Abstract
As all bacteria studied to date, the gastric pathogen Helicobacter pylori has an alternate lifestyle as a biofilm. H. pylori forms biofilms on glass surfaces at the air-liquid interface in stationary or shaking batch cultures. By light microscopy, we have observed attachment of individual, spiral H. pylori to glass surfaces, followed by division to form microcolonies, merging of individual microcolonies, and growth in the third dimension. Scanning electron micrographs showed H. pylori arranged in a matrix on the glass with channels for nutrient flow, typical of other bacterial biofilms. To understand the importance of biofilms to the H. pylori life cycle, we tested the effect of mucin on biofilm formation. Our results showed that 10% mucin greatly increased the number of planktonic H. pylori while not affecting biofilm bacteria, resulting in a decline in percent adherence to the glass. This suggests that in the mucus-rich stomach, H. pylori planktonic growth is favored over biofilm formation. We also investigated the effect of specific mutations in several genes, including the quorum-sensing gene, luxS, and the cagE type IV secretion gene. Both of these mutants were found to form biofilms approximately twofold more efficiently than the wild type in both assays. These results indicate the relative importance of these genes to the production of biofilms by H. pylori and the selective enhancement of planktonic growth in the presence of gastric mucin.
Helicobacter pylori, the primary etiological agent of gastritis, colonizes both the surface of gastric epithelial cells and the mucus gel layer. However, several recent studies have also identified H. pylori in water distribution systems (18, 29, 34, 38), surface wells or groundwater (16), rivers (11), and wastewater systems (17, 25, 29, 30) in several countries. In addition, H. pylori has been found existing on the inner surface of water storage earthenware in Gambia (3). While the mechanism of transmission of H. pylori is thought to be primarily by the fecal-oral route, the possibility that it may exist as a biofilm (most likely mixed species) on surfaces exposed to water may provide another route of infection.
Biofilms have been intensively studied as communities of bacteria able to survive in environments unfavorable to planktonic (free-living) bacteria (7, 24). In response to quorum-sensing signals, bacteria migrate and adhere to a surface, divide to form microcolonies, and expand laterally and vertically (8, 40, 48). Channels are formed in the biofilm to allow nutrients to reach bottom layers of bacteria. The mature biofilm is more resistant to killing by biocides and antibiotics, presumably due to the differentiation and survival of persister cells or slow growth rate (42). To further expand, sections of the biofilm shear off for colonization elsewhere (6).
Two studies have alluded to the ability of H. pylori to form a biofilm. A polysaccharide-containing biofilm has been observed at the air-liquid interface when H. pylori is grown in a glass fermenter (43). H. pylori is also capable of binding to a heterotrophic mixed species biofilm grown on stainless steel coupons (26). However, beyond determination of the sugars in the polymeric matrix, no further characterization of H. pylori biofilms has been done.
In the stomach, H. pylori exists primarily as swimming, planktonic bacteria, with a minority of bacteria adherent to the epithelium. Because mucin prevents H. pylori from binding to epithelial cells (12, 46) and prebound bacteria can be detached by washing with mucin (41), we were interested in the effect of mucin on H. pylori biofilm formation. We found that increasing concentrations of mucin significantly enhanced planktonic growth over biofilm formation.
H. pylori has a type IV secretion system (the cag pathogenicity island), which is known to be important for induction of interleukin-8 (IL-8) by gastric epithelial cells, as well as the secretion of bacterial proteins into host cells. We tested the effect of a specific mutation of this secretion system (cagE), which prevents assembly of the secretory apparatus, on biofilm formation by H. pylori. In addition, quorum-signaling systems are noted for their role in establishing communication among bacteria in a biofilm. H. pylori contains one such system, luxS, which has been shown to play a role in biofilm formation in Streptococcus mutans (31). We tested the effect of a mutation in this gene in H. pylori in two quantitative systems. Mutations in either cagE or luxS affected biofilm formation by H. pylori. Mutations in genes known to affect biofilm formation in other bacteria (response regulator, Clp protease, polyphosphokinase, and fibrillar hemagglutinin) did not affect biofilm formation.
MATERIALS AND METHODS
Growth and microscopy of H. pylori.
Table 1 lists the H. pylori strains used in this study. H. pylori strain SD14 is a self-aggregating cagA+ strain from a duodenal ulcer patient and was described previously (4, 5). The aggregates produced by strain SD14 grown in broth are visible to the naked eye. Strains SD3, SD4, and SD7 were similarly isolated from duodenal ulcer patients; however, these strains do not appreciably self-aggregate in liquid culture.
TABLE 1.
