Abstract
A recently published procedure to enrich for efficient competitive root tip colonizers (I. Kuiper, G. V. Bloemberg, and B. J. J. Lugtenberg, Mol. Plant-Microbe Interact. 14:1197-1205) after bacterization of seeds was applied to isolate efficient competitive root tip colonizers for both the dicotyledenous plant tomato and the monocotyledenous plant grass from a random Tn5luxAB mutant bank of the good root colonizer Pseudomonas fluorescens WCS365. Unexpectedly, the best-colonizing mutant, strain PCL1286, showed a strongly enhanced competitive root-tip-colonizing phenotype. Sequence analyses of the Tn5luxAB flanking regions showed that the transposon had inserted in a mutY homolog. This gene is involved in the repair of A · G mismatches caused by spontaneous oxidation of guanine. We hypothesized that, since the mutant is defective in repairing its mismatches, its cells harbor an increased number of mutations and therefore can adapt faster to the environment of the root system. To test this hypothesis, we constructed another mutY mutant and analyzed its competitive root tip colonization behavior prior to and after enrichment. As a control, a nonmutated wild type was subjected to the enrichment procedure. The results of these analyses showed (i) that the enrichment procedure did not alter the colonization ability of the wild type, (ii) that the new mutY mutant was strongly impaired in its colonization ability, but (iii) that after three enrichment cycles it colonized significantly better than its wild type. Therefore it is concluded that both the mutY mutation and the selection procedure are required to obtain an enhanced root-tip-colonizing mutant.
Competitive root tip colonization by Pseudomonas strains can play an important role in the efficient control of soilborne crop diseases caused by fungi (17, 30, 36, 40). Inadequate colonization is often the limiting factor in biocontrol (3, 30, 39). Pseudomonas fluorescens WCS365 is an excellent colonizer of various plant root systems. To study competitive root tip colonization, we decided to characterize colonization traits and genes. The results of multiple studies have recently been reviewed by Lugtenberg et al. (18). Among the colonization mutants a mutant was found which is impaired in competitive root tip colonization and mutated in a gene with homology to the sss gene from Pseudomonas aeruginosa (7). This gene encodes a protein belonging to the λ integrase family of site-specific recombinases involved in DNA rearrangements. The role of the sss homologue in colonization is proposed to be through genetic rearrangements causing different phenotypes. This colonization mutant is assumed to be locked in a phase that is not suitable for competitive colonization in the rhizosphere. Introduction of the sss gene into the poor colonizer P. fluorescens WCS307 and into the good colonizer P. fluorescens F113 improved competitive root-tip-colonizing abilities of these strains (6). These results show that it is possible to improve colonization through genetic engineering (6).
Efficient colonizers can be obtained after inoculation of seedlings with a mixture of different bacteria followed by growth of the seedling in a gnotobiotic sand system (32). The root tip contains the most efficient colonizing bacteria (32). Repeated use of this method yields enhanced root-tip-colonizing wild-type bacteria (14).
In the present study we applied this procedure to a random collection of Tn5luxAB-marked mutants of the efficient colonizer P. fluorescens WCS365 in an attempt to isolate mutants that are efficient in competitive root tip colonization of both the dicotyledenous plant tomato and the monocotyledenous plant grass. Surprisingly, we found a mutant that proved to be a superior competitive colonizer compared to its parental strain. Its isolation and its genetic and functional analyses are described in this paper.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas strains (Table 1) were grown in liquid King's medium B (KB) (13) at 28°C with vigorous shaking. The medium was solidified with 1.8% agar (Select agar; Gibco BRL, Life Technologies, Paisley, United Kingdom) and when appropriate, kanamycin (Sigma-Aldrich BV, Zwijndrecht, The Netherlands) or tetracycline was added in final concentrations of 50 and 40 μg/ml, respectively. Escherichia coli was grown at 37°C on solidified Luria-Bertani (LB) medium (29).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmid | Relevant characteristicsa | Reference or Source |
|---|---|---|
| P. fluorescens | ||
| WCS365 | Wild-type biocontrol strain; efficient competitive colonizer of tomato, potato, and wheat roots; Nalr | 5, 30 |
| PCL1285 | Kmr derivative of WCS365 not impaired in competitive root colonization on tomato and grass; Kmr | This study |
| PCL1286 | WCS365 Tn5luxAB mutant with an impaired mutY gene and enhanced competitive colonizing abilities on tomato and grass; Kmr | This study |
| PCL1805 | WCS365 mutant with an impaired mutY gene obtained by single homologous recombination; impaired in competitive colonizing abilities on tomato and grass; Kmr | This study |
