Abstract
Idiopathic pulmonary fibrosis (IPF) is a progressive disease of the middle aged and elderly with a prevalence of one million persons worldwide. The fibrosis spreads from affected alveoli into contiguous alveoli, creating a reticular network that leads to death by asphyxiation. Lung fibroblasts from patients with IPF have phenotypic hallmarks, distinguishing them from their normal counterparts: pathologically activated Akt signaling axis, increased collagen and α-smooth muscle actin expression, distinct gene expression profile, and ability to form fibrotic lesions in model organisms. Despite the centrality of these fibroblasts in disease pathogenesis, their origin remains uncertain. Here, we report the identification of cells in the lungs of patients with IPF with the properties of mesenchymal progenitors. In contrast to progenitors isolated from nonfibrotic lungs, IPF mesenchymal progenitor cells produce daughter cells manifesting the full spectrum of IPF hallmarks, including the ability to form fibrotic lesions in zebrafish embryos and mouse lungs, and a transcriptional profile reflecting these properties. Morphological analysis of IPF lung tissue revealed that mesenchymal progenitor cells and cells with the characteristics of their progeny comprised the fibrotic reticulum. These data establish that the lungs of patients with IPF contain pathological mesenchymal progenitor cells that are cells of origin for fibrosis-mediating fibroblasts. These fibrogenic mesenchymal progenitors and their progeny represent an unexplored target for novel therapies to interdict fibrosis.
Progressive scarring of the heart, blood vessels, lung, liver, kidney, and brain is a leading cause of death worldwide.1 Characteristic of these diseases, idiopathic pulmonary fibrosis (IPF) is a prevalent and progressive process.2–6 In IPF, the fibroblast population expands within alveolar structures, resulting in scarred nonfunctional airspaces, progressive hypoxemia, and death by asphyxiation. As the disease process evolves, fibrosis spreads contiguously from affected alveoli into anatomically intact adjacent gas exchange units, resulting in an expanding reticular network of fibrotic tissue.7–12 IPF lung fibroblasts display hallmarks that distinguish them from normal lung fibroblasts. Aberrant integrin signaling in the IPF fibroblast leads to sustained activation of proliferation and survival signaling pathways13–15; when grafted into model organisms, IPF fibroblasts form fibrotic lesions.16,17 Despite their central role in mediating progressive fibrotic destruction of the lung, the origins of the IPF fibroblast are not known.
There is a well-established precedent for stem and progenitor cells as a source of the majority cell population in healthy and diseased organs. Normal lung tissue contains stem and progenitor cells that self-renew and give rise to transit-amplifying cells that maintain cell numbers in the steady state and mediate repair and regeneration in response to injury.18–26 Neoplastic tissue contains pathological progenitors that exhibit self-renewal capacity and divide asymmetrically to produce malignant daughter cells27–32; disease-mediating progenitor cells have been implicated in chronic lung allograft rejection.33,34
Here, we report the identification of a subpopulation of cells in the lungs of patients with IPF with the properties of mesenchymal progenitors. Gene expression profiling of IPF lung mesenchymal progenitors distinguished them from mesenchymal progenitors isolated in a similar manner from the lungs of patient controls, with enrichment of genes associated with disease-relevant ontologies. The cellular progeny of IPF mesenchymal progenitors displayed all of the diagnostic hallmarks of the IPF fibroblast, including increased levels of phospho-Akt, increased expression of α-smooth muscle actin and type I collagen, and the ability to form fibrotic lesions in two model organisms. Analysis of IPF lung specimens revealed mesenchymal progenitor cells and cells bearing determinants of their progeny throughout the fibrotic reticulum. This is the first report in any progressive fibrotic disorder that documents diseased tissue harboring mesenchymal progenitor cells that are cells of origin for fibrosis-mediating fibroblasts.
Materials and Methods
Study Approval
De-identified patient samples were obtained under a waiver of informed consent from the University of Minnesota Institutional Review Board. Animal protocols were approved and conducted in accordance with the University of Minnesota Institutional Animal Care and Use Committee regulations.
Primary Mesenchymal Cell Lines
Eleven primary lung mesenchymal cell lines were established from patients who fulfilled diagnostic criteria for IPF, including a pathological diagnosis of usual interstitial pneumonia.8 Patient controls were selected to be similar in age to patients with IPF with nonfibrotic lung disorders. On the basis of these criteria, we established 10 nonfibrotic primary control fibroblast lines from lung tissue uninvolved by the primary disease process: adenocarcinoma (n = 4), squamous cell carcinoma (n = 1), carcinoid tumor (n = 2), fibrosarcoma (n = 1), leiomyosarcoma (n = 1), or bronchiectasis (n = 1). Cell lines were derived from lungs, characterized as mesenchymal cells, and were cultivated as previously described.35,36
A significant challenge when studying primary cells from patient samples is the need for ex vivo preparation and in vitro expansion, procedures that introduce uncontrolled variables into the system. In addition, the relative difficulty of acquiring such samples prevents exact matching of demographic variables. To address these issues, standard technical variables (eg, subcultivation number, patient age, preparation batch) were tracked to minimize the bias they introduced into our results. Although this prevented introduction of systematic bias in our experiments, it does increase the measurement variance. Despite this, a principal components analysis (performed in R using the prcomp function) of the RNA-Seq data revealed the first principle component to partition the samples between mesenchymal progenitor cells and their progeny and the second principal component to partition the samples between IPF and control. We did not observe partitioning on the basis of any other technical variable we tracked. Although we cannot exclude the possibility that hidden technical confounders influenced the data, this analysis supports the robustness of our results.
Isolation of Mesenchymal Progenitor Cells
IPF and control mesenchymal progenitor cells were isolated from primary IPF and control mesenchymal cell cultures at passage 0 (initial isolate before subcultivation) through passage 4. To isolate progenitors, primary IPF and control mesenchymal cells were labeled with mouse anti–stage-specific embryonic antigen 4 (SSEA4) antibody conjugated to Alexa Fluor 647 and mouse anti-SSEA1 conjugated to phycoerythrin (BD Biosciences, San Jose, CA). Cells were sorted on a FACSAria Cell Sorter (BD Biosciences). Cells that were SSEA4+ and SSEA1− (relative to mouse IgG3 κ isotype control conjugated to Alexa Fluor 647 and phycoerythrin) and <12 μm (designated small cells; forward and side calibrated using a 12-μm mesh; Millipore, Temecula, CA) were collected.
Multiparameter Flow Cytometry
Primary IPF and control mesenchymal cells were subjected to cell surface antigen phenotyping with the use of fluorescein isothiocyanate-, phycoerythrin-, or peridinin chlorophyll protein complex-cyanine 5.5–conjugated antibodies (BD Biosciences) against SSEA1, SSEA4, CD90, CD73, CD105, CD45, and CD34. Isotype-matched fluorophore-conjugated IgG antibodies were used as negative controls to set the gates. Cells were analyzed on a BD Biosciences FACSCalibur flow cytometer with the use of FlowJo Flow Cytometry Analysis software version 7.6.5 (TreeStar Inc, Ashland, OR).
Plastic-Adherent Clonal Growth Assay
Single-cell suspensions of SSEA4+/SSEA1−/small cells were sparsely plated on plastic tissue culture dishes and maintained in Dulbecco’s modified Eagle’s medium (DMEM) + 10% fetal bovine serum (FBS; 37°C, 10% CO2, 2 weeks). Enumeration of colony-forming unit fibroblast adherent colonies was performed microscopically after fixing cells with methanol and staining with crystal violet.
Tri-Lineage Differentiation Assay
IPF and control SSEA4+/SSEA1−/small cells were analyzed for tri-lineage differentiation capacity by using the following assay kits: StemPro Osteogenesis Differentiation Kit, catalog number A10072-01; Adipogenesis Differentiation Kit, catalog number A10070-01; Chondrogenesis Differentiation Kit, catalog number A10071-01; Gibco, Grand Island, NY). After 21 days in differentiation culture conditions, cells were fixed and labeled with antibodies against fatty acid binding protein 4 (adipocytes), osteocalcin (osteocytes), or aggrecan (chondrocytes) and visualized by immunofluorescence (all antibodies from R&D Systems, Minneapolis, MN).
RT-PCR
RT-PCR was performed as previously described.36 PCR reactions were subjected to gel electrophoresis, and only primers yielding a single product of the appropriate length were used. We took special care to validate octamer-binding transcription factor 4 (Oct4A) because of our intent to use it for immunohistochemical lung tissue analysis. This included using primer sequences documented to be specific37 and sequence-verifying the gel-purified PCR product.
Western Blot Analysis
Blots were performed as previously described.13–15
Derivation of Progeny from Mesenchymal Progenitor Cells
IPF and control progeny (ie, daughter cells) were derived from SSEA4+/SSEA1−/small cells that had undergone fluorescence-activated cell sorting (FACS).
Progeny from a Population of Flow-Sorted Progenitor Cells
IPF and control progenitors were isolated from IPF and control primary mesenchymal cell cultures by flow cytometry that selected for SSEA4hi/SSEA1−/small cells. One hundred progenitors were placed into 3.8-cm2 wells of 12-well tissue culture clusters and allowed to propagate and differentiate under tightly standardized conditions (DMEM + 10% FBS, 37°C, 10% CO2) for 21 days. The resultant cell population was designated IPF or control progeny.
Progeny from a Single IPF or Control Mesenchymal Progenitor Cell
Individual SSEA4+/SSEA1−/small cells (98.88% pure by postsort analysis) that had undergone FACS from a primary IPF and control mesenchymal cell culture (subcultivation 1) were placed at a concentration of one cell per well into 96-well clusters coated with STEMPRO MSC SFM CTS (Gibco) that contained 2.5% methylcellulose. Cultures were continued (DMEM + 10% FBS, 37°C, 10% CO2, 14 days) until a small number of colonies formed. The two largest IPF colonies and four largest control colonies were transferred to separate tissue culture dishes (35 mm) and allowed to propagate and differentiate (DMEM + 20% FBS, 37°C, 10% CO2) for 21 days.
Immunohistochemistry
Immunohistochemistry was performed on 4-μm paraffin-embedded IPF and control lung tissue and mounted on polylysine-coated slides. The sections were deparaffinized in xylene, rehydrated through a graded methanol series, quenched with 0.3% hydrogen peroxide in methanol, and immersed in a 98°C water bath for 30 minutes in Citrate Buffer (pH 6.0) for antigen retrieval. Sections were placed in 5% normal horse serum (Jackson ImmunoResearch Laboratories, West Grove, PA) to block nonspecific binding of secondary antibodies. Endogenous avidin and biotin binding sites were blocked by sequential incubation for 15 minutes each with an Avidin/Biotin Blocking Kit (Vector Laboratories, Burlingame, CA) and incubated overnight (18 to 20 hours, 4°C) in the monoclonal human primary antibodies, Oct4 clone 10H11·2 (1:500; Millipore), SSEA-4 (1:100; Abcam, Cambridge, MA), and α-smooth muscle actin (α-SMA; 1:100; Vector Laboratories). Sections were rinsed with PBS, placed in biotinylated horse anti-mouse IgG secondary antibody (1:500) for 30 minutes at room temperature, followed by R.T.U. Horseradish Peroxidase Streptavidin Complex (Vector Laboratories) for 30 minutes. Specific antibody binding was detected by using a 3,3′ diaminobenzidine peroxidase kit (DAB; Vector Laboratories). For double antigen labeling, we followed the protocol described above in a sequential manner with two modifications: the reagent used to prevent nonspecific binding was 5% normal horse serum/5% normal goat serum in a 1:1 ratio solution for the second antibody, and DAB/Ni was used as the detection reagent. All sections were counterstained with hematoxylin (Invitrogen, Frederick, MD) for 2 minutes, and PBS was applied to blue for 30 minutes. Specimens were coverslipped with a Prolong Antifade Kit (Invitrogen/Molecular Probes) and stored overnight at room temperature without light before image analysis.