H. pylori strains
| Strain | Phenotype | Origin | Reference | Source |
|---|---|---|---|---|
| SD3 | Cag+ | Duodenal ulcer | 5 | San Diego |
| SD4 | Cag+ | Duodenal ulcer | 4 | San Diego |
| SD7 | Cag+ | Duodenal ulcer | This study | San Diego |
| SD14 | Cag+ | Duodenal ulcer | 4 | San Diego |
| SD14ppk | Ppk− | SD14 | This study | San Diego |
| SD14luxS | LuxS− | SD14 | This study | San Diego |
| SD14clpA | ClpA− | SD14 | This study | San Diego |
| SD14hpaA | HpaA− | SD14 | This study | San Diego |
| SD14HP1365 | Response regulator− | SD14 | This study | San Diego |
| SPM314a | Cag− | Mouse adapted | 27 | R. Rappuoli |
| SPM326a | Cag+ | Mouse adapted | 27 | R. Rappuoli |
| SS1 | Cag+b | Mouse adapted | 23 | A. Lee |
| 43504 | Cag+ | Human antrum | 28 | ATCCc |
| 26695 | Cag+ | Gastritis | 45 | ATCCc |
| N6 | Piglet adapted | 10 | K. Eaton | |
| N6flaA | FlaA− | N6 | 10 | K. Eaton |
| N6flaB | FlaB− | N6 | 10 | K. Eaton |
| N6flaAB | FlaA,B− | N6 | 10 | K. Eaton |
| UA948fucT | Lex−d | UA948 | 37 | D. Taylor |
Bacteria were grown from frozen stocks on Columbia agar plates containing 5% sheep or horse blood and 1% Fungizone (amphotericin B; Omega Scientific) in an atmosphere of 10% CO2, 5% O2, and 85% N2. Bacteria were harvested at 2 to 3 days by scraping into brain heart infusion (BHI) broth containing 0.1% β-cyclodextrin (CD; Sigma) or 5% heat-inactivated (56°C, 20 min) fetal calf serum (ΔFCS). Because strain SD14 forms aggregates in broth culture, bacterial clumps of this strain were fragmented by multiple passages through a 25-gauge needle, and single bacteria were enumerated using a Petroff-Hauser counter. Our investigators have found previously that passage through this size needle did not alter the ability of the organism to adhere to gastric epithelial cells or to induce IL-8 production in these cells (4, 5). Cultures were adjusted to 5 × 107 cells ml−1, and 1 to 2 ml was inoculated per well into six-well microtiter trays. Each well also contained a sterile 25-mm borosilicate coverslip that had been placed at an angle in the chamber in order to allow biofilm formation at the air-liquid interface, and the cultures were gently shaken on an orbital shaker for 2 to 6 days. Alternatively, stationary bacteria were grown in eight-chamber slides (300 μl/well; Lab-TekII; Nalge Nunc) with 12-mm borosilicate circular coverslips inserted perpendicular to the slide surface.
Coverslips containing H. pylori biofilms were fixed for 1 to 2 h in 5% formalin and stained with carbol-fuchsin for light microscopy. Unfixed coverslips were also stained with the Live/Dead BacLight kit (Molecular Probes) as described by the manufacturer. Samples on glass slides were prepared for scanning electron microscopy (SEM) by fixation in 5% formalin and successive dehydrations through a graded series of alcohol, followed by coating with gold-palladium. SEM micrographs were obtained from the Electron Microscopy Core Facility at San Diego State University on a Hitachi S2700 instrument.
Quantitation of biofilm formation.
Because H. pylori grows slowly, becomes nonculturable but viable with extended culture, does not adhere appreciably to plastic, and only forms a biofilm at the air-liquid interface, new methods were devised for quantitation of biofilm formation.