| PCL1808 | WCS365 mutant with an impaired mutY gene obtained by single homologous recombination (PCL 1805) and 3 cycles of enrichment on tomato; enhanced in competitive colonization on tomato and grass; Kmr | This study |
| WCS365(3X) | WCS365 variant obtained after 3 enrichment cycles on tomato; wild-type colonizing abilities | This study |
| PCL1285(3X) | PCL1285 variant obtained after 3 enrichment cycles on tomato; wild-type colonizing abilities | This study |
| PCL1814 | PCL1286 variant harboring pMP5573; Kmr Tcr | This study |
| PCL1815 | PCL1805 variant harboring pMP5573; Kmr Tcr | This study |
| PCL1816 | PCL1808 variant harboring pMP5573; Kmr Tcr | This study |
| PCL1817 | WCS365 variant harboring pMP5573; Tcr | This study |
| E. coli | ||
| XL1-Blue | Used for transformation and propagation of plasmids | Stratagene, La Jolla, Calif. |
| DH5α | Used for transformation and propagation of plasmids | 10 |
| Plasmids | ||
| pRL1063A | Plasmid harboring promoterless Tn5luxAB; Kmr | 41 |
| pRK2013 | Helper plasmid for triparental mating; Nmr Kmr | 8 |
| pGEM-T-easy | Cloning vector for Taq-amplified PCR products; Apr | Promega |
| pMP4655 | pME6010 derivative expressing egfp under the control of the lac promoter; Tcr | 2 |
| pMP4662 | pME6010 derivative expressing rfp under the control of the lac promoter; Tcr | 2 |
| pMP5285 | pME3049 (38) derivative; suicide plasmid for Pseudomonas spp.; used for homologous recombination; Kmr Cbr | 15 |
| pMP5421 | pRL1063A derivative recovered from chromosomal DNA of PCL1286 after digestion with EcoRI; Kmr | This study |
| pMP5465 | pGEM-T Easy containing 1,187-bp PCR fragment of mutY; Apr | This study |
| pMP5564 | pMP5285 harboring part of PCR fragment of mutY from pMP5465 used for homologous recombination; Kmr | This study |
| pMP5572 | pGEM-T Easy containing 1,742-bp PCR fragment including complete mutY gene; Apr | This study |
| pME6031 | Stable shuttle vector for gram-negative bacteria based on pVS1 replicon (34), contains extra transcriptional terminator; Tcr | 11 |
| pMP5573 | pME6031 derivative containing complete mutY gene, used for complementation; Tcr | This study |
Resistance phenotypes; Apr, ampicillin; Nalr, nalidixic acid; Cbr, carbenicillin; Nmr, neomycin; Kmr, kanamycin; Tcr, tetracycline.
To determine growth rates of individual Pseudomonas strains, the optical density at 620 nm (OD620) was followed during growth. For growth of wild type and mutants in competition, overnight-grown KB cultures were diluted to an OD620 of 0.1 and suspensions of wild type and mutant were mixed in a 1:1 ratio. After overnight growth the culture was diluted 1,000-fold in fresh KB. The ratio between the wild type (Kms) and mutant (Kmr) was determined after dilutions were plated on KB and KB with kanamycin to distinguish between the wild type and mutant derivative. For competitive growth in exudate, tomato root exudate was collected as described below. Competitive growth was performed as described previously with some slight modifications. Cell suspensions were diluted in root exudate to approximately 104 cells per ml. Cultures were grown overnight, the number of viable cells was determined, and the culture was diluted 1,000-fold. This procedure was repeated three times.
Isolation of tomato root exudate.
Tomato root exudates (19) were isolated as described by Simons et al. (31). Briefly, 100 sterile seedlings were placed in 100 ml of plant nutrient solution (12) and allowed to grow in a climate-controlled growth chamber at 18°C, 70% relative humidity, and 16 h of daylight. After 7 days, root exudate was collected and tested on KB plates for infections. Sterile tomato root exudate was stored in 50-ml aliquots at −20°C.
Motility.
Motility was tested as described by Dekkers et al. (5). Briefly, KB was diluted 20-fold, semisolidified with 0.3% agar (Select agar), and poured into petri dishes. Samples of bacteria were applied in the middle of the agar plate, and after growth overnight, the diameter of the expanding colony was measured. Experiments were performed in triplicate.
Tn5 mutagenesis.
Mutants of P. fluorescens WCS365 were generated by random transposon mutagenesis. A triparental mating of WCS365 was performed with E. coli strains containing plasmid pRL1063A, which harbors a Tn5luxAB transposon and kanamycin marker gene (41), and helper plasmid pRK2013 (8). The transconjugants were selected for kanamycin resistance. Nalidixic acid was added to a final concentration of 15 μg/ml to reduce growth of E. coli helper strains.
To recover the regions flanking the transposon, chromosomal DNA of the mutant was isolated and digested with EcoRI. After recircularization of the plasmid the Tn5luxAB flanking chromosomal DNA was sequenced by Baseclear (Leiden, The Netherlands). Primers used were homologous to the left (oMP458, 5′-TACTAGATTCAATGCTATCAATGAG-3′) and right (oMP459, 5′-AGGAGGTCACATGGAATATCAGAT-3′) borders of Tn5luxAB of pRL1063A. Obtained sequences were analyzed using BLASTX in GenBank (1)
Enrichment of Tn5luxAB mutants.