In Vivo Fibrogenesis Assays
Zebrafish Xenotransplantation
Cells stained with a vital dye [either 5 μmol/L carboxyfluorescein succinimidyl ester (CFSE) or PKH26 (Sigma-Aldrich, St. Louis, MO)] were grafted into the central portion of the zebrafish embryo blastoderm at the oblong-sphere stages as previously described.16 Embryos were immobilized with Tricane after 48 hours. Fluorescent and corresponding bright field imaging of the CFSE-stained grafts in live embryos were performed with a Zeiss Axiovert upright microscope (Carl Zeiss, Thornwood, NY). After image acquisition, embryos were fixed with 4% paraformaldehyde, infiltrated with increasing concentrations of sucrose/PBS, and cryoembedded in OCT. For immunostaining, 5-μm sections were fixed in 100% methanol, rinsed in PBS, blocked with normal donkey serum, and incubated with the appropriate primary antibody. To identify engrafted human cells in zebrafish embryos, sections were incubated with CD59 [MEM-43] mouse monoclonal antibody (1:500; Abcam), followed by donkey anti-mouse cyanine 3 (1:500; Jackson ImmunoResearch Laboratories). Collagen type I was identified with human anti-procollagen type I antibody (1:200; Abcam) conjugated with horseradish peroxidase, and counterstained with H&E. The size of the fibrotic reticulum was quantified as previously described.16
Adoptive Transfer into Immunodeficient Mice
Cells were suspended in PBS (106 cells/mL) and injected via the tail vein into nonobese diabetic/severe combined immunodeficient/IL-2 receptor γ/β2 microglobulin mice (The Jackson Laboratory, Bar Harbor, ME) according to a published protocol.17 Mice were euthanized 60 days after adoptive transfer of human cells. Histological analysis of lung tissue was performed on paraffin-embedded and frozen lung tissue after H&E and trichrome staining. Cells positive for human β2-microglobulin by immunohistochemistry (anti-human β2 microglobulin antibody; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) were identified as human. The presence of lung fibrotic lesions by histological analysis served as the primary end point.17
Transcriptional Profiling and Analysis of RNA Sequencing Data
RNA was isolated from freshly sorted SSEA4+/SSEA1−/small cells or their progeny (IPF and control, n = 4; ie, 16 samples total) and sequenced (50-bp paired-end read run) on an Illumina Hiseq 2000. Sequence reads were aligned to hg19 with Tophat 2.0.8 (Bowtie 2.1.0.0, allowing one mismatch). Only reads that aligned uniquely were kept. Counts were mapped to genes by using the R package GenomicRanges (Aboyoun, no date). The count data were log2 transformed and normalized with quantile normalization. Differential expression was identified with the random variance model modified t-test.38 We used GAGE version 2.8.039 and annotation from the Gene Ontology Consortium40 to identify enriched cellular processes. Input data were signed (direction of regulation) –log10 (random variance model P values).
Validity testing was conducted by real-time quantitative PCR (qPCR) as previously described.41 The four genes chosen (FLT1, MAP3K8, IGF2BP1, YBX1) were selected from the 117 genes that met the following two criteria: they were assigned to the most significant ontologies that distinguished IPF from control, and among all genes that were altered (P < 0.05) in both IPF mesenchymal progenitors and IPF progeny relative to their control counterparts, the genes chosen displayed co-regulation between IPF mesenchymal progenitor cells and their progeny (ie, up in both progenitors and progeny or down in both progenitors and progeny). Total RNA was isolated and reverse transcribed by using a Taqman Reverse Transcriptase Reagent Kit (Roche, Indianapolis, IN) and primed with random hexamers. Primer sequences for selected genes were selected by using National Center for Biotechnology Information (NCBI) Primer-BLAST. qPCR was performed with a LightCycler FastStart DNA MasterPLUS SYBR Green I Kit (Roche). Primer sequences were as follows: FLT1, 5′-ACCAAAGCAATTCCCATGCC-3′ (forward), 5′-CAGCTACGGTTTCAAGCACC-3′ (reverse); MAP3K8, 5′-GCTTCCCTGGAGAGAAACCC-3′ (forward), 5′-ATTCCTCGGTGCTTCCTGTG-3′ (reverse); IGF2BP1, 5′-ACCTCCATTTACGGCCTCTTT-3′ (forward), 5′-TCTCCCCATTTTCCCCTCTTC-3′ (reverse); and YBX1, 5′-TCATCGCAACGAAGGTTTTGG-3′ (forward), 5′-GCCTTGAACTGGAACACCAC-3′ (reverse).
Samples were quantified at the log-linear portion of the curve by using LightCycler analysis software version 3.5 and compared with an external calibration standard curve.
Data Analysis
For zebrafish data, IPF was represented as an indicator variable (0 for controls, 1 for patients with IPF). A repeated measures analysis of variance was used to compare cells from patients with IPF with cells of controls, with the length of migration as the outcome variable, IPF as the variable of interest, and an interaction term for IPF with embryo. For the mouse data, fibrosis was coded as 0 (absent) or 1 (present), and the relationship between IPF and fibrosis was evaluated with Fisher exact test. Quantitative data are expressed as means ± SD. P < 0.05 was considered significant. Principal components analysis was performed in R by using the prcomp function.
Results
Mesenchymal Progenitor Cells Can Be Isolated from the Lungs of Patients with IPF
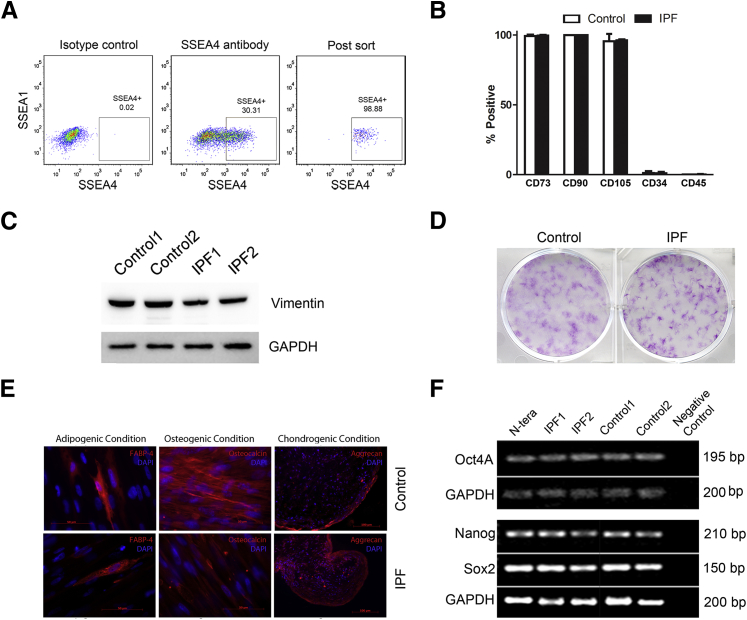
We established primary mesenchymal cell lines from IPF and control lung tissue.35 From these primary cell lines, we isolated mesenchymal progenitors by FACS on the basis of expression of SSEA4,42 lack of SSEA1, and size (<12 μm) (Figure 1A). No consistent differences were found in yields of SSEA4+ cells from IPF or control primary mesenchymal cell lines. To characterize the SSEA4+/SSEA1−/small cell population, we tested for signature mesenchymal stem/stromal cell (MSC) determinants.43,44 Similar to MSCs, control and IPF SSEA4+/SSEA1−/small cells expressed CD73, CD90, and CD105; both lacked expression of the fibrocyte determinants CD34 and CD45 (Figure 1B); both expressed vimentin (Figure 1C); and both were negative for the hematopoetic cell determinants CD11, CD19, and human leukocyte antigen DR (data not shown). One defining property of MSCs is the ability of a single precursor cell, termed a colony-forming unit fibroblast, to generate plastic-adherent colonies.45 Both IPF and control SSEA4+/SSEA1−/small cells displayed this property (Figure 1D). In common with MSCs, both IPF and control SSEA4+/SSEA1−/small cells exhibited tri-lineage mesenchymal differentiation capacity (Figure 1E); expressed mRNA encoding the progenitor transcription factors Oct4A, sex determining region Y (SRY)-box 2 (Sox2), and Nanog (Figure 1F); both expressed active β-catenin; and neither expressed Kruppel-like factor 4 (data not shown). Thus, our studies indicate that primary mesenchymal cell cultures derived from IPF and control lungs contain mesenchymal progenitor cells that share properties with MSCs.
Figure 1.
Recovery and characterization of SSEA4+/SSEA1−/small cells from IPF and control primary mesenchymal cell lines. A: Representative of SSEA4+/SSEA1−/small cells that had undergone FACS. Primary mesenchymal cell lines were sorted by FACS to isolate cells with relatively high surface expression of SSEA4 and low surface expression of SSEA1. Shown are the gates used (solid lines) to define the SSEA4+/SSEA1− cell population. The boxed area indicates the region defined as SSEA4+/SSEA4−. Cells were also sorted on the basis of size (forward and side scatter were calibrated with cells passing through a 12-μm mesh). B: Flow cytometric analysis of SSEA4+/SSEA1−/small cells for expression of cell surface determinants used to define MSCs (CD73, CD90, CD105) and fibrocytes (CD34 and CD45). Data shown are from SSEA4+/SSEA1−/small cells from two control and two IPF primary mesenchymal cell lines (means ± SD). C: SSEA+/SSEA1−/small cells were examined for vimentin expression by Western blot analysis. GAPDH is shown as a loading control. D: Colony-forming unit fibroblast assay: SSEA4+/SSEA1−/small cells were seeded at clonal density onto plastic tissue culture dishes (500 cells/9.5 cm2) and maintained in culture for 14 days. Shown are representative images of crystal violet-stained colonies formed by SSEA4+/SSEA1−/small cells sorted from two control and two IPF primary mesenchymal cell lines, each assayed in triplicate. E: SSEA4+/SSEA1−/small cells were investigated for tri-lineage differentiation capacity: i) adipocytes as determined by immunoreactivity to anti-FABP4 and the presence of lipid within the cells; ii) osteocytes by immunoreactivity to anti-osteocalcin; and iii) chondrocytes, as determined by the presence of aggrecan. Data shown are representative of SSEA4+/SSEA1−/small cells sorted from three control and three IPF primary mesenchymal cell lines. F: RT-PCR analysis of IPF and control SSEA4+/SSEA1−/small cells for progenitor transcription factors: Oct4A, Nanog, and Sox2, with GAPDH serving as a loading control. Human testicular embryonal carcinoma cells (N-TERA) were used as a positive control and PCR with primers but no cDNA as a negative control. Data shown are representative of SSEA4+/SSEA1−/small cells sorted from four IPF and four control primary mesenchymal cell lines. The Oct4A band was sequenced from one IPF and one control to verify its identity. Scale bars: 50 μm (E, left and middle panels); 100 μm (E, right panels). FABP4, fatty acid binding protein 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
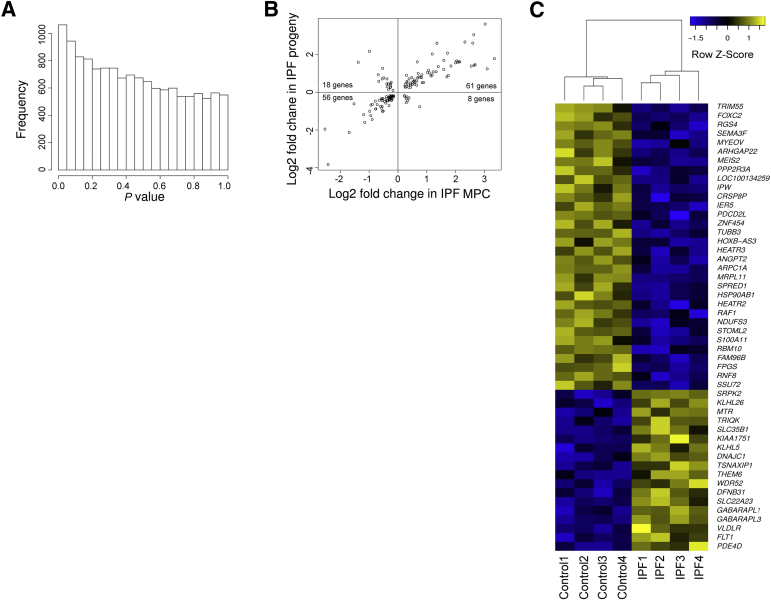
Transcriptional Profiling Identifies Key Differences That Distinguish IPF from Control Mesenchymal Progenitor Cells
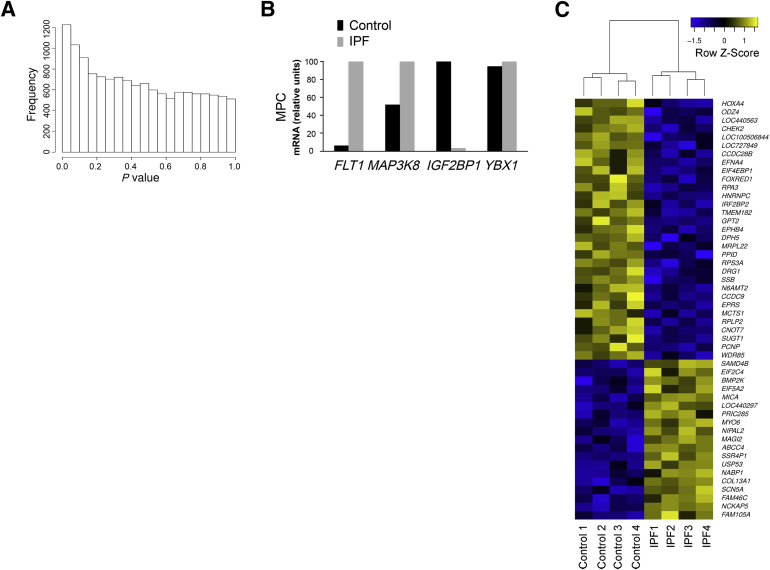
To elucidate molecular processes that distinguish control and IPF mesenchymal progenitor cells, we defined steady state mRNA levels genomewide. There were significant differences in gene expression between IPF and control mesenchymal progenitor cells on the basis of enrichment of genes with low P values (Figure 2A). A heatmap of the 50 most significant genes (P < 0.002) revealed a distinct IPF molecular signature (Figure 2C). To identify significant differences in disease-relevant functions, we conducted a gene set enrichment analysis.39,40 IPF mesenchymal progenitor cells displayed significant differences from controls in several disease-relevant ontologies, with a large number of gene ontology terms related to control of gene expression and proliferation (Supplemental Table S1). To test the validity of our transcriptional profiling, we directly quantified four genes that differed between IPF and control mesenchymal progenitor cells (for detailed selection criteria, see Materials and Methods): FLT1 and MAP3K8 (both genes showed elevated expression in IPF mesenchymal progenitors) and YBX1 and IGF2BP1 (both genes showed elevated expression in control mesenchymal progenitors). qPCR validated the transcriptional profiling for three of four genes (FLT1, MAP3K8, IGF2BP1), with YBX1 showing no significant difference (Figure 2B). Thus, IPF mesenchymal progenitors display a pattern of gene expression that distinguishes them from their control counterparts.
