Abstract
Oxygenases are promising biocatalysts for performing selective hydroxylations not accessible by chemical methods. Whereas toluene 4-monooxygenase (T4MO) of Pseudomonas mendocina KR1 hydroxylates monosubstituted benzenes at the para position and toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4 hydroxylates at the ortho position, toluene 3-monooxygenase (T3MO) of Ralstonia pickettii PKO1 was reported previously to hydroxylate toluene at the meta position, producing primarily m-cresol (R. H. Olsen, J. J. Kukor, and B. Kaphammer, J. Bacteriol. 176:3749-3756, 1994). Using gas chromatography, we have discovered that T3MO hydroxylates monosubstituted benzenes predominantly at the para position. TG1/pBS(Kan)T3MO cells expressing T3MO oxidized toluene at a maximal rate of 11.5 ± 0.33 nmol/min/mg of protein with an apparent Km value of 250 μM and produced 90% p-cresol and 10% m-cresol. This product mixture was successively transformed to 4-methylcatechol. T4MO, in comparison, produces 97% p-cresol and 3% m-cresol. Pseudomonas aeruginosa PAO1 harboring pRO1966 (the original T3MO-bearing plasmid) also exhibited the same product distribution as that of TG1/pBS(Kan)T3MO. TG1/pBS(Kan)T3MO produced 66% p-nitrophenol and 34% m-nitrophenol from nitrobenzene and 100% p-methoxyphenol from methoxybenzene, as well as 62% 1-naphthol and 38% 2-naphthol from naphthalene; similar results were found with TG1/pBS(Kan)T4MO. Sequencing of the tbu locus from pBS(Kan)T3MO and pRO1966 revealed complete identity between the two, thus eliminating any possible cloning errors. 1H nuclear magnetic resonance analysis confirmed the structural identity of p-cresol in samples containing the product of hydroxylation of toluene by pBS(Kan)T3MO.
Monooxygenases catalyze the introduction of an oxygen atom into a substrate while the second oxygen atom is reduced to water with electrons from NAD(P)H. This complex reaction often requires a metal center, and electron transfer from the reduced cofactors is mediated by additional proteins (15, 17). Oxidizing enzymes are of great potential to the chemical and pharmaceutical industries due to their high regio-, chemo-, and stereoselectivity and their ability to facilitate reactions with chemically stable substrates (1, 2). Due to their complexity, biological oxidation reactions are often performed using growing or resting cells (17).
One important group of bacterial oxygenases includes the toluene monooxygenases, which have additional potential applications in bioremediation (23, 30, 32). The aerobic biodegradation of toluene is well studied, and the pathway is available at the University of Minnesota databank (9). Xylene monooxygenase of Pseudomonas putida mt-2 hydroxylates toluene at the methyl side chain, resulting in benzyl alcohol (1). Toluene ortho-monooxygenase (TOM) of Burkholderia cepacia G4 hydroxylates the benzene ring at the ortho position to form o-cresol, which is further oxidized to 3-methylcatechol (20, 29). Toluene 3-monooxygenase (T3MO) of Ralstonia pickettii PKO1 was reported to hydroxylate toluene at the meta position (21), resulting in m-cresol, and toluene 4-monooxygenase (T4MO) of Pseudomonas mendocina KR1 is specific for para hydroxylations, producing primarily p-cresol (35).
Within this group of toluene monooxygenases, T3MO and T4MO share the highest similarity. The tbu operon of T3MO and the tmo operon of T4MO are identical in their organization and exhibit greater than 60% identity on the DNA level (16). Both T3MO (3) and T4MO (31) contain four components: a hydroxylase protein composed of three subunits in a (αβγ)2 quaternary structure (tbuA1A2U and tmoAEB, respectively), an NADH oxidoreductase (tbuC and tmoF, respectively), a small effector protein with no prosthetic groups (tbuV and tmoD, respectively), and a Rieske-type ferredoxin protein (tbuB and tmoC, respectively). The α subunit of the hydroxylase enzyme in T4MO (TmoA), which has been shown to control the regiospecificity of this enzyme (19, 25; A. Fishman, Y. Tao, W. E. Bentley, and T. K. Wood, unpublished data), shares 67.3% amino acid identity with the TbuA1 hydroxylase of T3MO (3).
Although much work has been done on the regulation of the T3MO operon (4, 13, 14, 16), little work on its catalytic activity on aromatic substrates has been reported. Olsen and coworkers initially isolated the T3MO operon from the R. pickettii PKO1 chromosome and cloned it into plasmid pRO1966 (21). Pseudomonas aeruginosa PAO1 cells containing this plasmid were reported to form m-cresol from toluene oxidation and were reported to not convert the cresol to methylcatechol (21). However, Tao et al. (33) have recently described the successive hydroxylation of benzene to phenol, phenol to catechol, and catechol to 1,2,3-trihydroxybenzene by Escherichia coli TG1 cells expressing T3MO. Here, we report the regiospecificity of T3MO on various aromatic compounds, show that it is capable of forming 4-methylcatechol from toluene, and indicate that the enzyme originally named T3MO is actually a para-hydroxylating enzyme, not a meta-oxidizing enzyme.