Enrichment cycles were performed as described by Kuiper et al. (14) with some slight modifications. Briefly, the Tn5luxAB mutant bank of WCS365 was grown overnight in KB at 28°C. The cells were washed, resuspended in phosphate-buffered saline (PBS), and used to inoculate germinated sterile seedlings of tomato (Lycopersicon esculentum mill. cv. Carmello; Novartis Seeds B.V., Enkhuizen, The Netherlands) or grass (Lolium multiflorum cv. Barmultra; Barenbrug Research, Wolfheze, The Netherlands) for the enrichment procedure. The seedlings were placed in a gnotobiotic system and grown as described below. Bacteria were isolated from the root tip after 7 days (32) and grown on KB plates supplemented with kanamycin. Subsequently, all colonies were scraped together from the plate and the mixture was used to inoculate fresh KB. After overnight growth the cells were washed in PBS and seedlings were inoculated as mentioned previously. These cycles of enrichment were carried out four times, alternating on tomato and grass.
Root tip colonization assays in a gnotobiotic sand system.
Root colonization assays were performed as described by Simons et al. (32), using a gnotobiotic system containing sterile quartz sand, to which 10% (vol/wt) plant nutrient solution was added to moisten the sand. For competitive colonization experiments, sterile germinated tomato or grass seeds were inoculated with a 1:1 mixture of the parental strain and the corresponding mutant strain. The seedlings were placed in tubes and allowed to grow in a climate-controlled growth chamber. After 7 days the plants were removed, root tips were isolated with adhering sand, and bacteria were removed from the root tips (32). The ratio of parental and mutant cells was determined by plating dilutions on solidified KB and on KB supplemented with kanamycin.
All results were statistically analyzed using the nonparametric Wilcoxon-Mann-Whitney test (33). To avoid log 0 cases, calculations were carried out using log (CFU + 1)/cm of root tip.
Construction of a single homologous recombinant and of a mutY construct for complementation.
A fragment of 1,187 bp of mutY was obtained by performing PCR on chromosomal DNA of WCS365 (using oMP515, 5′-GGCATGGACTATCGCGTCG-3′; and oMP516, 5′-CCAACTGGAAGTGGCTGAAGG-3′). After the 1,187-bp PCR fragment was cloned into pGEM-T Easy (Promega, Leiden, The Netherlands) (pMP5465) the construct was digested with SmaI and EcoRI made blunt by treatment with mung bean nuclease (New England Biolabs, Westburg, Leusden, The Netherlands). The obtained fragment (588 bp) (Fig. 1) was cloned into the Pseudomonas suicide vector pMP5285 (digested with KpnI treated with mung bean nuclease), resulting in pMP5564 (Fig. 1). This construct was electroporated into WCS365. After growth in KB and selection on KB with kanamycin a single homologous mutY recombinant, referred to as PCL1805, was isolated.
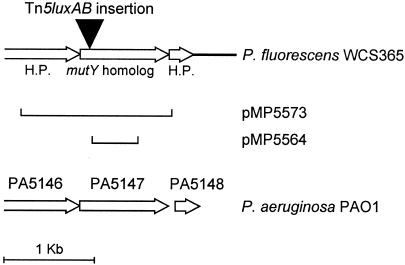
FIG. 1.
Map of DNA fragment (2,746 bp) isolated from the genome of P. fluorescens WCS365 with the help of Tn5luxAB. The fragment used for single homologous recombination and complementation and the genomic organization in P. aeruginosa PAO1 are shown. H.P., gene encoding a hypothetical protein.
For complementation the complete mutY gene (Fig. 1) was obtained from the genomic DNA of WCS365 by PCR (using oMP714 5′-GCAATTGTGCAAAGGCATCG-3′; and oMP715, 5′-AGCGTTCATCATGTTCAGGC-3′). After ligation of the 1,724-bp PCR fragment in pGEM-T Easy, resulting in pMP5572, the construct was digested with EcoRI and ligated into pME6031 (11). This construct, pMP5573, was electroporated into WCS365, PCL1286, PCL1805, and PCL1808. Cells harboring the plasmid were selected on KB supplemented with tetracycline, resulting in PCL1814, PCL1815, PCL1816, and PCL1817 (Table 1).
Mutator assay.
The mutator assay was performed as described previously (9, 20, 25, 28). Briefly, for a quantitative screening of the frequency of mutations remaining in wild-type P. fluorescens WCS365 and its mutant derivatives, we used only results for colonies which were rifampin sensitive at the beginning of the experiment. A rifampin-sensitive colony was resuspended in 1 ml of PBS, and the OD620 was set to 0.05. These cultures were diluted and plated on LB plates to determine the number of CFU at time point zero. The number of rifampin-resistant mutants present in this start mix was determined by plating on LB medium containing 100 μg of rifampin/ml. Cultures were grown overnight at 28°C; subsequently, dilutions were made and samples were plated on LB plates and LB plates containing rifampin. After 2 to 3 days of growth at 28°C, CFU were counted and mutation frequencies as number of mutations per cell per generation were determined.