Figure 2.
Transcriptional profiling identifies key differences that distinguish IPF from control mesenchymal progenitor cells. A: Histogram of gene-by-gene P values that compare IPF and control mesenchymal progenitor cells. Data shown are from eight independent mesenchymal progenitor cell lines. B: Validation testing for FLT1, MAP3K8, IGF2BP1, and YBX1. Shown are relative expression levels of each mRNA by qPCR. Data shown represent the average of two cell lines. C: Heatmap of the 50 most significantly altered genes that distinguish IPF from control mesenchymal progenitor cells. Colors represent per gene z-score (expression difference normalized for SD). n = 4 (IPF and control).
IPF Mesenchymal Progenitor Cells Are Cells of Origin for IPF Fibroblasts
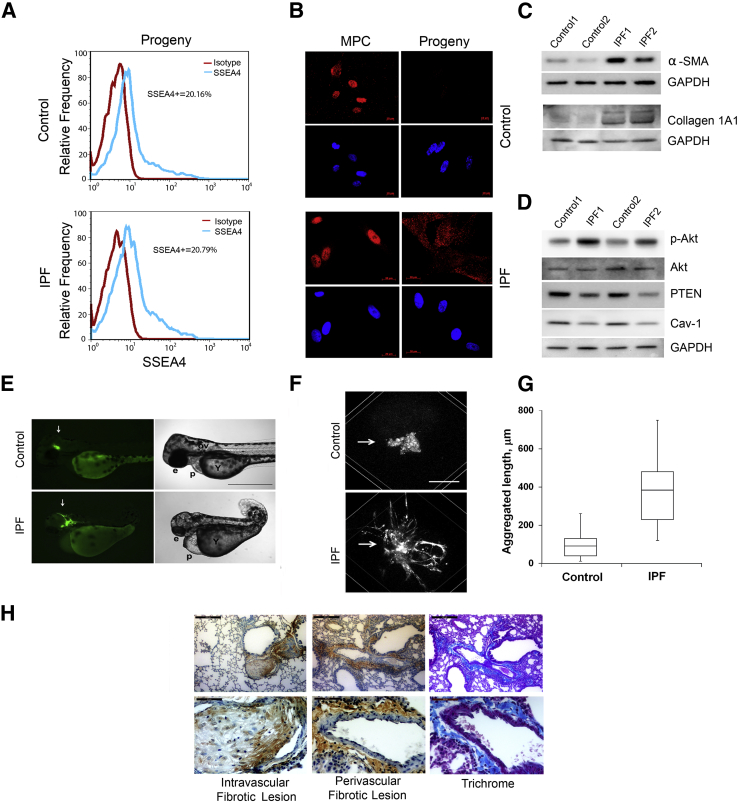
IPF lung fibroblasts have phenotypic hallmarks that distinguish them from controls.13–15,46 They express higher levels of α-SMA and type I collagen, express lower levels of caveolin-1 and phosphatase and tensin homologue (PTEN), and manifest pathological activation of the phosphatidylinositol 3-kinase/Akt signaling pathway, resulting in aberrantly high levels of phospho-Akt. To analyze the daughter cells of mesenchymal progenitors for these hallmarks, we used FACS to sort SSEA4+/SSEA1−/small cells from IPF and control primary mesenchymal cell cultures and allowed them to propagate and differentiate under tightly standardized conditions (Supplemental Figure S1). Prior work indicates that as mesenchymal progenitors propagate and differentiate in vitro, they display typical fibroblast morphology and lose expression of SSEA4, and Oct4 relocates from the nucleus to the cytoplasm where it persists for a period of time before being degraded.47 In accord with this, irrespective of origin, the progeny of mesenchymal progenitors displayed typical fibroblast morphology and lost SSEA4 expression (representative examples shown in Figure 3A); and Oct4 transited from the nucleus to the cytoplasm in IPF progeny and was at the limit of detection in controls (Figure 3B). However, only the daughter cells of IPF mesenchymal progenitors displayed IPF fibroblast hallmarks: increased levels of α-SMA, type I collagen, and phospho-Akt and decreased levels of PTEN and caveolin-1 (Figure 3, C and D). These data showthat IPF mesenchymal progenitors produce SSEA4−/nuclear Oct4−/cytoplasmic Oct4+ daughter cells with the biochemical hallmarks of IPF fibroblasts.
Figure 3.
The progeny of IPF MPCs display IPF fibroblast hallmarks. A: SSEA4 expression of progeny by flow cytometry. For reference, shown are isotype controls and a representative SSEA4 expression distribution of MPCs (ie, SSEA4+/SSEA1−/small cells that have undergone FACS). Data shown are from progeny derived from four independent MPC lines [IPF (n = 2), control (n = 2)]. B: Oct4 localization by immunofluorescence, DAPI to stain nuclei. C: α-SMA and collagen I expression. IPF and control progeny were analyzed by Western blot analysis. GAPDH is shown as a loading control. D: Akt signaling. IPF and control progeny were released from dishes with trypsin, cultured on 2-mg/mL polymerized collagen matrices for 48 hours, and cell extracts were examined for expression of caveolin-1, phospho-Akt, and PTEN by Western blot analysis. GAPDH is shown as a loading control. E–G: The progeny from four independent MPC lines were grafted into zebrafish embryos. Zebrafish xenograft assay (E). The progeny of control and IPF progenitors were stained with CFSE, engrafted into zebrafish embryos, and microscopically analyzed in live embryos after 48 hours. Shown are fluorescence (left panels) and bright field (right panels) images representative of at least 58 embryos per cell line. Arrows point to graft location. Graft-related embryonic deformities ranged from severe (microcephaly, microphthalmia, pericardial edema, upper right panel) to mild (head lump, lower right panel). Embryo orientation in the lateral view was from head to the left. F: Zebrafish xenograft assay with three-dimensional reconstruction. Control and IPF progeny were labeled with PKH26 vital dye. Fluorescent grafts were analyzed by single-photon confocal scanning. Shown are representative three-dimensional computer-generated reconstructions of the grafts 48 hours after grafting (arrows point to grafts). G: Shown is quantification of aggregated length of all processes in IPF and control grafts. H: Mouse xenograft assay. The progeny from seven independent MPC lines were injected into the tail vein of mice. Shown are representative images of the lungs (received the progeny of IPF progenitor cells). Human cells were identified with anti-human β2 microglobulin antibody. Sections shown indicate intravascular and perivascular fibrotic lesions, and trichrome stains of perivascular fibrotic lesions. Not shown here (Supplemental Figure S5) are normal lungs from mice that received the progeny of control progenitors. G: Data are expressed as follows: the horizontal line in the middle of each box indicates the median, and the top and bottom borders of the box mark the 75th and 25th percentiles; the whiskers above and below the box mark the 90th and 10th percentiles. n = 2 (A and E–G, IPF and control); n = 26 (G, IPF grafts); n = 27 (G, control grafts); n = 4 (H, IPF); n = 3 (H, control). P = 0.001, IPF grafts versus control grafts (G); P < 0.0001, composite reticulum length IPF versus control (G). Scale bars: 250 μm (E); 50 μm (F); 200 μm (H, left panels); 50 μm (H, right panels). Cav-1, caveolin-1; Collagen 1A1, type I collagen; e, eye; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MPC, mesenchymal progenitor cell; ov, otic vesicle; p, pericardium; Y, yolk sack.
When grafted into developing zebrafish embryos or immunocompromised mice, only IPF fibroblasts form fibrotic lesions.16,17 As a test of their fibrogenic potential, we examined whether IPF mesenchymal cell progenitors or their progeny displayed this behavior in both model organisms. Numerous studies have indicate that stem/progenitor cells integrate into the tissues of developing embryos after xenogenic transplantation.48–54 Consistently, we found that normal human bone marrow MSCs, when injected into the developing zebrafish, formed multiple small grafts that were incorporated into anatomically normal-appearing tissues and did not disrupt development. Similar to bone marrow MSCs, both IPF and control mesenchymal progenitors incorporated into the developing tissues of phenotypically normal embryos (Supplemental Figure S2). In sharp contrast, the properties of the mesenchymal progenitor progeny depended on their origin. Control progeny formed a small mass of nonmotile cells in the zebrafish, whereas IPF progeny formed an extensive fibrotic reticulum (Table 1, Figure 3, E–G, and Supplemental Figure S3, A and B) and expressed human pro-collagen type I (Supplemental Figure S3C). Results in the mouse assay paralleled results in the zebrafish, with the outcome depending on the origin of the mesenchymal progenitor progeny. The lungs of mice receiving the progeny of control mesenchymal progenitors were anatomically normal; whereas fibrotic lung lesions formed in all mice receiving the daughter cells of IPF progenitors (P < 0.0022) (Table 2, Figure 3H, and Supplemental Figure S4). Thus, only the progeny of IPF mesenchymal progenitors form fibrotic lesions in vivo–a defining hallmark of the IPF fibroblast.
Table 1.
Zebrafish Xenograft Data
| Cell lines∗ | Graft-bearing embryos, N | Normal embryos, n (%) | Abnormal embryos, n (%) | Fibrotic reticulum |
|---|---|---|---|---|
| IPF primary mesenchymal cells† | 35 | 4 (11.4) | 31 (88.6) | Yes |
| IPF MPCs | 43 | 36 (83.7) | 7 (16.3) | No |
| IPF MPC-progeny | 65 | 7 (10.8) | 58 (89.2) | Yes |
| Control primary mesenchymal cells† | 48 | 3 (6.3) | 45 (93.7) | No |
| Control MPCs | 15 | 12 (80) | 3 (20) | No |
| Control MPC-progeny | 95 | 14 (14.7) | 81 (85.3) | No |
MPCs, mesenchymal progenitor cells.