MATERIALS AND METHODS
Chemicals.
Nitrobenzene (NB) and naphthalene were purchased from Fisher Scientific Co. (Fair Lawn, N.J.), and p-cresol, methoxybenzene, 3-methoxyphenol, o-nitrophenol, m-nitrophenol, and p-nitrophenol were obtained from Acros Organics (Morris Plains, N.J.). o-Cresol, m-cresol, 1-naphthol, 2-naphthol, 4-methoxyphenol, methoxyhydroquinone, and guaiacol were obtained from Aldrich Chemical Co. (Milwaukee, Wis.). All materials used were of the highest purity available and were used without further purification.
Bacterial strains and growth conditions.
E. coli TG1 {supE hsdΔ5 thi Δ(lac-proAB) F′ [traD36 proAB+ lacIq lacZΔM15]} with the plasmid constructs was routinely cultivated at 37°C with shaking at 250 rpm on a C25 incubator shaker (New Brunswick Scientific Co., Edison, N.J.) in Luria-Bertani (LB) medium (27) supplemented with kanamycin at 100 μg/ml to maintain the plasmids. All experiments were conducted by diluting overnight cells to an optical density at 600 nm (OD) of 0.1 to 0.2 and growing them to an OD of 1.2. The exponentially grown cells were centrifuged at 13,000 × g for 8 min at 25°C in a Beckman J2-HS centrifuge (Palo Alto, Calif.) and resuspended in Tris-HNO3 buffer (50 mM, pH 7.0) or potassium phosphate buffer (50 mM, pH 7.0). Expression of wild-type T3MO or T4MO from pBS(Kan)T3MO [henceforth TG1(T3MO)] or pBS(Kan)T4MO [henceforth TG1(T4MO)] within E. coli strains produced blue cells on agar plates and in broth cultures. The blue color is indicative of indigo, formed by oxidation of indole from tryptophan (8).
P. aeruginosa PAO1 carrying pRO1966 (21) (henceforth PAO1/pRO1966) was grown overnight at 30°C on either LB medium or minimal medium M9 (27) containing either 0.6% lactate or 50 mM succinate as carbon source, supplied with 750 μg of ampicillin/ml. Cells were diluted with the same medium to an OD of 0.1 to 0.2, and toluene was added as an inducer for monooxygenase expression in the vapor phase by placing 100 μl in a small tube suspended from a rubber stopper. The exponentially grown cells were centrifuged and resuspended in potassium phosphate buffer (50 mM, pH 7.0). P. aeruginosa PAO1 was grown on LB medium and used as a negative control.
Construction of expression vectors.
To stably and constitutively express the toluene monooxygenase genes from the same promoter, the expression vectors pBS(Kan)T3MO and pBS(Kan)T4MO were constructed as described earlier by Tao et al. (33). The protein content of TG1(T3MO) and TG1(T4MO) was 0.24 mg of protein/ml/OD unit (33).
DNA sequencing and restriction enzyme analysis.
A dideoxy chain termination technique (28) with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Wellesley, Mass.) and a PE Biosystems ABI 373 DNA sequencer (Perkin-Elmer) was used to determine the nucleotide sequences of TG1(T3MO) and PAO1/pRO1966. Plasmids were isolated using a Mini or Midi Kit (Qiagen, Inc., Chatsworth, Calif.). Restriction enzymes XhoI, BclI, EcoRI, and AatII were used to digest both plasmids, and digestion products were analyzed on an 0.6% (wt/vol) agarose gel. Nine primers (Table 1) of 19 to 29 bp were designed based on the wild-type T3MO of R. pickettii (GenBank accession no. U04052) (3) for sequencing the T3MO locus tbuA1UBVA2C. The upstream region was sequenced using primer T3MOR1 (Table 1). Sequence data generated were analyzed using Vector NTI software (InforMax, Inc., Bethesda, Md.).
TABLE 1.
Primers used for sequencing the T3MO tbuA1UBVA2C locus of both TG1(T3MO) and P. aeruginosa PAO1/pRO1966
| Primer | Nucleotide sequence |
|---|---|
| T3MOBam HIfront | 5′-TGAGGGATCCCGCCAAGCAAAAAACACTAC-3′ |
| T3MotbuA1 | 5′-ATTTCCCGCACGACTATTGC-3′ |
| T3MotbuA2 | 5′-AACGCACCTTGATCGACCTG-3′ |
| T3MotbuA3 | 5′-GCTTCAGTACATGAACCTGGC-3′ |
| T3MotbuB1 | 5′-TGCGTTCCAGGGAATCTGCC-3′ |
| T3MotbuV1 | 5′-CGACTACGTCCGCTTCTAC-3′ |
| T3MotbuA4 | 5′-ATCTCGAACTGCGCGGCGTAC-3′ |
| T3MotbuA5 | 5′-GGTGCGCGAGTTCCGGCAC-3′ |
| T3MotbuC1 | 5′-AGTACGCGCTGCTGTATCCG-3′ |
| T3MOR1 | 5′-GGTGATGTCTCCTTGGTAGGG-3′ |
Enzymatic activity.