CLSM.
Sterile germinated tomato seedlings were inoculated with WCS365 cells harboring plasmid pMP4662 (rfp, encoding DsRed) and PCL1286 harboring plasmid pMP4655 (egfp, encoding enhanced green fluorescent protein [GFP]) (2) and grown in the gnotobiotic system. After 7 days of plant growth, roots were washed in PBS to remove sand particles and transferred to a cover slip. Samples were examined using an inverted Leica confocal laser scanning microscope (CLSM) (DMIRBE-SP) equipped with an argon laser for eGFP visualization (excitation, 488 nm; emission, 501 to 540 nm) and a krypton laser (excitation, 568 nm; emission, 575 to 600 nm) for DsRed visualization.
Nucleotide sequence accession number.
The GenBank accession number for the mutY homolog sequence and surrounding hypothetical genes is AY429340.
RESULTS
Isolation of PCL1286, an enhanced root-tip-colonizing Tn5luxAB derivative of P. fluorescens WCS365.
A random Tn5luxAB mutant bank of P. fluorescens WCS365 (Table 1) was constructed and used to inoculate seedlings, which were subsequently allowed to grow. The bacteria, which had reached the 1-cm root tip, were collected, allowed to multiply, and used to inoculate a new seedling. In order to select for a good competitive root-tip-colonizing phenotype on both the dicotyledenous plant tomato and on the monocotyledenous plant grass, the mutants were subjected to four cycles of enrichment, alternating on tomato and grass. After every enrichment step only the bacteria able to reach the 1-cm root tip were used in the next step. Mutant PCL1285 was isolated and showed competitive root-tip-colonizing abilities equal to those of wild-type WCS365. Mutant PCL1286 was isolated as the best competitive colonizing mutant. When tested for its competitive colonization abilities against its parental strain, WCS365, it appeared to colonize the tomato root tip 100-fold better and grass root tips 1,000-fold better than the wild type (Table 2). Furthermore, PCL1286 analyzed in competition with another wild type, Pseudomonas chlororaphis strain PCL1391, on the tomato root system showed a more than 1,000-fold better root-tip-colonizing ability (Table 2).
TABLE 2.
Competitive root tip colonization ability of PCL1286 in a gnotobiotic sand systema
| Competing strains | Competitive root tip colonization [log10 (CFU + 1)/cm of root tip]b
|
|
|---|---|---|
| Wild type | PCL1286 | |
| WCS365 vs. PCL1286 (tomato) | 3.3 ± 0.9a | 5.1 ± 0.1b |
| WCS365 vs. PCL1286 (grass) | 2.8 ± 0.7a | 5.7 ± 0.5b |
| PCL1391 vs. PCL1286 (tomato) | 2.0 ± 0.8a | 5.4 ± 0.1b |
Mutant strains were inoculated on seedlings in a 1:1 ratio with the wild-type strain. Plant roots were analyzed after 7 days.
In every experiment 10 seedlings were inoculated and plants were individually processed after growth. When values in the same row are followed by a different letter, they are significantly different at P = 0.05 for nonparametric multiple comparisons by the Wilcoxon-Mann-Whitney test.
To analyze whether the superior colonizing ability of PCL1286 could be due to enhanced growth or motility, these traits were analyzed both for individual strains and in competition. In none of the above-mentioned traits did PCL1286 differ from the wild type, WCS365 (data not shown). In order to test whether PCL1286 grows faster on exudate, e.g., as a result of an increased rate of uptake of nutrients in a nutrient-poor environment, growth in competition in tomato root exudate (see Materials and Methods) as the carbon source was tested for wild-type WCS365 and its superior-colonizing mutant. After 30 generations, no significant difference in CFU was observed between wild-type WCS365 and mutant PCL1286.
In order to test whether the enhanced colonizing abilities of PCL1286 are due to colonization of other root parts, CLSM studies were performed. Strains WCS365 (containing pMP4662, expressing eGFP) and PCL1286 (containing pMP4655, expressing DsRed) were analyzed alone and in competition on tomato. The visualization results showed that PCL1286 colonizes the same root parts as WCS365, namely cell-cell junctions, when analyzed on the tomato root alone. CFU determination at the root tip showed no significant difference in cell numbers, with log10 values per cm of root tip of 5.1 ± 0.4 and 4.8 ± 0.8 CFU for WCS365 and PCL1286, respectively. Analyses of competitive colonization also showed no difference in colonization sites between the strains but the mutant was present in much higher numbers (Table 2). Mixed microcolonies of WCS365 and PCL1286 were formed (http://www.ibl.leidenuniv.nl/index.php3?m=149&c=116&garb=0.027470871862106116&session=).
Genetic characterization of mutant PCL1286.
