor-Robinson S.D. p53 Mutations in human cholangiocarcinoma: a review. Liver Int. 2005;25:704–716. doi: 10.1111/j.1478-3231.2005.01106.x. [DOI] [PubMed] [Google Scholar]
- 15.Edlund K., Larsson O., Ameur A., Bunikis I., Gyllensten U., Leroy B., Sundström M., Micke P., Botling J., Soussi T. Data-driven unbiased curation of the TP53 tumor suppressor gene mutation database and validation by ultradeep sequencing of human tumors. Proc Natl Acad Sci U S A. 2012;109:9551–9556. doi: 10.1073/pnas.1200019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girgin C., Tarhan H., Hekimgil M., Sezer A., Gürel G. P53 mutations and other prognostic factors of renal cell carcinoma. Urol Int. 2001;66:78–83. doi: 10.1159/000056575. [DOI] [PubMed] [Google Scholar]
- 17.Ohgaki H., Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Hoesel A.Q., van de Velde C.J., Kuppen P.J., Liefers G.J., Putter H., Sato Y., Elashoff D.A., Turner R.R., Shamonki J.M., de Kruijf E.M., van Nes J.G., Giuliano A.E., Hoon D.S. Hypomethylation of LINE-1 in primary tumor has poor prognosis in young breast cancer patients: a retrospective cohort study. Breast Cancer Res Treat. 2012;134:1103–1114. doi: 10.1007/s10549-012-2038-0. [DOI] [PubMed] [Google Scholar]
- 19.Sunami E., de Maat M., Vu A., Turner R.R., Hoon D.S. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Formeister E.J., Tsuchiya M., Fujii H., Shpyleva S., Pogribny I.P., Rusyn I. Comparative analysis of promoter methylation and gene expression endpoints between tumorous and non-tumorous tissues from HCV-positive patients with hepatocellular carcinoma. Mutat Res. 2010;692:26–33. doi: 10.1016/j.mrfmmm.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho N.Y., Kim B.H., Choi M., Yoo E.J., Moon K.C., Cho Y.M., Kim D., Kang G.H. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211:269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 22.Iskow R.C., McCabe M.T., Mills R.E., Torene S., Pittard W.S., Neuwald A.F., Van Meir E.G., Vertino P.M., Devine S.E. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shukla R., Upton K.R., Munoz-Lopez M., Gerhardt D.J., Fisher M.E., Nguyen T., Brennan P.M., Baillie J.K., Collino A., Ghisletti S., Sinha S., Iannelli F., Radaelli E., Dos Santos A., Rapoud D., Guettier C., Samuel D., Natoli G., Carninci P., Ciccarelli F.D., Garcia-Perez J.L., Faivre J., Faulkner G.J. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belgnaoui S.M., Gosden R.G., Semmes O.J., Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L., Dahlstrom J.E., Chandra A., Board P., Rangasamy D. Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res Treat. 2012;136:129–142. doi: 10.1007/s10549-012-2246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsunoki A., Kawakami K., Kotake M., Kaneko M., Kitamura H., Ooi A., Watanabe G., Minamoto T. LINE-1 methylation shows little intra-patient heterogeneity in primary and synchronous metastatic colorectal cancer. BMC Cancer. 2012;12:574. doi: 10.1186/1471-2407-12-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morikawa T., Kuchiba A., Liao X., Imamura Y., Yamauchi M., Qian Z.R., Nishihara R., Sato K., Meyerhardt J.A., Fuchs C.S., Ogino S. Tumor TP53 expression status, body mass index and prognosis in colorectal cancer. Int J Cancer. 2012;131:1169–1178. doi: 10.1002/ijc.26495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris C.R., Dewan A., Zupnick A., Normart R., Gabriel A., Prives C., Levine A.J., Hoh J. p53 Responsive elements in human retrotransposons. Oncogene. 2009;28:3857–3865. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace N.A., Belancio V.P., Deininger P.L. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419:75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasior S.L., Wakeman T.P., Xu B., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coufal N.G., Garcia-Perez J.L., Peng G.E., Marchetto M.C., Muotri A.R., Mu Y., Carson C.T., Macia A., Moran J.V., Gage F.H. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc Natl Acad Sci U S A. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haoudi A., Semmes O.J., Mason J.M., Cannon R.E. Retrotransposition-competent human LINE-1 induces apoptosis in cancer cells with intact p53. J Biomed Biotechnol. 2004;2004:185–194. doi: 10.1155/S1110724304403131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonova K.I., Brodsky L., Lipchick B., Pal M., Novototskaya L., Chenchik A.A., Sen G.C., Komarova E.A., Gudkov A.V. p53 Cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci U S A. 2013;110:E89–E98. doi: 10.1073/pnas.1216922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The IHC (A) and Western blot (B) validation of exogenous LINE-1 ORF1p detection, using polyclonal anti-ORF1p antibody. Tet-On HeLa cells were stably transfected doxycycline (Dox)–inducible LINE-1 (LINE-1 RP) retrotransposon expression vector and either treated or not treated with 1 μg/mL Dox. Photomicrographs in A are shown in pairs, with the H&E-stained sections and IHC sections showing LINE-1-ORF1p immunoreactivity. ORF1p expression is seen as a brown signal; the counterstain is blue. Identical results were obtained using a mouse monoclonal anti–LINE-1 ORF1p antibody (data not shown). Original magnifications: ×160 (A and B). Scale bars: 20 μm (A and B).
The IHC validation of endogenous LINE-1 ORF1p immunolabeling. A representative LINE-1–ORF1p–positive embryonal carcinoma sample (A and B) and histologically normal adult testis, where LINE-1 ORF1p highlights infrequent spermatogonial cells (C). Original magnifications: ×160 (A and C); ×260 (B). Scale bar = 20 μm.
LINE-1 ORF1p is not expressed in proliferating benign human cells. Undetectable LINE-1 immunolabeling in proliferating compartments of a reactive lymph node (A), skin (C), and colon (E). Infrequent colonic crypt base cells exhibited a non-specific reactivity qualitatively consistent with neuroendocrine vacuoles is shown in E. Ki-67 cell cycle antigen is expressed in proliferating cells, specifically reactive lymphocytes (B), epidermal basal cells (D), and colonic mucosa cells lining the crypts (F).
Scoring system for intensity of LINE-1 IHC staining. Photomicrographs show semiquantitative intensity of LINE-1 immunoreactivity in representative TMA cores. Original magnification, ×20. Scale bar = 100 μm.
Representative photomicrographs showing side-by-side comparison of two pancreatic cancer cases immunolabeled with polyclonal and monoclonal LINE-1 ORF1p antibodies. A representative LINE-1–ORF1p low to negative pancreatic carcinoma case immunolabeled with polyclonal (A and B) and monoclonal (C and D) LINE-1 ORF1p antibodies. A representative LINE-1 ORF1p high positive pancreatic carcinoma case immunolabeled with polyclonal (E and F) and monoclonal (G and H) antibodies. Original magnifications: ×64 (A, C, E, and G); ×260 (B, D, F, and H).
LINE-1 immunolabeling intensity in TP53 wild-type (wt) and TP53 mutated tumors, related to Figure 3. Increased LINE-1 immunolabeling intensity was associated with deficient TP53 in lung carcinomas (A), pancreatic carcinomas (B), and secondary GBMs (C). ∗P < 0.05, based on a two-sided Student’s t-test.