Oxidation of NB, naphthalene, methoxybenzene, and cresols was determined using high-pressure liquid chromatography (HPLC). Two milliliters of concentrated cell suspensions (OD of 5 to 8) was mixed with substrate concentrations specified in the text (from a 100 mM stock solution in ethanol) in 15-ml serum vials sealed with a Teflon-coated septum and aluminum crimp seal. The specific initial reaction rate was constant over this range of cell biomass. The negative controls used in these experiments contained the same monooxygenase without substrates (plus solvent) as well as TG1/pBS(Kan) with substrates (no-monooxygenase control). The inverted vials were shaken at room temperature at 300 rpm on an IKA-Vibrax-VXR shaker (Cincinnati, Ohio) for 2.5 to 30 min, and then 1 ml of the cell suspension was removed and centrifuged in a 16 M Labnet Spectrafuge (Edison, N.J.) for 1 to 2 min. The supernatant was filtered and analyzed.
For toluene oxidation, the cells [E. coli TG1 harboring pBS(Kan)T3MO or pBS(Kan)T4MO or P. aeuroginosa PAO1 harboring pRO1966] were prepared as described earlier and phosphate buffer (50 mM, pH 7) was used for washing and resuspending the cells. The serum vials containing exponentially grown cells were sealed, and then 250 to 1,930 μM toluene was added with a syringe, calculated as if all the toluene were in the liquid phase (actual initial liquid concentration was 90 to 700 μM, based on Henry's law) (7). The reaction was stopped by adding 2 ml of 500 μM hexadecane in ethyl acetate to the vial with a syringe, and the vial was vortexed thoroughly to ensure full extraction of the toluene. The organic phase was separated from the aqueous phase by centrifugation, and 2 to 3 μl was injected into the gas chromatography (GC) column. At least two independent experiments were performed to characterize each strain with each substrate described in this paper.
Analytical methods.
Oxidation of toluene (to methylcatechol), NB, methoxybenzene, o-cresol, m-cresol, p-cresol, and naphthalene was measured using reverse-phase HPLC. Filtered samples were injected into a Zorbax SB-C8 column (Agilent Technologies; 5 μm, 4.6 by 250 mm) with a Waters Corporation (Milford, Mass.) 515 solvent delivery system coupled to a photodiode array detector (Waters 996). The gradient elution for detecting NB and cresol oxidation products was performed with H2O (0.1% formic acid) and acetonitrile (70:30 for 0 to 8 min, gradient to 40:60 at 15 min, and gradient to 70:30 at 20 min) as the mobile phase at a flow rate of 1 ml/min. The gradient elution for detecting methoxybenzene products was 80:20 for 0 to 17 min, gradient to 50:50 at 22 min, and gradient to 80:20 at 30 min and for naphthalene oxidation products was 65:35 for 0 to 5 min, gradient to 35:65 at 12 min, and gradient to 65:35 at 20 min. Compounds were identified by comparison of retention times and UV-visible spectra to those of authentic standards as well as by coelution with standards.
Oxidation of toluene to cresol was measured by GC with a Hewlett-Packard 6890N gas chromatograph equipped with an EC-WAX capillary column (30 m by 0.25 mm, 0.25-μm thickness; Alltech Associates, Inc., Deerfield, Ill.) and a flame ionization detector. The injector and detector were maintained at 250 and 275°C, respectively, and a split ratio of 3:1 was used. The He carrier gas flow rate was maintained at 0.8 ml/min. The temperature program was 80°C for 5 min and 80 to 205°C at a rate of 5°C/min, 205 to 280°C at 15°C/min, and 280°C for 5 min. Under these conditions, the retention times for toluene and o-, p-, and m-cresols were 4.2, 27.2, 28.9, and 29.1 min, respectively. Hexadecane was used as an internal standard. Retention times were determined by comparisons to authentic standards as well as by coelution with standards.
1H nuclear magnetic resonance (NMR) analysis was performed on a DRX-400 (400.144-MHz) instrument (Bruker BioSpin Corp., Billerica, Mass.) with a standard 30° pulse and 1.5-s relaxation delay. CDCl3 was used as a solvent.
RESULTS
Toluene oxidation rate and product distribution by T3MO.