Isolation of chromosomal DNA of PCL1286, followed by digestion with EcoRI and religation of the digested fragments, resulted in plasmid pMP5421 containing Tn5luxAB and flanking regions. By using primers based on the border sequences of Tn5luxAB, the chromosomal DNA adjacent to the transposon insertion was sequenced. Homology studies of the predicted protein sequence showed that Tn5luxAB is inserted in an open reading frame (ORF) with homology to mutY from Pseudomonas putida KT2440, which encodes an A · G-specific adenine glycosylase (24) (77% identity and 83% homology at the amino acid level) and to PA5147, a mutY homolog of P. aeruginosa PAO1 (73% identity and 80% homology at the amino acid level) (35) (Fig. 1). Tn5luxAB in PCL1286 appeared to be inserted 15 bp downstream of the start codon of the mutY gene. The mutY genes of P. aeruginosa PAO1 and P. fluorescens WCS365 are 1,068 and 1,077 bp in size, respectively. In WCS365 an ORF encoding a hypothetical protein is located upstream of mutY. This ORF is also present in P. aeruginosa PAO1 (Fig. 1). Downstream of mutY, an ORF encoding a small hypothetical protein was found in WCS365. This ORF is also present at the same location in P. aeruginosa, as PA5148 (Fig. 1). No clear promoter sequence could be identified upstream of the mutY homolog in WCS365. In P. aeruginosa there is no indication that mutY is part of an operon. For E. coli it is known that mutY is a monocistronic gene (23, 26). Results presented in Fig. 2, which will be discussed later in this report, show that also in WCS365 mutY is a single operating gene and that a promoter sequence must be present but is apparently difficult to identify.
FIG. 2.
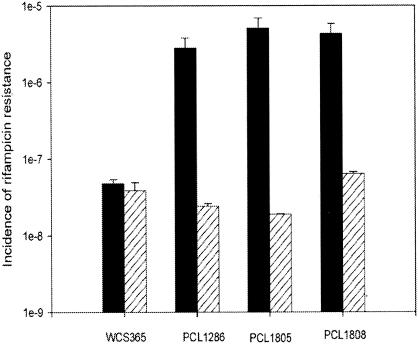
Incidence of rifampin resistance mutations in WCS365 and its mutY derivatives, PCL1286, PCL1805, and PCL1808 (black bars). Hatched bars represent the same strains containing pMP5573, harboring wild-type mutY. The rifampin concentration used was 100 μg/ml. The results are plotted as the incidence of rifampin resistance mutations per cell per generation.
Reconstruction of events in the history of PCL1286 by generation of a new mutY mutant and analysis of the subsequent effects of enrichment cycles on competitive root tip colonization.
For the construction of a new mutY mutant a PCR fragment of mutY was cloned into pGEM-T Easy, resulting in pMP5465. After digestion an internal fragment not containing a start or stop codon was cloned into the Pseudomonas suicide vector pMP5258, resulting in pMP5564 (Table 1). After electroporation to WCS365 and single homologous recombination, mutant strain PCL1805 was isolated by using selection for kanamycin resistance. To check the recombination, PCR on genomic DNA from PCL1805 was performed. By using one primer specific for pMP5285 and one primer specific for mutY a product was obtained demonstrating that the insertion of the plasmid into the genome of WCS365 was correct.
Analyses of the competitive root-tip-colonizing abilities of PCL1805 in quartz sand in competition with parental strain WCS365 prior to enrichment revealed an almost 1,000-fold-decreased ability for PCL1805 on tomato (Table 3). On grass root tips a phenotype impaired fivefold was observed (Table 3). Subsequently, three cycles of enhanced root tip colonization after tomato seedling inoculation were performed for PCL1805 and for wild-type WCS365 and its Kmr derivative PCL1285. PCL1805 obtained after enrichment on tomato was named PCL1808. Analyzing its root-tip-colonizing ability in competition with parental strain WCS365 showed that the colonizing abilities of PCL1808 are much better than those of PCL1805; PCL1808 colonizes six- and twofold better on tomato and grass, respectively (Table 3). In contrast, the colonizing abilities of WCS365 and its Kmr derivative PCL1285 were not improved by the enrichment procedure on tomato (Table 3).
TABLE 3.
Competitive tomato and grass root tip colonization abilities of PCL1805, PCL1808, PCL1285(3X), and WCS365(3X) in a gnotobiotic sand systema
| Competing strains | Competitive root tip colonization [log10 (CFU + 1)/cm of root tip]b
|
|||||
|---|---|---|---|---|---|---|
| Tomato
|
Grass
|
Tomato
|
||||
| Wild type | Mutant | Wild type | Mutant | Before enrichment | After enrichment | |
| WCS365 vs. PCL1805 | 4.8 ± 0.9a | 1.9 ± 1.7b | 5.3 ± 0.2a | 4.6 ± 0.6b | ||
| WCS365 vs. PCL1808 | 5.0 ± 0.2a | 5.8 ± 0.2b | 5.5 ± 0.3a | 5.8 ± 0.2b | ||
| WCS365 vs. PCL1285(3×)c | 5.0 ± 0.3a | 5.1 ± 0.3a | ||||
| PCL1285 vs. WCS365(3×) | 5.4 ± 0.2a | 5.5 ± 0.2a | ||||
Mutant strains were inoculated on seedlings in a 1:1 ratio with the wild-type strain. Plant roots were analyzed after 7 days.