Data shown are from two IPF cell lines and two control cell lines.
In accord with our published data.16
Table 2.
Mouse Xenograft Data
| Cell line | Subject | Fibrotic lesions∗ |
|---|---|---|
| IPF 1 | Mouse 1 | Yes |
| IPF 1 | Mouse 2 | Yes |
| IPF 1 | Mouse 3 | Yes |
| IPF 2 | Mouse 4 | Yes |
| IPF 3 | Mouse 5 | Yes |
| IPF 4 | Mouse 6 | Yes |
| Control 1 | Mouse 7 | No |
| Control 2 | Mouse 8 | No |
| Control 3 | Mouse 9 | No |
| Control 3 | Mouse 10 | No |
| Control 3 | Mouse 11 | No |
P < 0.0022 compares fibrotic lesions resulting from injection of daughter cells derived from IPF progenitor cells with daughter cells derived from control progenitor cells.
Although FACS efficiently isolates a nearly pure population of SSEA4+/SSEA1−/small cells (>98% pure), if even a small minority subpopulation included differentiated primary fibroblasts that had a proliferative advantage, after 21 days in culture these cells rather than the progeny of SSEA4+/SSEA1−/small cells could be the dominant cell type. To test for this possibility, we labeled the SSEA4+/SSEA1−/small cell population that had undergone FACS with CSFE, a vital dye that covalently binds to intracellular proteins and is stoichiometrically diluted as cells divide. When we quantified dye intensity during a 6-day interval, we observed a single, uniform pattern of CFSE dilution characteristic of a uniformly proliferating population (Supplemental Figure S5). This result excluded the possibility that the progeny of SSEA4+/SSEA1−/small cells were contaminated by a rapidly proliferating subpopulation of differentiated IPF fibroblasts.
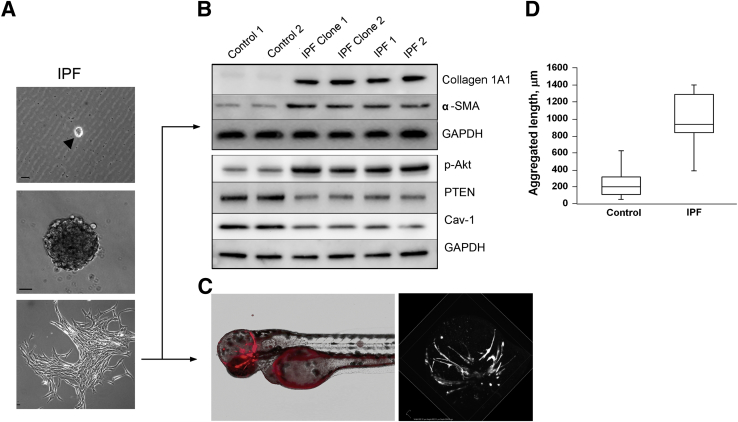
As direct proof of concept, we generated daughter cells from a single FACS-sorted IPF mesenchymal progenitor cell (Figure 4A). When analyzed for IPF fibroblast hallmarks, the clonal progeny of a single IPF progenitor cell displayed increased levels of α-SMA and type I collagen, low levels of caveolin 1 and PTEN accompanied by increased levels of phospho-Akt (Figure 4B), and formed an extensive fibrotic reticulum in the zebrafish assay (Figure 4, C and D, and Supplemental Table S2). Interestingly, the cell population resulting from a single control progenitor cell was much smaller than the population from an identically cultured single IPF progenitor (Supplemental Figure S6A). As a result there were only enough control cells to perform a limited Western blot analysis. IPF clonal progeny displayed higher collagen I and α-SMA expression than did control clonal progeny (Supplemental Figure S6B). However, a sufficient number of control clonal progeny were available to perform the zebrafish xenograft assay. In marked contrast to IPF clonal progeny, the clonal progeny of a single control mesenchymal progenitor cell did not form a fibrotic reticulum; engrafted cells were predominantly nonmotile in the zebrafish (Figure 4D, Supplemental Figure S6C, and Supplemental Table S2). These data proved that IPF mesenchymal progenitors can produce progeny manifesting the phenotypic hallmarks of the IPF fibroblast.
Figure 4.
The progeny of a single IPF mesenchymal progenitor cell displays the IPF fibroblast phenotype. A: Derivation of progeny from a single progenitor. Upper panel: SSEA4+ cells (99% pure) that had undergone FACS were seeded at one cell per well in 96-well dishes coated with methylcellulose and were observed for colony formation. The arrow denotes a single SSEA4 expressing IPF mesenchymal progenitor cell (MPC). Middle panel: Shown is a phase-contrast image of a colony formed by the progeny of a single progenitor cell. Bottom panel: Expansion of the colony on a tissue culture dish. B: IPF clonal progeny have the biochemical hallmarks of IPF fibroblasts. Western blots of the progeny of mesenchymal progenitors with each lane representing the clonal progeny of a single progenitor, designated IPF clone 1 (lane 3) and IPF clone 2 (lane 4) for collagen 1A1, α-SMA, GAPDH, p-Akt, PTEN, and Cav-1. Results for two primary control mesenchymal cell lines (designated control 1 and 2; lanes 1 and 2) and two primary IPF mesenchymal cell lines (designated IPF 1 and 2; lanes 5 and 6) are shown as a reference. C: IPF clonal progeny form a fibrotic reticulum in zebrafish. An example representative of seven zebrafish embryos grafted with the progeny of a single IPF progenitor. Graft forming a fibrotic reticulum (red) appears in head region with nonspecific autofluorescence outlining the yolk sac (left panel). A representative three-dimensional computer-generated reconstruction of the xenograft 48 hours after grafting shows the formation of a fibrotic reticulum (right panel). D: Quantification of aggregated length of all processes in IPF and control clonal progeny grafts. Data are expressed as follows: the horizontal line in the middle of each box indicates the median, and the top and bottom borders of the box mark the 75th and 25th percentiles; the whiskers above and below the box mark the 90th and 10th percentiles (D). n = 10 (D, IPF grafts); n = 25 (D, control). P < 0.001 (D). Scale bars: 10 μm (A). Cav-1, caveolin-1; Collagen 1A1, type I collagen; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; p-Akt, phospho-Akt; α-SMA, α-smooth muscle actin.
IPF Fibrotic Reticulum Contains Mesenchymal Cells Expressing Progenitor Determinants
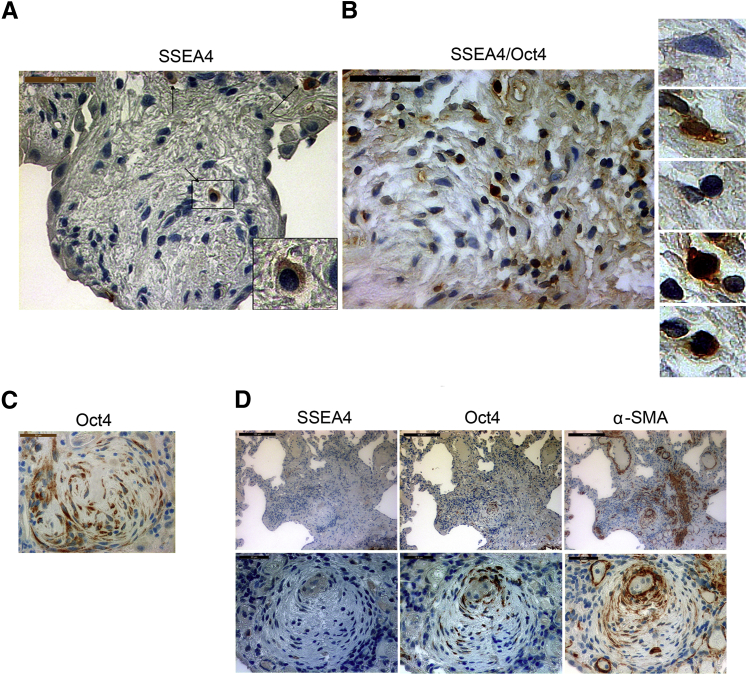
Next, we sought to determine whether cells with mesenchymal progenitor determinants or their progeny resided in IPF fibrotic lesions by analyzing lung pathological specimens from patients with IPF (n = 12) and patient controls (n = 5). For this purpose, on the basis of our results and prior knowledge,47,55 we used SSEA4 expression combined with localization of Oct4 to identify IPF mesenchymal progenitors (SSEA4+/nuclear Oct4+/cytoplasmic Oct4−) and their progeny (SSEA4−/nuclear Oct4−/cytoplasmic Oct4+) in lung pathological specimens from patients with IPF (n = 12).
SSEA4+ cells were interspersed throughout the IPF fibrotic reticulum in all 12 specimens examined (Figure 5A and Supplemental Figure S7). To more completely characterize the SSEA4+ cells, we developed a procedure to immunohistochemically double-stain pathological specimens for SSEA4 and Oct4. Our analysis found numerous examples of solitary SSEA4+ cells with nuclear Oct4 scattered throughout the fibrotic reticulum of each specimen analyzed (n = 6) (Figure 5B and Supplemental Figure S8). In addition, fibroblastic foci in all 12 specimens were heavily populated by clusters of SSEA4− cells that fell into the following three groups: i) a majority population that expressed only cytoplasmic Oct4; ii) some cells expressing both nuclear and cytoplasmic Oct4; and iii) a few cells displaying only nuclear Oct4 (Figure 5C and Supplemental Figure S9, A and B).
Figure 5.
Mesenchymal progenitor cells and their progeny are present in the IPF fibrotic reticulum. A: Analysis of IPF lung specimens for cells that express SSEA4, Oct4, and α-SMA. SSEA4-expressing cells in and around the perimeter of an IPF fibroblastic focus. Arrows point to SSEA4 expressing cells. The inset highlights an SSEA4 immunoreactive cell. B, left panel: Double staining for SSEA4 and Oct4 identified SSEA4+ cells with a nuclear Oct4 staining pattern within the IPF fibrotic reticulum. SSEA4 is shown in brown (DAB) and Oct4 in black (DAB/Ni). Right panels: High magnification images of cells within the fibroblastic focus shows a nonimmunoreactive cell, a cell immunoreactive for SSEA4 only, a cell immunoreactive for Oct4 only, and two cells immunoreactive for both SSEA4 and Oct4 are shown (top to bottom, respectively). C: An IPF fibroblastic focus heavily populated with cells displaying cytoplasmic Oct4. D: Serial sections of an IPF fibroblastic focus. Several cells express SSEA4 in and around the focus; however, most cells within the focus do not display SSEA4 immunoreactivity (left panels). Cells display cytoplasmic Oct4 and α-smooth muscle actin in a similar distribution (middle and right panels). n = 12 (A). Scale bars: 50 μm (A–C); 100 μm (D, top left and right panels); 99.8 μm (D, top middle panel); 20 μm (D, bottom left and right panels); 19.8 μm (D, bottom middle panel). α-SMA, α-smooth muscle actin.
Mesenchymal cells comprising fibroblastic foci express high levels of α-SMA.5 When analyzing serial sections of fibroblastic foci, we found many cells expressing cytoplasmic Oct4 and α-SMA (n = 4) (Figure 5D). Thus, a small number of cells with progenitor determinants (SSEA4+/nuclear Oct4+) populated the IPF fibrotic reticulum, accompanied by a larger number of mesenchymal cells with characteristics of their progeny (α-SMA+/SSEA4−/cytoplasmic Oct4+). Of note, relatively intact alveoli adjacent to fibrotic regions did not contain Oct4 immunoreactive cells (Supplemental Figure S9C). The alveolar walls of control lung tissue contained no cells with Oct4 or SSEA4 immunoreactivity (Supplemental Figure S10, A and B). However, SSEA4-expressing cells were observed in small and large airways and vascular structures of control lung tissue (Supplemental Figure S10C). Thus, IPF mesenchymal progenitor cells and cells bearing determinants of their daughter cells populate the fibrotic reticulum in IPF.