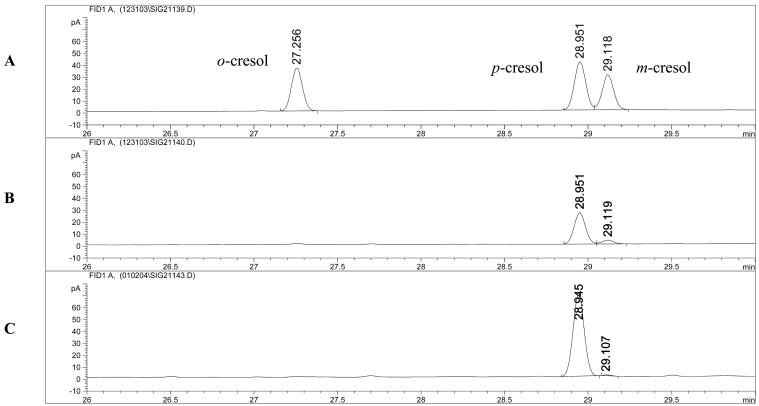
TG1 cells expressing T3MO were recently shown by our group to oxidize successively benzene to phenol, phenol to catechol, and catechol to 1,2,3-trihydroxybenzene (33). While studying the oxidation of toluene by these cells using GC, we discovered that they produce 90% p-cresol and 10% m-cresol (Fig. 1B; Table 2), in contrast to the 100% m-cresol reported by Olsen et al. (21). These results were obtained at least 10 times in independent experiments and were verified by comparing the retention times with authentic standards (Fig. 1A) and by performing coelution with m-cresol. In addition, we compared the results with those of TG1(T4MO) (Fig. 1C), which has been shown to oxidize toluene to primarily p-cresol (19; Fishman et al., unpublished), and TG1 cells expressing pBS(Kan)TOM [TG1(TOM)], which oxidize toluene to 100% o-cresol (20, 26). TG1/pBS(Kan) cells did not oxidize toluene at all, indicating that the oxidation of toluene is related to the expression of the monooxygenase.
FIG. 1.
GC chromatogram of a mixture of o-cresol, p-cresol, and m-cresol standards (A); products formed from toluene oxidation by TG1(T3MO) (B); and products formed by TG1(T4MO) (C).
TABLE 2.
Saturation kinetic values (apparent Vmax and Km) and product distribution for toluene oxidation by E. coli TG1 cells expressing T3MO and T4MO from pBS(Kan)T3MO and pBS(Kan)T4MO, respectively, and toluene product distribution by P. aeruginosa PAO1/pRO1966 expressing T3MO grown under different conditions
| Strain | Growth mediumc | Kinetics of toluene oxidationa
|
Regiospecificity of toluene oxidationb (%)
|
||||
|---|---|---|---|---|---|---|---|
| Apparent Vmax (nmol/min/mg of protein) | Apparent Km (μM) | Vmax/Km | o-Cresol | m-Cresol | p-Cresol | ||
| TG1/pBS(Kan) | LB KAN | 0 | 0 | 0 | |||
| TG1/pBS(Kan)T3MO | LB KAN | 11.5 ± 0.3 | 250 | 0.046 | 0 | 10 | 90 |
| TG1/pBS(Kan)T4MO | LB KAN | 15.1 ± 0.8 | 110 | 0.137 | 0 | 3 | 97 |
| P. aeruginosa PAO1 | LB + toluene | 0 | 0 | 0 | |||
| PAO1/pRO1966 | LB + AMP + toluene | 0 | 10 | 90 | |||
| PAO1/pRO1966 | M9 + AMP + lactate + toluene | 0 | 9 | 91 | |||
| PAO1/pRO1966 | M9 + AMP + succinate + toluene | 0 | 11 | 89 | |||
The toluene concentration in the liquid phase was 90 to 700 μM based on Henry's law constant of 0.27 (7) (250 to 1,920 μM added if all the toluene is in the liquid phase). Seven different toluene concentrations were used for each strain.
Based on GC analysis over a 20-min period. The initial toluene concentration in the liquid was 90 μM.
KAN, kanamycin; AMP, ampicillin.
To corroborate these results, we performed toluene oxidation experiments using PAO1/pRO1966, the strain that was originally used by Olsen et al. in their characterization of T3MO (21). The cells were grown in a medium similar to that described in the original paper (minimal medium with lactate as a carbon source) and in two other media: minimal medium with sodium succinate as a carbon source and LB medium. With the use of GC, it was found that P. aeruginosa PAO1 expressing T3MO oxidized toluene to ∼90% p-cresol and ∼10% m-cresol under all growth conditions examined (Table 2), which matched the product spectrum of TG1/pBS(Kan)T3MO. Furthermore, cells without a plasmid were unable to oxidize toluene, indicating that the cresol formation was associated with the expression of the monooxygenase. Analysis of the toluene oxidation products by HPLC revealed one cresol peak (p- and m-cresol elute together, so they cannot be distinguished) and one 4-methylcatechol peak, demonstrating that T3MO can successively hydroxylate toluene to methylcatechol.
Analysis of the rates of toluene oxidation by TG1(T3MO) and TG1(T4MO) at different substrate concentrations showed that these enzymes followed typical saturation kinetics. The apparent Vmax value was 30% higher for T4MO, accompanied by a lower Km value, resulting in an overall threefold increase in the Vmax/Km value (Table 2). This indicates that, for toluene, TG1(T4MO) is not only a more regiospecific enzyme than TG1(T3MO), it is a more efficient catalyst, too.
Analysis of TG1(T3MO) sequence.