In every experiment 10 seedlings were inoculated and plants were individually processed after growth. When values in the same row are followed by a different letter, they are significantly different at P = 0.05 for nonparametric multiple comparisons by the Wilcoxon-Mann-Whitney test.
3× refers to the fact that this strain has been subjected to three enrichment cycles on tomato.
Frequency of rifampin resistance in P. fluorescens WCS365 and its mutY derivatives.
Bacteria with a mutY mutation are unable to repair their A · G mismatches, which results in a higher number of mutations per cell (22). To test whether this was the case here, the frequency of rifampin resistance mutations was determined for wild-type WCS365 and its mutY derivatives PCL1286 (Tn5luxAB), PCL1805 (single homologous recombinant, prior to enrichment), and PCL1808 (PCL1805 after enrichment). The frequencies of rifampin-resistant cells per cell per generation was 4.8 × 10−8 (Fig. 2) for the wild-type strain and 2.8 × 10−6, 5.1 × 10−6, and 4.3 × 10−6 for mutants PCL1286, PCL1805, and PCL1808, respectively (Fig. 2). By using PCR, the complete mutY gene was isolated from the genome of WCS365 (Fig. 1) and cloned into the rhizosphere stable plasmid pME6031, resulting in pMP5573 (Table 1). This construct was electroporated into the four mentioned strains, resulting in PCL1814, PCL1815, PCL1816, and PCL1817 (Table 1). This resulted in a strong and significant decrease of rifampin resistance to wild-type levels in the three mutY mutants, whereas it had hardly any or no influence on the wild type (Fig. 2).
DISCUSSION
Kuiper et al. (14) recently described a method to isolate the best competitive root tip colonizer from a mixture of wild-type strains. We applied this method to a Tn5luxAB mutant bank of P. fluorescens strain WCS365 in order to test whether it is possible to isolate a mutant that is a good competitive colonizer on the roots of both tomato and grass plants. To our surprise, the best-colonizing mutant, strain PCL1286, was a much better competitive root tip colonizer than its wild type on both tomato and grass (Table 3). No differences were observed between mutant PCL1286 and wild-type WCS365 with respect to the major competitive colonization traits motility (data not shown) and growth in competition in KB or tomato root exudate. Similarly, no differences were observed in colonization strategies between the two strains as judged from CLSM studies with derivatives marked with autofluorescent proteins (http://www.ibl.leidenuniv.nl/index.php3?m=149&c=116&garb=0.027470871862106116&session=). We therefore have no physiological explanation for the superior colonizing character of mutant PCL1286.
Genetic analyses of PCL1286 showed that Tn5luxAB is inserted in a mutY homologue. This gene encodes a mismatch correction glycosylase, which is capable of removing the adenine paired with and oxidized guanine, 8-oxoG (37). It is known that a mutY mutant of E. coli, a so-called mutator, can adapt much more quickly to a certain environment than the wild type due to an increased mutation frequency. In this way the mutant can outcompete the wild type and compose up to 100% of the population (27). Downstream of mutY in PCL1286 a gene encoding a small hypothetical protein is located (Fig. 1) with no known function. Since cloned mutY can restore the number of mutant cells to the wild-type level (Fig. 2), MutY and not the small hypothetical protein is responsible for the mutant's colonization phenotype.
In order to investigate whether the enhanced competitive colonization ability of mutant PCL1286 is due to the mutY mutation, to the enrichment procedure, or to a combination of these two, we constructed a new mutY derivative of WCS365, designated PCL1805. It appeared that this new mutant reached the root tip of tomato and grass 700- and 5-fold less frequently than its wild type (Table 3), indicating that a mutY mutation alone dramatically decreases a strain's colonizing ability. Three enrichment cycles on tomato did not influence the competitive colonization behavior of WCS365 (Table 3), showing that the enrichment procedure alone did not yield enhanced colonizers. However, strain PCL1808, a derivative of the mutY mutant PCL1805 obtained from the tomato root after three enrichment cycles, appeared not only to be a better colonizer than its parental strain, PCL1805, but also was recovered from tomato and grass root tips six- and twofold more frequently than WCS365, respectively. It thus appeared that PCL1808, a derivative of PCL1805 isolated from the root tip after three enrichment cycles on tomato, was approximately 4,200- and 10-fold improved in competitive root tip colonization on tomato and grass, respectively, relative to the original mutant, PCL1805. These results indicate that the combination of mutY mutation and enrichment procedure results in the superior colonizing phenotype.