Genes and Pathways That Distinguish IPF Mesenchymal Progenitor Cells and Their Progeny from Their Control Counterparts Are Co-Regulated
To determine whether IPF and control progeny displayed disparate patterns of gene expression similar to their progenitors, we defined their steady state mRNA levels genomewide. In accord with their distinct in vivo phenotypes, IPF and control progeny displayed significant gene expression differences (Figure 6A). Validation testing of the daughter cell gene expression profiling data set for the same four genes examined in the progenitor data set (FLT1, MAP3K8, IGF2BP1, YBX1) confirmed the profiling data for all four genes (Supplemental Figure S11). A heatmap of the 50 most significant genes (P < 0.002) indicated that IPF progeny displayed a distinct molecular signature (Figure 6C), and gene set enrichment analysis indicated major differences in many of the same ontologies that distinguish IPF from control progenitors (Supplemental Table S3). To determine whether there was genomewide concordance between IPF mesenchymal progenitor cells and their progeny, all genes that were altered (P < 0.05) in both IPF mesenchymal progenitors and IPF progeny relative to their control counterparts were collected. On the basis of the hypothesis that some of the defining gene expression characteristics of IPF mesenchymal progenitors should be retained in their progeny, we analyzed whether these genes were co-regulated (ie, up-regulated in both IPF mesenchymal progenitors and IPF progeny or down-regulated in both IPF mesenchymal progenitors and IPF progeny). Our data indicated a striking degree of co-regulation among the genes that distinguish IPF from control (P = 1.9 × 10−15, Fisher exact test) (Figure 6B and Table 3). Thus, the molecular underpinnings of the IPF fibroblast are reflected by genomewide changes in the transcriptome that can be traced from its cell of origin.
Figure 6.
Genes and pathways that distinguish IPF mesenchymal progenitor cells and their progeny from their control counterparts are co-regulated. Steady state mRNA profiles identify activation of cellular processes in IPF progeny consistent with their fibrotic phenotype. A: Histogram of gene-by-gene P values that compare IPF and control progeny. Data shown are from progeny derived from eight independent mesenchymal progenitor cell lines. B: Shown are log2 fold changes of genes altered in both IPF mesenchymal progenitors and IPF progeny. Co-regulated genes appear in the right upper and left lower quadrants of the plot. C: Heatmap of the 50 most significantly altered genes between IPF and control progeny. Colors represent per gene z-score (expression difference normalized for SD). Co-regulation of genes distinguishing IPF mesenchymal progenitor cells and IPF progeny from their control counterparts. P = 1.9 × 10−15, Fisher exact test (B). n = 4 (A, IPF and control).
Table 3.
List of Co-Regulated Disease-Associated Genes in IPF Mesenchymal Progenitors and IPF Progeny
| Genes co-regulated up in both IPF MPC and progeny | Genes co-regulated down in both IPF MPC and progeny | ||||||
|---|---|---|---|---|---|---|---|
| 1 | FAM225B | 32 | A4GALT | 1 | SNORD116-20 | 32 | LOC440563 |
| 2 | MICA | 33 | SNRK | 2 | MIR1244-1 | 33 | NDP |
| 3 | LHFP | 34 | ETNK2 | 3 | PQBP1 | 34 | NDUFS3 |
| 4 | GPNMB | 35 | PRKACB | 4 | RPP38 | 35 | NEFH |
| 5 | RAB31 | 36 | TMEM184C | 5 | IGF2BP1 | 36 | YBX1 |
| 6 | BVES | 37 | MCTP2 | 6 | WDR6 | 37 | MBIP |
| 7 | FICD | 38 | PSEN1 | 7 | RPL35 | 38 | ATP6V1G2 |
| 8 | CYGB | 39 | PLSCR4 | 8 | INO80C | 39 | GNB1L |
| 9 | GJD3 | 40 | PTGFRN | 9 | NUDT10 | 40 | FOXRED1 |
| 10 | MAP3K8 | 41 | SERINC1 | 10 | EEF1B2 | 41 | DDX28 |
| 11 | EDNRB | 42 | PTPRM | 11 | FBL | 42 | DANCR |
| 12 | EIF2C4 | 43 | SSR4P1 | 12 | FAU | 43 | MRPS26 |
| 13 | A2M | 44 | TNFSF4 | 13 | DNAJC8 | 44 | MRPL11 |
| 14 | EPAS1 | 45 | OSGIN2 | 14 | EXOSC7 | 45 | MRPL9 |
| 15 | STOM | 46 | BEST1 | 15 | PPRC1 | 46 | SNRPA |
| 16 | FLT1 | 47 | TMEM204 | 16 | PES1 | 47 | SRP68 |
| 17 | SASH1 | 48 | GALNT12 | 17 | RPL36 | 48 | SNORA24 |
| 18 | GFRA1 | 49 | CCDC68 | 18 | ODZ4 | 49 | SNORA40 |
| 19 | ZADH2 | 50 | ZNF611 | 19 | SNORD52 | 50 | C1QBP |
| 20 | GPR37 | 51 | TMTC1 | 20 | PRPF19 | 51 | C11orf83 |
| 21 | FAM225A | 52 | TM2D1 | 21 | GNAZ | 52 | FSD1 |
| 22 | OSTM1 | 53 | SGIP1 | 22 | ZNF454 | 53 | PRR3 |
| 23 | TRHDE | 54 | ARHGAP5-AS1 | 23 | CHMP4A | 54 | TUBB1 |
| 24 | ICAM1 | 55 | RGS9 | 24 | HNRNPC | 55 | SYNJ2 |
| 25 | ACER2 | 56 | CABLES1 | 25 | B4GALNT4 | 56 | WDR85 |
| 26 | NCKAP5 | 57 | ACVRL1 | 26 | IMPDH2 | ||
| 27 | GVINP1 | 58 | SCARB2 | 27 | IPW | ||
| 28 | RND3 | 59 | KIAA0247 | 28 | RNF138P1 | ||
| 29 | PPAPDC2 | 60 | GAB2 | 29 | KIF5A | ||
| 30 | MITF | 61 | CLEC2B | 30 | C1orf31 | ||
| 31 | NQO2 | 31 | ZNF880 | ||||
Discussion
The majority fibroblast population derived from the IPF lung manifests a distinct pathological phenotype, but its origin remains unknown.5 Here, we report the discovery of mesenchymal progenitor cells recovered from the IPF lung that generate fibroblasts displaying the IPF phenotype. We demonstrate that IPF mesenchymal progenitor cells are present in the IPF fibrotic reticulum, can be isolated and cultivated from IPF lungs, and that the daughter cells of these IPF mesenchymal progenitors generate fibrotic lesions when grafted into two model organisms. These findings suggest that the fibrogenic mesenchymal progenitor cells we have identified may be causally implicated in the relentless fibroproliferation characteristic of IPF. Our studies provide the first evidence that pathological mesenchymal progenitor cells can serve as cells of origin for IPF fibroblasts. This finding has major implications in efforts to develop therapeutics to interdict the fibrotic process.
In accord with established protocols, we isolated mesenchymal progenitor cells by FACS from primary cultures of mesenchymal cells by using an antibody to SSEA4, a cell surface protein expressed by stem cells.42 However, analysis of the sort did not indicate a discrete subpopulation of SSEA4+ cells. Instead, it revealed a gradient of SSEA4-expressing cells. The spectrum ranged from a small subpopulation of cells strongly expressing SSEA4, merging into a distribution of cells displaying a diminishing amount of SSEA4, that in turn merged with a majority population of cells lacking SSEA4 expression. Our studies indicate that the cells strongly expressing SSEA4 also express nuclear Oct4 and share properties with MSCs. As these mesenchymal progenitors propagated and differentiated, their daughter cells lost SSEA4 expression, and Oct4 shifted from a nuclear to cytoplasmic location. Phenotypic analysis of this SSEA4−/nuclear Oct4−/cytoplasmic Oct4+ daughter cell population reveals properties associated with their origin–IPF or control. We interpret these data to indicate that the SSEA4− population of IPF mesenchymal cells represents a differentiation spectrum that ranges from early generations of progenitor cell progeny up to activated myofibroblasts. It will be important to identify surface determinants to enable accurate classification of these different differentiation states to gain a full understanding of mesenchymal cell population dynamics in the IPF lung.
Although healthy tissues contain MSCs that function to repair tissue after injury,22,56 and great enthusiasm has been expressed for their therapeutic potential in many diseases, including IPF,57 there is precedent for mesenchymal progenitors to participate in disease pathogenesis. With the use of animal models of disease, several studies report that normal MSCs and bone marrow progenitor cells can differentiate into fibroblasts that contribute to pathological processes.56,58,59 Normal MSCs can also produce transforming growth factor-β and Wnt proteins that can stimulate fibroblast proliferation.60 In human disease, bone marrow MSCs from patients with multiple myeloma manifest durable abnormalities that promote malignant cell maintenance and progression,61 and mesenchymal progenitors can be identified at sites of chronic lung allograft rejection.33,34 Consistently, we identify pathological mesenchymal progenitor cells in the IPF lung that generate progeny that manifest the IPF fibroblast phenotype. However, it is important to note that our data do not reveal the origin of the pathological IPF MPCs we have identified; currently, we lack the tools to determine whether they are derived from normal resident lung MSCs, bone marrow MSCs, a pathological de-differentiation event, or another source. It also remains to be determined whether the pathway we have discovered in IPF from pathological mesenchymal progenitors to diseased fibroblasts represents a more general paradigm for progressive fibrosis in other fibrotic lung disorders or for fibrosis of other organs.
Our study does not directly address the mechanisms leading to the stable acquisition of a pathological state by IPF mesenchymal progenitors or their progeny; however, an elegant study that examined the ability of extracellular matrix to durably shape fibroblast phenotype provides insight into how this might occur.62 In that report, lung fibroblasts cultured on stiff matrices become activated and continue to express an activated phenotype even when returned to pliable matrices, providing proof of concept for durable reprogramming of mesenchymal cells on the basis of the mechanics of the matrix microenvironment. This finding raises the possibility that a fibrotic extracellular matrix as is found in IPF could stably skew the phenotype of mesenchymal progenitors and their progeny.
To our knowledge, our study is the first to show that mesenchymal progenitor cells derived from a naturally occurring human fibrotic organ can generate progeny with the biological properties and molecular hallmarks of fibrotic fibroblasts. Transcriptional profiling revealed a distinct gene expression profile that distinguished IPF mesenchymal progenitor cells and their daughter cells from controls. Consistent with the idea that the hallmarks of the IPF fibroblast have their roots in pathological mesenchymal progenitor cells, we observed striking concordance among the genes and pathways that were significantly altered in both IPF mesenchymal progenitor cells and their progeny relative to controls. Moreover, we found that most of these genes and pathways were co-regulated. Thus, our data indicate that IPF mesenchymal progenitor cells display a pathological gene expression pattern and provide initial mechanistic insight into gene pathways that define the IPF phenotype. However, we recognize that this is only the first step. In a disease process as complex as IPF, unraveling the detailed mechanism(s) leading to the genesis of IPF mesenchymal progenitor cells and their differentiation to IPF fibroblasts will likely be unveiled slowly and systematically over many years.
Progressive scarring of the heart, blood vessels, lung, liver, kidney, and brain leads to millions of deaths each year worldwide. Despite decades of intensive investigation, the origin of the fibroblasts that mediate fibrotic organ destruction remained unknown. Several recent publications that used state-of-the-art lineage tracing technology to examine the origin of fibrosis-generating fibroblasts after injury in model organisms have reached definitive, but different conclusions.63–67 What has been definitively established in a mouse model is that several different stromal populations can contribute to lung fibrogenesis.67 A lingering problem in this field has been the paucity of direct knowledge about human organ fibrosis. Here, we report the discovery and molecular characterization of mesenchymal progenitor cells, derived from a human fibrotic organ, that are capable of generating daughter cells that have an activated phenotype and autonomously form a fibrotic reticulum in vivo. If confirmed by other investigators, our discovery provides strong impetus for a therapeutic paradigm shift focused on developing approaches that target fibrogenic mesenchymal progenitor cells before they generate fibrogenic fibroblasts that mediate organ failure.