The entire T3MO tbuA1UBV1A2C locus from plasmid pBS(Kan)T3MO and the entire tbuA1tbuBtbuC locus of the original sequencing vector pRO1966 (3) were sequenced as well as the upstream region of both vectors. There were no changes in the DNA sequence due to cloning into pBS(Kan); however, four sequencing errors were identified in the published T3MO sequences (3): codons 33 and 34 of tbuB should be changed from CGA AGG to GAA GGG (tbuB R33E and R34G) and codons 28 and 29 of tbuC should be changed from CGG GGC to CGC GCC (tbuC R28R and G29A). These changes were deposited in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov) along with a note indicating that this is a para-hydroxylating enzyme (GenBank accession no. AY541701). Each of the changes was verified twice for each plasmid by the dideoxy chain termination technique. In addition, sequencing the upstream region of pBS(Kan)T3MO showed clearly that this plasmid contained a lac promoter region and multiple cloning sites originally from the pBS(Kan) plasmid. Sequencing the region upstream of pRO1966 confirmed the presence of the T3MO promoter region (ptbuA1) (3).
In addition, the identities of pBS(Kan)T3MO and the original vector pRO1966 were confirmed with the restriction enzymes XhoI, BclI, EcoRI, and AatII. The results for digestion of pRO1966 by the restriction enzymes gave the correct restriction map as reported in the literature (3), and the digestion results for pBS(Kan)T3MO were consistent, too. Thus, these DNA analyses confirmed that the correct T3MO enzyme isolated from R. pickettii PAO1 was used.
Oxidation of different aromatic compounds by TG1(T3MO).
The regiospecificity of TG1 cells expressing T3MO was further evaluated using different aromatic substrates, and TG1(T4MO) was used for comparison (Table 3). Both enzymes oxidized methoxybenzene to 100% p-methoxyphenol, whereas TG1(TOM) produced mainly guaiacol (o-methoxyphenol; results not shown). The products formed from oxidation of the different cresols were also identical for TG1(T3MO) and TG1(T4MO), although the rates were significantly lower for TG1(T3MO). The rate of specific initial formation of 4-methylcatechol from 1 mM m-cresol by TG1(T3MO) was 0.6 ± 0.2 versus 9.5 ± 1.3 nmol/min/mg of protein by TG1(T4MO), a 16-fold difference in activity. The rate of formation of this product from 1 mM p-cresol oxidation was only sixfold higher for T4MO (6 versus 0.97 nmol/min/mg of protein).
TABLE 3.
Regiospecificity of oxidation of various aromatic substrates by E. coli TG1 cells expressing T3MO and T4MO from pBS(Kan)T3MO and pBS(Kan)T4MO, respectively
| Substratea | Concn (mM) | Product | T3MO (%) | T4MO (%) |
|---|---|---|---|---|
| Methoxybenzene | 0.5 | p-Methoxyphenol | 100 | 100 |
| NB | 0.2 | p-Nitrophenol | 66 | 90 |
| m-Nitrophenol | 34 | 10 | ||
| Naphthalene | 5b | 1-Naphthol | 62 | 52 |
| 2-Naphthol | 38 | 48 | ||
| p-Cresol | 1 | 4-Methylcatechol | 100 | 100 |
| m-Cresol | 1 | 4-Methylcatechol | 100 | 100 |
| o-Cresol | 1 | 3-Methylcatechol methylhydroquinone | 91 | 91 |
| 9 | 9 |
The incubation time of the cells with the substrate was 30 min, except for naphthalene, which was incubated for 3 h.
Naphthalene solubility in water is 0.27 mM (6).
NB was the only substituted benzene tested in which T3MO showed a noteworthy preference for the meta position, forming 34% m-nitrophenol. T4MO, however, remained conservative, producing primarily p-nitrophenol. TG1(T4MO) produced nearly equal amounts of 1- and 2-naphthol from 5 mM naphthalene whereas TG1(T3MO) produced 62% 1-naphthol and 38% 2-naphthol. These rather similar results were substantially different from those for TG1(TOM), which gave predominantly 1-napthanol (98%) (5). It is therefore concluded that T3MO does not possess meta-hydroxylating properties but is a para-hydroxylating enzyme.
NMR analysis.