We hypothesized that the appearance of excellent competitive colonizers in a mutY background is due to the higher number of unrepaired mutations. To test this, we used the observation that the frequency of unrepaired mutations is higher in a mutY than in a wild-type population (25). The results indeed show that mutY mutant populations harbor a higher number of rifampin-resistant mutations than wild-type WCS365. The observation that complementation with a plasmid harboring the cloned mutY gene restores this frequency to wild-type levels shows that the mutY mutation is responsible for the high incidence of rifampin resistance mutations. This makes it likely that a mutY population is also rich in other mutations and that combinations of these mutations present in some mutant cells are responsible for the enhanced colonization ability.
It appeared that the higher number of mutations present in a mutY cell not only leads to poor colonizers (e.g., PCL1805) but that specific combinations of mutations are beneficial for colonization (e.g., in PCL1808). The power of mutator cells and the enrichment procedure is shown by the different colonization behavior of the two mutY mutants obtained after applying the enrichment procedure. Alternating selection on tomato and grass results in another quantitative competitive root tip colonization phenotype (PCL1286) (Table 2) different from that resulting from enrichment for tomato colonizers only (PCL1808) (Table 3). The results suggest that different combinations of mutations are required for superior colonization on grass plus tomato (PCL1286) than for colonization of tomato alone (PCL1808). The observation that by selecting PCL1808 on tomato, its colonization on grass is also improved suggests that one or more mutations are beneficial for colonization on both plants.
A mutY population contains cells with combinations of mutations which allow fast adaptation to a certain environment such as the rhizosphere. Similar adaptation behavior has been shown for mutY mutants under different conditions in cultures (21) and also in nature, e.g., in cystic fibrosis patients (28). Poor colonization is considered a limiting factor for many applications of beneficial bacteria in the rhizosphere (3, 30, 39). Our results open opportunities to develop derivatives of beneficial bacteria which are highly adapted to the rhizosphere of certain plant species or even plant cultivars, e.g., for biocontrol based on antibiosis (4) or phytoremediation (16).
Acknowledgments
We thank Pepijn T. Boerée for technical assistance.
REFERENCES
- 1.Altschul, S. E., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bloemberg, G. V., A. H. M. Wijfjes, G. E. M. Lamers, N. Stuurman, and B. J. J. Lugtenberg. 2000. Simultaneous imaging of Pseudomonas fluorescens WCS365 populations expressing three different autofluorescent proteins in the rhizosphere: new perspectives for studying microbial communities. Mol. Plant-Microbe Interact. 13:1170-1176. [DOI] [PubMed] [Google Scholar]
- 3.Bull, C. T., D. M. Weller, and L. S. Thomashow. 1991. Relationship between root colonization and suppression of Gaeumannomyces graminis var. triciti by Pseudomonas fluorescens strain 2-79. Phytopathology 81:954-959. [Google Scholar]
- 4.Chin-A-Woeng, T. F. C., G. V. Bloemberg, I. H. M. Mulders, L. C. Dekkers, and B. J. J. Lugtenberg. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 13:1340-1345. [DOI] [PubMed] [Google Scholar]
- 5.Dekkers, L. C., C. J. Bloemendaal, L. A. de Weger, C. A. Wijffelman, H. P. Spaink, and B. J. J. Lugtenberg. 1998. A two-component system plays an important role in the root-colonizing ability of Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 11:45-56. [DOI] [PubMed] [Google Scholar]
- 6.Dekkers, L. C., I. H. M. Mulders, C. C. Phoelich, T. F. C. Chin-A-Woeng, A. H. M. Wijfjes, and B. J. J. Lugtenberg. 2000. The sss colonization gene of the tomato-Fusarium oxysporum f. radicis-lycopersici biocontrol strain Pseudomonas fluorescens WCS365 can improve root colonization of other wild-type Pseudomonas spp. bacteria. Mol. Plant-Microbe Interact. 13:1177-1183. [DOI] [PubMed] [Google Scholar]
- 7.Dekkers, L. C., C. C. Phoelich, L. van der Fits, and B. J. J. Lugtenberg. 1998. A site-specific recombinase is required for competitive root colonization by Pseudomonas fluorescens WCS365. Proc. Natl. Acad. Sci. USA 95:7051-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helsinki. 1980. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorazi, R., J. J. Lingutla, and M. Z. Humayun. 2002. Expression of mutant alanine tRNAs increases spontaneous mutagenesis in Escherichia coli. Mol. Microbiol. 44:131-141. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13:232-237. [DOI] [PubMed] [Google Scholar]
- 12.Hoffland, E., G. R. Findenegg, and J. A. Nelemans. 1989. Solubilization of rock phosphate by rape. Plant Soil 113:161-165. [Google Scholar]
- 13.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 14.Kuiper, I., G. V. Bloemberg, and B. J. J. Lugtenberg. 2001. Selection of a plant-bacterium pair as a novel tool for rhizostimulation of polycyclic aromatic hydrocarbon-(PAH)-degrading bacteria. Mol. Plant-Microbe Interact. 14:1197-1205. [DOI] [PubMed] [Google Scholar]