Acknowledgments
We thank Dr. Dan Kaufman (Stem Cell Institute, University of Minnesota, Minneapolis, MN) for Oct4, Nanog, and Sox2 PCR primers; iPS cells; and for technical advice; Dr. Angela Panoskaltsis-Mortari (University of Minnesota) for technical advice; the Flow Cytometry Core Facility of the Masonic Cancer Center, a comprehensive cancer center designated by the National Cancer Institute; and University of Minnesota Core Facilities: the Biomedical Genomics Center; The Minnesota Supercomputing Institute; and BIONET for technical support.
Footnotes
Supported by National Heart, Lung, and Blood Institute grants R01 HL074882 and P01 HL91775 (C.A.H.) and R01 HL089249 (P.B.B.); funds provided by the O’Brien family; the Swedish Research Council (O.L.); the National Center for Advancing Translational Sciences of the National Institutes of Health award8UL1TR000114-02; and the Flow Cytometry Core Facility of the Masonic Cancer Center, a comprehensive cancer center designated by the National Cancer Institute, supported in part by P30 CA77598 and the University of Minnesota Imaging Center.
H.X. and V.B. contributed equally to this work.
Disclosures: None declared.
Supplemental Data
Morphology of primary mesenchymal cells, mesenchymal progenitors (SSEA4+/SSEA4−), and their daughter cells. Shown are representative phase contrast images of primary mesenchymal cells derived from human lungs, mesenchymal progenitor cells isolated by FACS, and the progeny (daughter cells) of the mesenchymal progenitors. Scale bar = 10 μm.
Visualization of engrafted human mesenchymal progenitor cells in zebrafish embryos. Control or IPF mesenchymal progenitor cells that had undergone FACS were labeled with CFSE and engrafted into zebrafish embryos at the late blastula stage. A graft containing normal bone marrow mesenchymal stem cells (MSCs) labeled with CFSE and engrafted into zebrafish embryos is shown as a progenitor cell control. A: At 48 hours after grafting, multiple small grafts containing CFSE-positive cells were localized to the head and trunk regions in developmentally normal host embryos. Shown are representative fluorescent (left panels) and bright field (right panels) images of live embryos (lateral view, head to the left) with the bone marrow, control, or IPF grafts localized to the pericardium, heart, and skin (arrows point to grafts). B: Engrafted CFSE-labeled IPF mesenchymal progenitor cells (single or paired) were found in various tissues throughout the embryo and were morphologically similar to the surrounding host cells. Shown is a representative sagittal section of the head region from a 2-day-old graft-bearing embryo immunostained with anti-human CD59/Cy-3 antibody. Of note, this antibody has no cross-reactivity with zebrafish cells and is therefore useful for identifying human cells grafted into zebrafish.16 The data demonstrate the ability of primary human mesenchymal progenitor cells to blend with surrounding host cells in chimeric embryos. Upper left panel: Phase contrast (10×) image of the embryo. Upper right panel: Merged FITC/TRITC/DAPI image (10×) of the embryo showing the inset (dashed box) location. Middle left panel: TRITC staining delineating CD59/Cy-3+ cells. Middle right panel: FITC staining delineating CFSE+ human MPCs within the embryo. Lower left panel: DAPI staining delineating nuclei. Lower right panel: Merged FITC/TRITC/DAPI image demonstrating position of CD59/CFSE+ human MPCs within the embryo. FITC indicates CFSE-labeled cells; TRITC, CD59/Cy-3+ cells. Scale bars: 250 μm (A); 30 μm (B, inset B). Original magnifications: ×10 (B); ×40 (inset, B). Cy-3, cyanine 3; e, eye; FITC, fluorescein isothiocyanate; OV, otic vesicle; P, pericardium; TRITC, tetramethylrhodamine siuothiocyanate; Y, yolk sac.
The progeny of IPF mesenchymal progenitor cells form an extensive fibrotic reticulum and express procollagen type I in vivo. Control or IPF mesenchymal progenitor cell progeny were labeled with CFSE and engrafted into zebrafish embryos. A: At 48 hours after transplantation, IPF progeny produced an elaborate fibrotic reticular network, whereas control cells formed an amorphous mass. Shown is representative three-dimensional computer-generated reconstruction of the IPF and control progeny xenografts 48 hours after grafting, showing the formation of an extensive fibrotic reticulum by the IPF progeny. B: Shown are representative images of sections from 2-day-old embryos with control or IPF grafts immunostained with anti-human CD59/Cy-3 antibodies. Shown are DAPI, merged images; FITC, CFSE+ cells; TRITC, CD59/Cy-3+ cells. The fibrotic reticulum is shown bound by a dashed oval. C: Human procollagen (type I) is expressed in IPF mesenchymal progenitor cell progeny. Shown are representative images of sections from control and IPF graft-bearing embryos that were immunostained with a human anti-procollagen type I antibody and horseradish peroxidase and counterstained with H&E. Sham-treated embryos with no graft at 48 hours after transplantation served as a negative control. Human IPF lung tissue displaying procollagen type I immunoreactive cells in a fibroblastic focus served as a positive control for the procollagen type I staining procedure. Immunoreactive cells were detected in the IPF graft-bearing embryos and human fibrotic tissue (detailed in insets). No cross-reactivity was detected in zebrafish embryonic cells. Scale bars: 50 μm (B); 200 μm (C); 50 μm (insets; dashed box). Cy-3, cyanine 3; FITC, fluorescein isothiocyanate; TRITC, tetramethylrhodamine isothiocyanate.
Control mesenchymal progenitor cell progeny do not form fibrotic lesions in the lungs of immunodeficient mice. The progeny of control mesenchymal progenitor cells were injected via tail vein into immunodeficient mice. Sixty days after adoptive transfer, mice were sacrificed. Immunohistochemical analysis with an anti-human β2 microglobulin antibody was performed to identify human cells within the mouse lungs. The lungs were anatomically normal.
Examination of mesenchymal progenitor cells that had undergone FACS for proliferative heterogeneity. SSEA4+/SSEA1−/small cells that had undergone FACS were labeled with CFSE, seeded into six-well tissue culture dishes, and cultured overnight in complete medium. CFSE-labeled cells were released from the dish and analyzed for fluorescence intensity 1, 2, 3, and 6 days after seeding. To minimize the effect of cell density on proliferation during the experiment, cells were seeded at 1 × 104 cells/cm2 for day 1 and day 2, 5 × 103 cells/cm2 for day 3, and 2 × 103 cells/cm2 for day 6. A: Shown is the population distribution of signal intensity as a function of time. B: The mean fluorescence for the cell population at each time point is fit to a single exponential curve (t1/2 = 1.1 days; R2 = 0.9808).
The progeny of a single control mesenchymal progenitor cell displays the control fibroblast phenotype. SSEA4+/SSEA1−/small cells (99% pure) that had undergone FACS were seeded at 1 cell per well in 96-well dishes coated with methylcellulose and observed for colony formation. Individual IPF (n = 3) and control (n = 3) progenitor cells formed separate colonies and were selected for further analysis. Cells from each individual colony were dispersed and cultured for 21 days as described in Materials and Methods. A: Quantification of IPF and control clonal progeny cell numbers is shown at day 21. B: Shown are Western blots of the clonal progeny derived from a single IPF and control mesenchymal progenitor cell for collagen 1A1, α-SMA, and GAPDH. Control clone 1 and 2 = lanes 2 and 3, and IPF clone 1 and 2 = lanes 4 and 5. Shown as a reference are the results for the primary IPF and control mesenchymal cell lines from which the progenitors were derived. Control parental fibroblasts = lane 1 and IPF parental fibroblasts = lane 6. C: Control clonal progeny were labeled with CFSE and grafted into zebrafish embryos. Shown is an example representative of nine zebrafish embryos with deformities caused by the progeny of a single control progenitor: left panel, grafted cells formed a compact, sessile mass (green, fluorescein isothiocyanate) located in head region with nonspecific autofluorescence outlining the yolk sac; middle panel, graft-bearing embryo with unilateral anophthalmia (bright field); and right panel, merged image. Embryo orientation: lateral view, head to the left. Scale bar = 200 μm. Collagen 1A1, type I collagen; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; α-SMA, α-smooth muscle actin.
Immunohistochemical analysis of IPF lung tissue. IPF lung tissue was stained with secondary antibody only (no SSEA4 primary antibody). Scale bar = 50 μm.
Immunohistochemical analysis of IPF lung tissue for co-expression of SSEA4 and Oct4. A: Representative images of double staining of IPF lung tissue sections (n = 6 patients with IPF) for SSEA4 and Oct4. SSEA4 is shown in brown (DAB) and Oct4 in black (DAB/Ni). B: Representative IPF (left panel) and control (right panel) lung tissue stained with secondary antibody only (no SSEA4 and Oct4 primary antibodies). Scale bars: 50 μm (A and B).
Immunohistochemical analysis of IPF lung tissue for Oct4. A: IPF fibroblastic focus demonstrating cells with a nuclear and cytoplasmic staining pattern. B: Representative IPF lung tissue stained with secondary antibody only (no Oct4 primary antibody). C: Relatively normal appearing IPF lung tissue adjacent to a fibrotic focus. DAB, brown. Scale bars: 50 μm (A–C).
Immunohistochemcial analysis of control lung tissue. Representative images of alveolar structures within control lung tissue stained with primary antibodies to SSEA4 (A) and Oct4 (B). A paucity of SSEA4 immunoreactive cells are seen in anatomically normal-appearing alveolar structures in control lung tissue. C: SSEA4 immunoreactive cell in a small vascular structure in control lung. D: SSEA4 immunoreactive cell in a larger vascular structure. E: Several SSEA4 immunoreactive cells in a large airway structure. Scale bars: 50 μm (A–C).
Validation testing of gene expression profiling data from the progeny of mesenchymal progenitor cells. Shown are relative expression levels of each mRNA by qPCR. Data shown represent the average of three cell lines. Genes and primers are as shown in Figure 2.