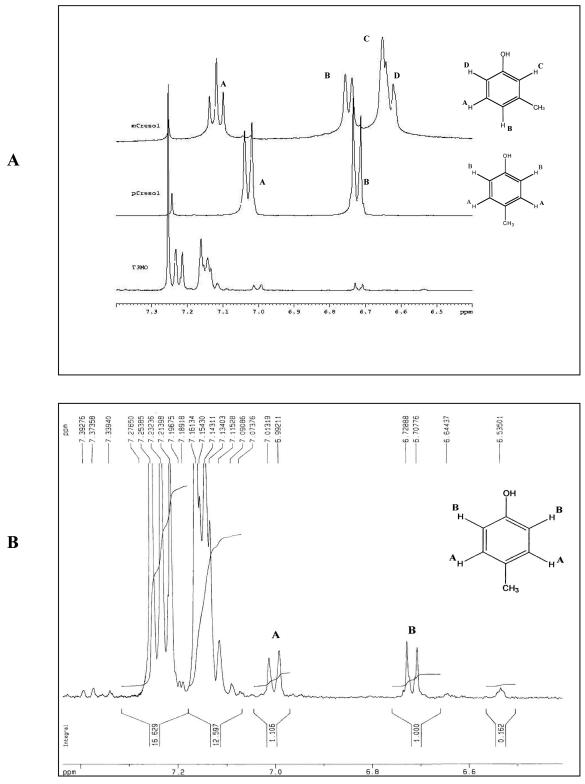
The 1H NMR spectra of p- and m-cresol are distinct (Fig. 2A) (http://www.aist.go.jp/RIODB/SDBS/menu-e.html). p-Cresol is a symmetrical molecule, with the four aromatic protons giving two doublets: δ = 7.028 (A doublet, 2H) and δ = 6.724 (B doublet, 2H). In contrast, the aromatic protons of m-cresol are all different, giving the following assignment: δ = 7.118 (A triplet, 1H), δ = 6.742 (B doublet, 1H), δ = 6.65 (C singlet, 1H), and δ = 6.64 (C doublet, 1H). Since the 1H NMR spectra of a reaction mixture following oxidation of toluene by TG1(T3MO) (Fig. 2B) contain two doublets at δ = 7.003 (doublet, 2H) and δ = 6.718 (doublet, 2H) and match the standard p-cresol, it is therefore evident that T3MO produces p-cresol rather than m-cresol from toluene and that the 1H NMR analysis corroborates the GC analysis.
FIG. 2.
(A) Partial 1H NMR spectra (7.4 to 6.4 ppm) of an m-cresol standard, a p-cresol standard, and a reaction mixture of E. coli TG1 expressing wild-type T3MO after oxidation of toluene. (B) Enlargement of the spectra of p-cresol formed from oxidation of toluene by TG1 expressing wild-type T3MO. The assignments of the four aromatic protons are also shown.
DISCUSSION
Toluene-oxidizing enzymes have been studied extensively due to their potential in degrading environmental pollutants (10, 22, 30). Among toluene monooxygenases, T3MO of R. pickettii is the only reported enzyme capable of hydroxylating toluene at the meta position (UM-BBD; http://umbbd.ahc.umn.edu/index.html). Besides the original publication by Olsen et al. describing the formation of m-cresol from toluene by this enzyme (21), there have been no other reports on the regiospecificity of T3MO. Our results indicate that this enzyme is actually a para-hydroxylating enzyme similar to T4MO of P. mendocina KR1 and that it can successively oxidize toluene to 4-methylcatechol. We suggest the new name toluene para-monooxygenase for this enzyme.
To distinguish the cresol isomers, a good analytical separation method is required. Reverse-phase HPLC with a standard C8 or C18 column did not allow us to obtain separation between p- and m-cresols under various H2O (0.1% formic acid)-acetonitrile gradients. GC was therefore the method of choice, which is also described in reports by Fox and coworkers (19, 24). The excellent base-to-base separation of the cresol mixture (Fig. 1A) enabled good assessment of TG1(T3MO) regiospecificity for toluene oxidation (Fig. 1B). Comparison with TG1(T4MO), a well-studied para-hydroxylating enzyme (25, 34, 35), further substantiated the fact that T3MO is not a meta-hydroxylating enzyme. The original source for the T3MO locus, pRO1966, was used to construct pBS(Kan)T3MO, and PAO1/pRO1966 also exhibited identical regiospecificity under various growth conditions (Table 2). Sequencing of the tbu operon in both plasmids, pBS(Kan)T3MO and pRO1966, revealed complete identity between the two, so there were no cloning errors. The 1H NMR spectra of the reaction mixture following oxidation of toluene by TG1(T3MO) (Fig. 2) also provided structural evidence that p-cresol was the major product (which exhibited the same two doublets [A2B2] as did the standard compound).
The similarity of T3MO and T4MO was evident from the oxidation of many substrates, not just toluene. Analysis of the product distribution of six additional aromatic compounds demonstrated the closeness in regiospecificity of these two enzymes (Table 3), although TG1(T3MO) exhibited lower specificity for NB oxidation and had lower formation rates for all of the compounds investigated. The superiority of TG1(T4MO) as a toluene catalyst was also obvious from the higher apparent Vmax and lower Km values (Table 2).
Byrne et al. had already indicated the significant overall homology between the Tbu proteins of T3MO of R. pickettii PKO1 and the Tmo proteins of T4MO of P. mendocina KR1 (3). A recent study of the structure and specificity of the effector protein (TmoD) of T4MO of P. mendocina KR1 showed that the closely related TbuV (effector protein of T3MO of R. pickettii PKO1) provided partial activation of the reconstituted T4MO complex, whereas the more distantly related TomA2 (effector protein of TOM of B. cepacia G4) did not (12). The authors also found unusual 1H NMR chemical shifts of the side chain amide signals of TmoD N34 and strict conservation of this residue in other diiron hydroxylase effector proteins. NMR studies of these proteins showed that the conserved Asn groups in TmoD and TbuV had spectra similar to each other but different from that of TbmC (effector protein of TOM of Pseudomonas sp. strain JS150), leading to speculation on the role of N34 regarding the specificity of the enzyme (12). Our results provide catalytic evidence corroborating this previously reported structural similarity between T3MO and T4MO.