- 15.Kuiper, I., G. V. Bloemberg, S. Noreen, J. E. Thomas-Oates, and B. J. J. Lugtenberg. 2001. Increased uptake of putrescine in the rhizosphere inhibits competitive root colonization by Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 14:1096-1104. [DOI] [PubMed] [Google Scholar]
- 16.Kuiper, I., L. Kravchenko, G. V. Bloemberg, and B. J. J. Lugtenberg. 2002. Pseudomonas putida strain PCL1444, selected for efficient root colonization and naphthalene degradation, effectively utilizes root exudate components. Mol. Plant-Microbe Interact. 15:734-741. [DOI] [PubMed] [Google Scholar]
- 17.Lugtenberg, B. J. J., L. A. de Weger, and J. W. Bennett. 1991. Microbial stimulation of plant growth and protection from disease. Curr. Opin. Biotechnol. 2:457-464. [Google Scholar]
- 18.Lugtenberg, B. J. J., L. C. Dekkers, and G. V. Bloemberg. 2001. Molecular Determinations of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 19.Lugtenberg, B. J. J., L. V. Kravchenko, and M. Simons. 1999. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1:439-446. [DOI] [PubMed] [Google Scholar]
- 20.Maliszewska-Tkaczyk, M., P. Jonczyk, M. Bialoskorska, R. M. Schaaper, and I. J. Fijalkowska. 2000. SOS mutator activity: unequal mutagenesis on leading and lagging strands. Proc. Natl. Acad. Sci. USA 97:12678-12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao, E. F., L. Lane, J. Lee, and J. H. Miller. 1997. Proliferation of mutators in a cell population. J. Bacteriol. 179:417-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1996. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol. 50:625-643. [DOI] [PubMed] [Google Scholar]
- 23.Neidhardt, F. C., R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.). 1996. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington D.C.
- 24.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, D. S. Martins, V. D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, L. P. Chris, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 25.Notley-McRobb, L., and T. Ferenci. 2000. Experimental analysis of molecular events during mutational periodic selections in bacterial evolution. Genetics 156:1493-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notley-McRobb, L., R. Pinto, S. Seeto, and T. Ferenci. 2002. Regulation of mutY and nature of mutator mutations in Escherichia coli populations under nutrient limitation. J. Bacteriol. 184:739-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notley-McRobb, L., S. Seeto, and T. Ferenci. 2002. Enrichment and elimination of mutY mutators in Escherichia coli populations. Genetics 162:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Schippers, B., A. W. Bakker, and P. A. H. M. Bakker. 1987. Interactions of deleterious and benificial rhizosphere microorganisms and the effect of cropping practices. Annu. Rev. Phytopathol. 25:339-358. [Google Scholar]
- 31.Simons, M., H. P. Permentier, L. A. de Weger, C. A. Wijffelman, and B. J. J. Lugtenberg. 1997. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 10:102-106. [Google Scholar]
- 32.Simons, M., A. J. van der Bij, J. Brand, L. A. de Weger, C. A. Wijffelman, and B. J. J. Lugtenberg. 1996. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol. Plant-Microbe Interact. 9:600-607. [DOI] [PubMed] [Google Scholar]
- 33.Sokal, R. R., and F. J. Rohlf. 1981. Biometry. W. H. Freeman & Co., San Francisco, Calif.
- 34.Stanisich, V. A., P. M. Bennett, and M. H. Richmond. 1977. Characterization of a translocation unit encoding resistance to mercuric ions that occurs on a nonconjugative plasmid in Pseudomonas aeruginosa. J. Bacteriol. 129:1227-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 36.Suslow, T. V., and M. N. Schroth. 1981. Role of deleterious rhizobacteria as minor pathogens in reducing crop growth. Phytopathology 72:111-115. [Google Scholar]
- 37.Urios, A., G. Herrera, V. Aleixandre, and M. Blanco. 1994. Processing of MucA protein is required for spontaneous and benzo[a]pyrene-induced reversion of the Escherichia coli trpA23 missense mutation by G.C-T.A transversions: effect of a deficiency in the MutY DNA glycosylase. Mutat. Res. 311:257-263. [DOI] [PubMed] [Google Scholar]
- 38.Voisard, C., C. T. Bull, C. Keel, J. Laville, M. Maurhofer, U. Schnider, G. Défago, and D. Haas. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHAO: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. N. Dowling, and N. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Verlagsgesellschaft, Weinheim, Germany.
- 39.Weller, D. M. 1988. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26:379-407. [Google Scholar]
- 40.Weller, D. M., and L. S. Thomashow. 1994. Current challenges in introducing beneficial microorganisms into the rhizosphere, p. 1-17. In F. O' Gara, D. N. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere organisms. VCH Verlagsgesellschaft, Weinheim, Germany.
- 41.Wolk, C. P., Y. Cai, and J.-M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]