References
- 1.Wynn T.A., Ramalingam T.R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez I.E., Eickelberg O. New cellular and molecular mechanisms of lung injury and fibrosis in idiopathic pulmonary fibrosis. Lancet. 2012;380:680–688. doi: 10.1016/S0140-6736(12)61144-1. [DOI] [PubMed] [Google Scholar]
- 3.King T.E., Jr., Pardo A., Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378:1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 4.Noble P.W. Idiopathic pulmonary fibrosis: natural history and prognosis. Clin Chest Med. 2006;27(1 suppl 1):S11–S16. doi: 10.1016/j.ccm.2005.08.003. v. [DOI] [PubMed] [Google Scholar]
- 5.Noble P.W., Barkauskas C.E., Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest. 2012;122:2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raghu G., Weycker D., Edelsberg J., Bradford W.Z., Oster G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2006;174:810–816. doi: 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- 7.King T.E., Costabel U., Cordier J.F., DoPico G.A., DuBois R.M., Lynch D., Lynch III J.P., Myers J., Panos R., Raghu G., Schwartz D., Smith C.M. American Thoracic Society. Idiopathic pulmonary fibrosis: diagnosis and treatment. International consensus statement. American Thoracic Society (ATS), and the European Respiratory Society (ERS) Am J Respir Crit Care Med. 2000;161:646–664. doi: 10.1164/ajrccm.161.2.ats3-00. [DOI] [PubMed] [Google Scholar]
- 8.Travis W.D., King T.E., Bateman E.D., Lynch D.A., Capron F., Center D., Colby T.V., Cordier J.F., DuBois R.M., Galvin J., Grenier P., Hansell D.M., Hunninghake G.W., Kitaichi M., Muller N.L., Myers J.L., Nagai S., Nicholson A., Raghu G., Wallaert B., American Thoracic Society. European Respiratory Society American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee, June 2001. Am J Respir Crit Care Med. 2002;165:277–304. doi: 10.1164/ajrccm.165.2.ats01. [DOI] [PubMed] [Google Scholar]
- 9.Cool C.D., Groshong S.D., Rai P.R., Henson P.M., Stewart J.S., Brown K.K. Fibroblast foci are not discrete sites of lung injury or repair: the fibroblast reticulum. Am J Respir Crit Care Med. 2006;174:654–658. doi: 10.1164/rccm.200602-205OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn C., McDonald J.A. The roles of the myofibroblast in idiopathic pulmonary fibrosis. Ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis. Am J Pathol. 1991;138:1257–1265. [PMC free article] [PubMed] [Google Scholar]
- 11.Noble P.W., Homer R.J. Idiopathic pulmonary fibrosis: new insights into pathogenesis. Clin Chest Med. 2004;25:749–758. doi: 10.1016/j.ccm.2004.04.003. vii. [DOI] [PubMed] [Google Scholar]
- 12.Selman M., King T.E., Pardo A., American Thoracic Society. European Respiratory Society. American College of Chest Physicians Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy. Ann Intern Med. 2001;134:136–151. doi: 10.7326/0003-4819-134-2-200101160-00015. [DOI] [PubMed] [Google Scholar]
- 13.Xia H., Diebold D., Nho R., Perlman D., Kleidon J., Kahm J., Avdulov S., Peterson M., Nerva J., Bitterman P., Henke C. Pathological integrin signaling enhances proliferation of primary lung fibroblasts from patients with idiopathic pulmonary fibrosis. J Exp Med. 2008;205:1659–1672. doi: 10.1084/jem.20080001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia H., Khalil W., Kahm J., Jessurun J., Kleidon J., Henke C.A. Pathologic caveolin-1 regulation of PTEN in idiopathic pulmonary fibrosis. Am J Pathol. 2010;176:2626–2637. doi: 10.2353/ajpath.2010.091117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia H., Seeman J., Hong J., Hergert P., Bodem V., Jessurun J., Smith K., Nho R., Kahm J., Gaillard P., Henke C. Low alpha(2)beta(1) integrin function enhances the proliferation of fibroblasts from patients with idiopathic pulmonary fibrosis by activation of the beta-catenin pathway. Am J Pathol. 2012;181:222–233. doi: 10.1016/j.ajpath.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benyumov A.O., Hergert P., Herrera J., Peterson M., Henke C., Bitterman P.B. A novel zebrafish embryo xenotransplantation model to study primary human fibroblast motility in health and disease. Zebrafish. 2012;9:38–43. doi: 10.1089/zeb.2011.0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierce E.M., Carpenter K., Jakubzick C., Kunkel S.L., Flaherty K.R., Martinez F.J., Hogaboam C.M. Therapeutic targeting of CC ligand 21 or CC chemokine receptor 7 abrogates pulmonary fibrosis induced by the adoptive transfer of human pulmonary fibroblasts to immunodeficient mice. Am J Pathol. 2007;170:1152–1164. doi: 10.2353/ajpath.2007.060649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giangreco A., Reynolds S.D., Stripp B.R. Terminal bronchioles harbor a unique airway stem cell population that localizes to the bronchoalveolar duct junction. Am J Pathol. 2002;161:173–182. doi: 10.1016/S0002-9440(10)64169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajstura J., Rota M., Hall S.R., Hosoda T., D'Amario D., Sanada F., Zheng H., Ogorek B., Rondon-Clavo C., Ferreira-Martins J., Matsuda A., Arranto C., Goichberg P., Giordano G., Haley K.J., Bardelli S., Rayatzadeh H., Liu X., Quaini F., Liao R., Leri A., Perrella M.A., Loscalzo J., Anversa P. Evidence for human lung stem cells. N Engl J Med. 2011;364:1795–1806. doi: 10.1056/NEJMoa1101324. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Kim C.F., Jackson E.L., Woolfenden A.E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 21.Majka S.M., Beutz M.A., Hagen M., Izzo A.A., Voelkel N., Helm K.M. Identification of novel resident pulmonary stem cells: form and function of the lung side population. Stem Cells. 2005;23:1073–1081. doi: 10.1634/stemcells.2005-0039. [DOI] [PubMed] [Google Scholar]
- 22.Ortiz L.A., Gambelli F., McBride C., Gaupp D., Baddoo M., Kaminski N., Phinney D.G. Mesenchymal stem cell engraftment in lung is enhanced in response to bleomycin exposure and ameliorates its fibrotic effects. Proc Natl Acad Sci U S A. 2003;100:8407–8411. doi: 10.1073/pnas.1432929100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stripp B.R., Shapiro S.D. Stem cells in lung disease, repair, and the potential for therapeutic interventions: state-of-the-art and future challenges. Am J Respir Cell Mol Biol. 2006;34:517–518. doi: 10.1165/rcmb.F315. [DOI] [PubMed] [Google Scholar]
- 24.Summer R., Fitzsimmons K., Dwyer D., Murphy J., Fine A. Isolation of an adult mouse lung mesenchymal progenitor cell population. Am J Respir Cell Mol Biol. 2007;37:152–159. doi: 10.1165/rcmb.2006-0386OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summer R., Kotton D.N., Liang S., Fitzsimmons K., Sun X., Fine A. Embryonic lung side population cells are hematopoietic and vascular precursors. Am J Respir Cell Mol Biol. 2005;33:32–40. doi: 10.1165/rcmb.2005-0024OC. [DOI] [PubMed] [Google Scholar]
- 26.Weiss D.J., Kolls J.K., Ortiz L.A., Panoskaltsis-Mortari A., Prockop D.J. Stem cells and cell therapies in lung biology and lung diseases. Proc Am Thorac Soc. 2008;5:637–667. doi: 10.1513/pats.200804-037DW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 29.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 30.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., Campbell L.L., Polyak K., Brisken C., Yang J., Weinberg R.A. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 32.Wu C., Amini-Nik S., Nadesan P., Stanford W.L., Alman B.A. Aggressive fibromatosis (desmoid tumor) is derived from mesenchymal progenitor cells. Cancer Res. 2010;70:7690–7698. doi: 10.1158/0008-5472.CAN-10-1656. [DOI] [PubMed] [Google Scholar]
- 33.Badri L., Murray S., Liu L.X., Walker N.M., Flint A., Wadhwa A., Chan K.M., Toews G.B., Pinsky D.J., Martinez F.J., Lama V.N. Mesenchymal stromal cells in bronchoalveolar lavage as predictors of bronchiolitis obliterans syndrome. Am J Respir Crit Care Med. 2011;183:1062–1070. doi: 10.1164/rccm.201005-0742OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lama V.N., Smith L., Badri L., Flint A., Andrei A.C., Murray S., Wang Z., Liao H., Toews G.B., Krebsbach P.H., Peters-Golden M., Pinsky D.J., Martinez F.J., Thannickal V.J. Evidence for tissue-resident mesenchymal stem cells in human adult lung from studies of transplanted allografts. J Clin Invest. 2007;117:989–996. doi: 10.1172/JCI29713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B., Polunovsky V., White J., Blazar B., Nakhleh R., Jessurun J., Peterson M., Bitterman P. Mesenchymal cells isolated after acute lung injury manifest an enhanced proliferative phenotype. J Clin Invest. 1992;90:1778–1785. doi: 10.1172/JCI116052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsson O., Diebold D., Fan D., Peterson M., Nho R.S., Bitterman P.B., Henke C.A. Fibrotic myofibroblasts manifest genome-wide derangements of translational control. PLoS One. 2008;3:e3220. doi: 10.1371/journal.pone.0003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rijlaarsdam M.A., van Herk H.A., Gillis A.J., Stoop H., Jenster G., Martens J., van Leenders G.J., Dinjens W., Hoogland A.M., Timmermans M., Looijenga L.H. Specific detection of OCT3/4 isoform A/B/B1 expression in solid (germ cell) tumours and cell lines: confirmation of OCT3/4 specificity for germ cell tumours. Br J Cancer. 2011;105:854–863. doi: 10.1038/bjc.2011.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright G.W., Simon R.M. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 39.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris M.A., Deegan J.I., Ireland A., Lomax J., Ashburner M., Tweedie S., Carbon S., Lewis S., Mungall C., Day-Richter J., Eilbeck K., Blake J.A., Bult C., Diehl A.D., Dolan M., Drabkin H., Eppig J.T., Hill D.P., Ni L., Ringwald M., Balakrishnan R., Binkley G., Cherry J.M., Christie K.R., Costanzo M.C., Dong Q., Engel SR., Fisk D.G., Hirschman J.E., Hitz B.C., Hong E.L., Krieger C.J., Miyasato S.R., Hash R.S., Park J., Skrzpek M.S., Gene Ontology Consortium The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson O., Perlman D.M., Fan D., Reilly C.S., Peterson M., Dahlgren C., Liang Z., Li S., Polunovsky V.A., Wahlestedt C., Bitterman P.B. Apoptosis resistance downstream of eIF4E: posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic Acids Res. 2006;34:4375–4386. doi: 10.1093/nar/gkl558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gang E.J., Bosnakovski D., Figueiredo C.A., Visser J.W., Perlingeiro R.C. SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood. 2007;109:1743–1751. doi: 10.1182/blood-2005-11-010504. [DOI] [PubMed] [Google Scholar]
- 43.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 44.Moore B.B., Kolodsick J.E., Thannickal V.J., Cooke K., Moore T.A., Hogaboam C., Wilke C.A., Toews G.B. CCR2-mediated recruitment of fibrocytes to the alveolar space after fibrotic injury. Am J Pathol. 2005;166:675–684. doi: 10.1016/S0002-9440(10)62289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Short B., Brouard N., Occhiodoro-Scott T., Ramakrishnan A., Simmons P.J. Mesenchymal stem cells. Arch Med Res. 2003;34:565–571. doi: 10.1016/j.arcmed.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Ramos C., Montano M., Garcia-Alvarez J., Ruiz V., Uhal B.D., Selman M., Pardo A. Fibroblasts from idiopathic pulmonary fibrosis and normal lungs differ in growth rate, apoptosis, and tissue inhibitor of metalloproteinases expression. Am J Respir Cell Mol Biol. 2001;24:591–598. doi: 10.1165/ajrcmb.24.5.4333. [DOI] [PubMed] [Google Scholar]
- 47.Ferro F., Spelat R., D'Aurizio F., Puppato E., Pandolfi M., Beltrami A.P., Cesselli D., Falini G., Beltrami C.A., Curcio F. Dental pulp stem cells differentiation reveals new insights in Oct4A dynamics. PLoS One. 2012;7:e41774. doi: 10.1371/journal.pone.0041774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estrada J., Li P., Mir B. Multiorgan engraftment of human somatic cells in swine foetuses after intra-blastocyst transplantation. Reprod Domest Anim. 2011;46:630–635. doi: 10.1111/j.1439-0531.2010.01718.x. [DOI] [PubMed] [Google Scholar]
- 49.Harder F., Henschler R., Junghahn I., Lamers M.C., Muller A.M. Human hematopoiesis in murine embryos after injecting human cord blood-derived hematopoietic stem cells into murine blastocysts. Blood. 2002;99:719–721. doi: 10.1182/blood.v99.2.719. [DOI] [PubMed] [Google Scholar]
- 50.Harder F., Kirchhof N., Petrovic S., Schmittwolf C., Durr M., Muller A.M. Developmental potentials of hematopoietic and neural stem cells following injection into pre-implantation blastocysts. Ann Hematol. 2002;81(suppl 2):S20–S21. [PubMed] [Google Scholar]
- 51.Harder F., Kirchhof N., Petrovic S., Wiese S., Muller A.M. Erythroid-like cells from neural stem cells injected into blastocysts. Exp Hematol. 2004;32:673–682. doi: 10.1016/j.exphem.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Parameswaran V., Laize V., Gavaia P.J., Leonor Cancela M. ESSA1 embryonic stem like cells from gilthead seabream: a new tool to study mesenchymal cell lineage differentiation in fish. Differentiation. 2012;84:240–251. doi: 10.1016/j.diff.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Prelle K., Zink N., Wolf E. Pluripotent stem cells–model of embryonic development, tool for gene targeting, and basis of cell therapy. Anat Histol Embryol. 2002;31:169–186. doi: 10.1046/j.1439-0264.2002.00388.x. [DOI] [PubMed] [Google Scholar]