We believe that the poor HPLC method of separation of p- and m-cresol (Fig. 2 in reference 21) led to the incorrect classification of T3MO as a meta-hydroxylating toluene monooxygenase. Olsen et al. also present mass spectrometry (MS) analysis of m-cresol produced from toluene by toluene-grown cells of PAO1/pRO1966 (Fig. 3A in reference 21). Although MS spectra of o-, m-, and p-cresol are very similar, as one would expect from chemical isomers, the fragmentation pattern of p-cresol is different in its ratio of m/z fragments, 77/79. Whereas this ratio is >1 for p-cresol (i.e., the intensity of m/z 77 is higher than that of m/z 79), it is <1 for o- and m-cresol (11, 18) (http://webbook.nist.gov/chemistry). The MS spectra presented by Olsen and coworkers (21) are comparable with those of standard p-cresol, rather than m-cresol as suggested by the authors. In addition, the authors found that m-xylene was oxidized by T3MO to 2,4-dimethylphenol (which is the product formed by T4MO) (25) rather than the expected 3,5-dimethylphenol and refer to this as surprising. So it seems that there are indications in the original publication that the monooxygenase expressed by PAO1/pRO1966 was not a genuine T3MO.
We have provided strong evidence that T3MO of R. pickettii PKO1 is a para-hydroxylating enzyme transforming toluene to p-cresol and 4-methylcatechol successively. Hence, there has yet to be discovered a true toluene meta-monooxygenase, but perhaps one may be made through protein engineering.
Acknowledgments
This research was supported by the National Science Foundation (BES-0124401) and the U.S. Environmental Protection Agency.
We are grateful for the help provided by James Stuart of the University of Connecticut in the GC-MS analysis and for the assistance provided by Martha Morton of the University of Connecticut in the NMR analyses.
REFERENCES
- 1.Buhler, B., B. Witholt, B. Hauer, and A. Schmid. 2002. Characterization and application of xylene monooxygenase for multistep biocatalysis. Appl. Environ. Microbiol. 68:560-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton, S. G. 2003. Oxidizing enzymes as biocatalysts. Trends Biotechnol. 21:543-549. [DOI] [PubMed] [Google Scholar]
- 3.Byrne, A. M., J. J. Kukor, and R. H. Olsen. 1995. Sequence analysis of the gene cluster encoding toluene-3-monooxygenase from Pseudomonas pickettii PKO1. Gene 154:65-70. [DOI] [PubMed] [Google Scholar]
- 4.Byrne, A. M., and R. H. Olsen. 1996. Cascade regulation of the toluene-3-monooxygenase operon (tbuA1UBVA2C) of Burkholderia pickettii PKO1: role of the tbuA1 promoter (PtbuA1) in the expression of its cognate activator, TbuT. J. Bacteriol. 178:6327-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canada, K. A., S. Iwashita, H. Shim, and T. K. Wood. 2002. Directed evolution of toluene ortho-monooxygenase for enhanced 1-naphthol synthesis and chlorinated ethene degradation. J. Bacteriol. 184:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean, J. A. 1985. Lange's handbook of chemistry, 13th ed. McGraw-Hill Book Co., New York, N.Y.
- 7.Dolfing, J., A. J. van den Wijngaard, and D. B. Janssen. 1993. Microbiological aspects of the removal of chlorinated hydrocarbons from air. Biodegradation 4:261-282. [DOI] [PubMed] [Google Scholar]
- 8.Eaton, R. W., and P. J. Chapman. 1995. Formation of indigo and related compounds from indolecarboxylic acids by aromatic acid-degrading bacteria: chromogenic reactions for cloning genes encoding dioxygenases that act on aromatic acids. J. Bacteriol. 177:6983-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, L. B. M., C. D. Hershberger, E. M. Bryan, and L. P. Wackett. 2001. The University of Minnesota biocatalysis/biodegradation database: emphasizing enzymes. Nucleic Acids Res. 29:340-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folsom, B. R., and P. J. Chapman. 1991. Performance characterization of a model bioreactor for the biodegradation of trichloroethylene by Pseudomonas cepacia G4. Appl. Environ. Microbiol. 57:1602-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heller, S. R., and G. W. A. Milne. 1978. EPA/NIH mass spectral data base, vol. 1. U.S. Government Printing Office, Washington, D.C.
- 12.Hemmi, H., J. M. Studts, Y. K. Chae, J. Song, J. L. Markley, and B. G. Fox. 2001. Solution structure of the toluene 4-monooxygenase effector protein (T4moD). Biochemistry 40:3512-3524. [DOI] [PubMed] [Google Scholar]
- 13.Kahng, H.-Y., A. M. Byrne, R. H. Olsen, and J. J. Kukor. 2000. Characterization and role of tbuX in utilization of toluene by Ralstonia pickettii PKO1. J. Bacteriol. 182:1232-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kukor, J. J., and R. H. Olsen. 1992. Complete nucleotide sequence of tbuD, the gene encoding phenol/cresol hydroxylase from Pseudomonas pickettii PKO1, and functional analysis of the encoded enzyme. J. Bacteriol. 174:6518-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leahy, J. G., P. J. Batchelor, and S. M. Morcomb. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 27:449-479. [DOI] [PubMed] [Google Scholar]
- 16.Leahy, J. G., G. R. Johnson, and R. H. Olsen. 1997. Cross-regulation of toluene monooxygenases by the transcriptional activators TbmR and TbuT. Appl. Environ. Microbiol. 63:3736-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Z., J. B. van Beilen, W. A. Duetz, A. Schmid, A. de Raadt, H. Grieng, and B. Witholt. 2002. Oxidative biotransformations using oxygenases. Curr. Opin. Chem. Biol. 6:136-144. [DOI] [PubMed] [Google Scholar]
- 18.Mass Spectrometry Data Centre.1983. Eight peak index of mass spectra, 3rd ed., vol. 3. The Mass Spectrometry Data Centre, The Royal Society of Chemistry, Nottingham, United Kingdom.
- 19.Mitchell, K. H., J. M. Studts, and B. G. Fox. 2002. Combined participation of hydroxylase active site residues and effector protein binding in a para to ortho modulation of toluene-4-monooxygenase regiospecificity. Biochemistry 41:3176-3188. [DOI] [PubMed] [Google Scholar]
- 20.Newman, L. M., and L. P. Wackett. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066-14076. [DOI] [PubMed] [Google Scholar]
- 21.Olsen, R. H., J. J. Kukor, and B. Kaphammer. 1994. A novel toluene-3-monooxygenase pathway cloned from Pseudomonas pickettii PKO1. J. Bacteriol. 176:3749-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parales, R. E., N. C. Bruce, A. Schmid, and L. P. Wackett. 2002. Biodegradation, biotransformation, and biocatalysis (B3). Appl. Environ. Microbiol. 68:4699-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, J., J. J. Kukor, and L. M. Abriola. 2002. Characterization of the adaptive response to trichloroethylene-mediated stresses in Ralstonia pickettii PKO1. Appl. Environ. Microbiol. 68:5231-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pikus, J. D., K. H. Mitchell, J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 2000. Threonine 201 in the diiron enzyme toluene-4-monooxygenase is not required for catalysis. Biochemistry 39:791-799. [DOI] [PubMed] [Google Scholar]
- 25.Pikus, J. D., J. M. Studts, K. McClay, R. J. Steffan, and B. G. Fox. 1997. Changes in the regiospecificity of aromatic hydroxylation produced by active site engineering in the diiron enzyme toluene 4-monooxygenase. Biochemistry 36:9283-9289. [DOI] [PubMed] [Google Scholar]
- 26.Rui, L., Y. Kwon, A. Fishman, K. F. Reardon, and T. K. Wood. Saturation mutagenesis of toluene ortho-monooxygenase of Burkholderia cepacia G4 for enhanced 1-naphthol synthesis and chloroform degradation. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheilds, M. S., M. J. Reagin, R. R. Gerger, R. Campbell, and C. Somerville. 1995. TOM, a new aromatic degradative plasmid from Burkholderia cepacia G4. Appl. Environ. Microbiol. 61:1352-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shim, H., D. Ryoo, P. Barbieri, and T. K. Wood. 2001. Aerobic degradation of mixtures of tetrachloroethylene, trichloroethylene, dichloroethylenes, and vinyl chloride by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1. Appl. Microbiol. Biotechnol. 56:265-269. [DOI] [PubMed] [Google Scholar]
- 31.Studts, J. M., K. H. Mitchell, J. D. Pikus, K. McClay, R. J. Steffan, and B. G. Fox. 2000. Optimized expression and purification of toluene 4-monooxygenase hydroxylase. Protein Expr. Purif. 20:58-65. [DOI] [PubMed] [Google Scholar]
- 32.Sun, A. K., and T. K. Wood. 1996. Trichloroethylene degradation and mineralization by pseudomonads and Methylosinus trichosporium OB3b. Appl. Microbiol. Biotechnol. 45:248-256. [DOI] [PubMed] [Google Scholar]
- 33.Tao, Y., A. Fishman, W. E. Bentley, and T. K. Wood. Oxidation of benzene to phenol, catechol, and 1,2,3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pickettii PKO1. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 34.Whited, G. M., and D. T. Gibson. 1991. Separation and partial characterization of the enzymes of the toluene-4-monooxygenase catabolic pathway in Pseudomonas mendocina KR1. J. Bacteriol. 173:3017-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yen, K.-M., M. R. Karl, L. M. Blatt, M. J. Simon, R. B. Winter, P. R. Fausset, H. S. Lu, A. A. Harcourt, and K. K. Chen. 1991. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J. Bacteriol. 173:5315-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]