- 54.Siqueira da Fonseca S.A., Abdelmassih S., de Mello Cintra Lavagnolli T., Serafim R.C., Clemente Santos E.J., Mota Mendes C., de Souza Pereira V., Ambrosio C.E., Miglino M.A., Visintin J.A., Abdelmassih R., Kerkis A., Kerkis I. Human immature dental pulp stem cells' contribution to developing mouse embryos: production of human/mouse preterm chimaeras. Cell Prolif. 2009;42:132–140. doi: 10.1111/j.1365-2184.2008.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Y., Yang Y., Li W., Chen Q., Li J., Pan X., Zhou L., Liu C., Chen C., He J., Cao H., Yao H., Zheng L., Xu X., Xia Z., Ren J., Xiao L., Li L., Shen B., Zhou H., Wang Y.J. Reciprocal regulation of Akt and Oct4 promotes the self-renewal and survival of embryonal carcinoma cells. Mol Cell. 2012;48:627–640. doi: 10.1016/j.molcel.2012.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee C.H., Shah B., Moioli E.K., Mao J.J. CTGF directs fibroblast differentiation from human mesenchymal stem/stromal cells and defines connective tissue healing in a rodent injury model. J Clin Invest. 2010;120:3340–3349. doi: 10.1172/JCI43230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toonkel R.L., Hare J.M., Matthay M.A., Glassberg M.K. Mesenchymal stem cells and idiopathic pulmonary fibrosis. Potential for clinical testing. Am J Respir Crit Care Med. 2013;188:133–140. doi: 10.1164/rccm.201207-1204PP. [DOI] [PubMed] [Google Scholar]
- 58.Sopel M.J., Rosin N.L., Lee T.D., Legare J.F. Myocardial fibrosis in response to Angiotensin II is preceded by the recruitment of mesenchymal progenitor cells. Lab Invest. 2011;91:565–578. doi: 10.1038/labinvest.2010.190. [DOI] [PubMed] [Google Scholar]
- 59.Spaeth E.L., Dembinski J.L., Sasser A.K., Watson K., Klopp A., Hall B., Andreeff M., Marini F. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS One. 2009;4:e4992. doi: 10.1371/journal.pone.0004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salazar K.D., Lankford S.M., Brody A.R. Mesenchymal stem cells produce Wnt isoforms and TGF-beta1 that mediate proliferation and procollagen expression by lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2009;297:L1002–L1011. doi: 10.1152/ajplung.90347.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reagan M.R., Ghobrial I.M. Multiple myeloma mesenchymal stem cells: characterization, origin, and tumor-promoting effects. Clin Cancer Res. 2012;18:342–349. doi: 10.1158/1078-0432.CCR-11-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balestrini J.L., Chaudhry S., Sarrazy V., Koehler A., Hinz B. The mechanical memory of lung myofibroblasts. Integr Biol (Camb) 2012;4:410–421. doi: 10.1039/c2ib00149g. [DOI] [PubMed] [Google Scholar]
- 63.Hashimoto N., Jin H., Liu T., Chensue S.W., Phan S.H. Bone marrow-derived progenitor cells in pulmonary fibrosis. J Clin Invest. 2004;113:243–252. doi: 10.1172/JCI18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Humphreys B.D., Lin S.L., Kobayashi A., Hudson T.E., Nowlin B.T., Bonventre J.V., Valerius M.T., McMahon A.P., Duffield J.S. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim K.K., Kugler M.C., Wolters P.J., Robillard L., Galvez M.G., Brumwell A.N., Sheppard D., Chapman H.A. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A. 2006;103:13180–13185. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moeller A., Gilpin S.E., Ask K., Cox G., Cook D., Gauldie J., Margetts P.J., Farkas L., Dobranowski J., Boylan C., O'Byrne P.M., Strieter R.M., Kolb M. Circulating fibrocytes are an indicator of poor prognosis in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;179:588–594. doi: 10.1164/rccm.200810-1534OC. [DOI] [PubMed] [Google Scholar]
- 67.Rock J.R., Barkauskas C.E., Cronce M.J., Xue Y., Harris J.R., Liang J., Noble P.W., Hogan B.L. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108:E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphology of primary mesenchymal cells, mesenchymal progenitors (SSEA4+/SSEA4−), and their daughter cells. Shown are representative phase contrast images of primary mesenchymal cells derived from human lungs, mesenchymal progenitor cells isolated by FACS, and the progeny (daughter cells) of the mesenchymal progenitors. Scale bar = 10 μm.
Visualization of engrafted human mesenchymal progenitor cells in zebrafish embryos. Control or IPF mesenchymal progenitor cells that had undergone FACS were labeled with CFSE and engrafted into zebrafish embryos at the late blastula stage. A graft containing normal bone marrow mesenchymal stem cells (MSCs) labeled with CFSE and engrafted into zebrafish embryos is shown as a progenitor cell control. A: At 48 hours after grafting, multiple small grafts containing CFSE-positive cells were localized to the head and trunk regions in developmentally normal host embryos. Shown are representative fluorescent (left panels) and bright field (right panels) images of live embryos (lateral view, head to the left) with the bone marrow, control, or IPF grafts localized to the pericardium, heart, and skin (arrows point to grafts). B: Engrafted CFSE-labeled IPF mesenchymal progenitor cells (single or paired) were found in various tissues throughout the embryo and were morphologically similar to the surrounding host cells. Shown is a representative sagittal section of the head region from a 2-day-old graft-bearing embryo immunostained with anti-human CD59/Cy-3 antibody. Of note, this antibody has no cross-reactivity with zebrafish cells and is therefore useful for identifying human cells grafted into zebrafish.16 The data demonstrate the ability of primary human mesenchymal progenitor cells to blend with surrounding host cells in chimeric embryos. Upper left panel: Phase contrast (10×) image of the embryo. Upper right panel: Merged FITC/TRITC/DAPI image (10×) of the embryo showing the inset (dashed box) location. Middle left panel: TRITC staining delineating CD59/Cy-3+ cells. Middle right panel: FITC staining delineating CFSE+ human MPCs within the embryo. Lower left panel: DAPI staining delineating nuclei. Lower right panel: Merged FITC/TRITC/DAPI image demonstrating position of CD59/CFSE+ human MPCs within the embryo. FITC indicates CFSE-labeled cells; TRITC, CD59/Cy-3+ cells. Scale bars: 250 μm (A); 30 μm (B, inset B). Original magnifications: ×10 (B); ×40 (inset, B). Cy-3, cyanine 3; e, eye; FITC, fluorescein isothiocyanate; OV, otic vesicle; P, pericardium; TRITC, tetramethylrhodamine siuothiocyanate; Y, yolk sac.
The progeny of IPF mesenchymal progenitor cells form an extensive fibrotic reticulum and express procollagen type I in vivo. Control or IPF mesenchymal progenitor cell progeny were labeled with CFSE and engrafted into zebrafish embryos. A: At 48 hours after transplantation, IPF progeny produced an elaborate fibrotic reticular network, whereas control cells formed an amorphous mass. Shown is representative three-dimensional computer-generated reconstruction of the IPF and control progeny xenografts 48 hours after grafting, showing the formation of an extensive fibrotic reticulum by the IPF progeny. B: Shown are representative images of sections from 2-day-old embryos with control or IPF grafts immunostained with anti-human CD59/Cy-3 antibodies. Shown are DAPI, merged images; FITC, CFSE+ cells; TRITC, CD59/Cy-3+ cells. The fibrotic reticulum is shown bound by a dashed oval. C: Human procollagen (type I) is expressed in IPF mesenchymal progenitor cell progeny. Shown are representative images of sections from control and IPF graft-bearing embryos that were immunostained with a human anti-procollagen type I antibody and horseradish peroxidase and counterstained with H&E. Sham-treated embryos with no graft at 48 hours after transplantation served as a negative control. Human IPF lung tissue displaying procollagen type I immunoreactive cells in a fibroblastic focus served as a positive control for the procollagen type I staining procedure. Immunoreactive cells were detected in the IPF graft-bearing embryos and human fibrotic tissue (detailed in insets). No cross-reactivity was detected in zebrafish embryonic cells. Scale bars: 50 μm (B); 200 μm (C); 50 μm (insets; dashed box). Cy-3, cyanine 3; FITC, fluorescein isothiocyanate; TRITC, tetramethylrhodamine isothiocyanate.
Control mesenchymal progenitor cell progeny do not form fibrotic lesions in the lungs of immunodeficient mice. The progeny of control mesenchymal progenitor cells were injected via tail vein into immunodeficient mice. Sixty days after adoptive transfer, mice were sacrificed. Immunohistochemical analysis with an anti-human β2 microglobulin antibody was performed to identify human cells within the mouse lungs. The lungs were anatomically normal.
Examination of mesenchymal progenitor cells that had undergone FACS for proliferative heterogeneity. SSEA4+/SSEA1−/small cells that had undergone FACS were labeled with CFSE, seeded into six-well tissue culture dishes, and cultured overnight in complete medium. CFSE-labeled cells were released from the dish and analyzed for fluorescence intensity 1, 2, 3, and 6 days after seeding. To minimize the effect of cell density on proliferation during the experiment, cells were seeded at 1 × 104 cells/cm2 for day 1 and day 2, 5 × 103 cells/cm2 for day 3, and 2 × 103 cells/cm2 for day 6. A: Shown is the population distribution of signal intensity as a function of time. B: The mean fluorescence for the cell population at each time point is fit to a single exponential curve (t1/2 = 1.1 days; R2 = 0.9808).
The progeny of a single control mesenchymal progenitor cell displays the control fibroblast phenotype. SSEA4+/SSEA1−/small cells (99% pure) that had undergone FACS were seeded at 1 cell per well in 96-well dishes coated with methylcellulose and observed for colony formation. Individual IPF (n = 3) and control (n = 3) progenitor cells formed separate colonies and were selected for further analysis. Cells from each individual colony were dispersed and cultured for 21 days as described in Materials and Methods. A: Quantification of IPF and control clonal progeny cell numbers is shown at day 21. B: Shown are Western blots of the clonal progeny derived from a single IPF and control mesenchymal progenitor cell for collagen 1A1, α-SMA, and GAPDH. Control clone 1 and 2 = lanes 2 and 3, and IPF clone 1 and 2 = lanes 4 and 5. Shown as a reference are the results for the primary IPF and control mesenchymal cell lines from which the progenitors were derived. Control parental fibroblasts = lane 1 and IPF parental fibroblasts = lane 6. C: Control clonal progeny were labeled with CFSE and grafted into zebrafish embryos. Shown is an example representative of nine zebrafish embryos with deformities caused by the progeny of a single control progenitor: left panel, grafted cells formed a compact, sessile mass (green, fluorescein isothiocyanate) located in head region with nonspecific autofluorescence outlining the yolk sac; middle panel, graft-bearing embryo with unilateral anophthalmia (bright field); and right panel, merged image. Embryo orientation: lateral view, head to the left. Scale bar = 200 μm. Collagen 1A1, type I collagen; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; α-SMA, α-smooth muscle actin.
Immunohistochemical analysis of IPF lung tissue. IPF lung tissue was stained with secondary antibody only (no SSEA4 primary antibody). Scale bar = 50 μm.
Immunohistochemical analysis of IPF lung tissue for co-expression of SSEA4 and Oct4. A: Representative images of double staining of IPF lung tissue sections (n = 6 patients with IPF) for SSEA4 and Oct4. SSEA4 is shown in brown (DAB) and Oct4 in black (DAB/Ni). B: Representative IPF (left panel) and control (right panel) lung tissue stained with secondary antibody only (no SSEA4 and Oct4 primary antibodies). Scale bars: 50 μm (A and B).
Immunohistochemical analysis of IPF lung tissue for Oct4. A: IPF fibroblastic focus demonstrating cells with a nuclear and cytoplasmic staining pattern. B: Representative IPF lung tissue stained with secondary antibody only (no Oct4 primary antibody). C: Relatively normal appearing IPF lung tissue adjacent to a fibrotic focus. DAB, brown. Scale bars: 50 μm (A–C).
Immunohistochemcial analysis of control lung tissue. Representative images of alveolar structures within control lung tissue stained with primary antibodies to SSEA4 (A) and Oct4 (B). A paucity of SSEA4 immunoreactive cells are seen in anatomically normal-appearing alveolar structures in control lung tissue. C: SSEA4 immunoreactive cell in a small vascular structure in control lung. D: SSEA4 immunoreactive cell in a larger vascular structure. E: Several SSEA4 immunoreactive cells in a large airway structure. Scale bars: 50 μm (A–C).
Validation testing of gene expression profiling data from the progeny of mesenchymal progenitor cells. Shown are relative expression levels of each mRNA by qPCR. Data shown represent the average of three cell lines. Genes and primers are as shown in Figure 2.