In the first method, individual respiratory activities of planktonic and biofilm bacteria grown in the same microtiter well were compared using a method adapted from quantification of biofilms of Candida dubliniensis (35). Wells in a 12-unit microtiter tray were filled approximately half full with autoclaved coarse glass frit (approximately 2- to 5-mm diameter; Bullseye Glass) to allow adherence of H. pylori at the air-liquid interface. H. pylori strains were grown for 2 days on agar, dispersed into BHI broth containing 0.1% CD (for the mucin experiments) or 5% ΔFCS (for mutant experiments), counted, and adjusted to 5 × 107 cells ml−1. For experiments with mucin, BHI broth was autoclaved together with type II (crude) mucin from porcine stomach (Sigma) at the percentages indicated below in Fig. 4, and 0.1% CD was added prior to incubation. One milliliter of bacteria was inoculated per well in 12-well microtiter trays containing the frit. Preliminary experiments indicated that maximal biofilm formation occurred at 5 days with CD and at 3 to 4 days with ΔFCS; therefore, cultures were incubated at 37°C for 3 to 5 days as appropriate for the additive, with or without gentle shaking (as indicated in the figure legends) in the same atmosphere as described above. For quantitation, planktonic bacteria were removed from the mixture and assayed separately from the remaining biofilm bacteria using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT; Sigma). In preliminary experiments, we found that incubation of H. pylori with MTT gave higher values and less background absorbance than other tetrazolium reagents in BHI broth (data not shown). We also found that coccoid H. pylori cells (obtained from 7- to 10-day-old agar plates) isolated on a 20-to-80% Percoll (Pharmacia) step gradient were approximately 40% as metabolically active as spiral forms isolated from the same gradient (data not shown). MTT is reduced during respiration to form the water-insoluble, purple formazan, which may be extracted from the bacteria with isopropanol for absorbance measurements. Nonadherent, planktonic bacteria were aspirated from the remaining biofilm bacteria colonizing the glass frit, and both populations were assayed separately, as follows. (i) For biofilms, the glass frit containing adherent H. pylori cells was washed twice with phosphate-buffered saline (PBS). A 1.5-ml aliquot of sterile MTT (0.5 mg ml−1) in BHI broth containing 0.1% CD was added to the glass frit, and the biofilms were incubated with gentle shaking in the same atmosphere as described above for 24 h. This amount of time was required since the growth rate of H. pylori is slow under these conditions (t1/2 > 12 h). The solution was aspirated from the frit containing the adherent, purple-stained bacteria, and 1.5 ml of isopropanol was added to the frit to extract the formazan. (ii) For planktonic bacteria, nonadherent H. pylori cells, including bacteria from the first wash of the glass frit with PBS, were pelleted (15,000 × g; 5 min), resuspended in 1 ml of MTT in broth as above, and incubated with shaking in a separate 12-well tray for 24 h. Planktonic bacteria were pelleted (15,000 × g; 5 min) and resuspended in 1 ml of isopropanol to extract the formazan for absorbance measurements.
FIG. 4.
Effect of the luxS mutation on biofilm formation by two different strains of H. pylori. (A) Three-day-old H. pylori SD3 and SD3luxS biofilms were quantitated using the respiratory assay, as described in the text. (B) SD14 and SD14luxS were grown in biofilms for 3 days and quantitated using the direct count assay, as described in Materials and Methods. Error bars represent means ± standard deviations.
The isopropanol suspensions from both biofilm and planktonic cultures were pelleted (15,000 × g; 5 min), supernatants were removed, and the absorbance of the supernatants at 550 nm was measured in an Ultrospec III spectrophotomer (Pharmacia LKB). Absorbance values were adjusted to reflect the differences in volumes of isopropanol-extracted planktonic and biofilm bacteria (1 ml versus 1.5 ml, respectively). Percent adherence was calculated as the (adjusted) A550 for the biofilm bacteria divided by the sum of the planktonic plus (adjusted) biofilm absorbances (35). All quantitative experiments were performed in quadruplicate, with at least two repetitions of the experiment.
The second method for quantitation involved direct counting of both planktonic and biofilm bacteria grown in the same well. For this, 1 ml of bacteria was inoculated at 5 × 107 ml−1 into six-well microtiter trays filled approximately half full with autoclaved glass frit. Bacteria were grown in BHI broth containing 0.1% CD, and the bacteria were grown with shaking for 5 days. Planktonic bacteria were aspirated, fixed with 2% formalin, and counted under polarized light using a Petroff-Hauser counter. Four squares were counted from replicates of four wells, and results were averaged. Biofilm bacteria remaining on the glass frit were washed twice with PBS, resuspended in PBS with 2% formalin, and sonicated for 3 min in a water bath. Bacteria released from the frit were aspirated and counted as above, and the percentage of adherent bacteria was calculated as for the respiratory assay.
Construction of the luxS mutant in H. pylori.
Two Escherichia coli clones were obtained from the American Type Culture Collection (GHPDN83 and GHPEF23) containing segments of the genome of H. pylori strain 26695 surrounding the luxS gene (gene fragment HP0104, nucleotides 112673 to 110931; fragment HP0106, nucleotides 114475 to 113336; both minus strand [45]). The chloramphenicol resistance (cat) gene of Campylobacter jejuni (kindly provided by H. Mobley, originally from Diane Taylor) was engineered to remove the transcriptional stop site and ligated in the same orientation between the two clones. Primers used for the PCR with the cat gene were 5′-CGGGATCCTCGGCGGTGTTCCTTTCCAAG-3′ (forward primer) and 5′-GCTCTAGACTCGAGCGCCCTTTAGTTCCTAAAGGGT-3′ (reverse primer). Insertion of the cat gene resulted in a deletion of 162 bp starting 15 bp downstream of the luxS start site. Because of the removal of the transcriptional stop site in the cat gene, this deletion was not expected to disrupt the metB or cpdB genes located upstream or downstream, respectively, of the luxS gene. In addition, a construct with the opposite orientation of the cat gene with respect to the luxS coding sequence was made. Plasmid DNA was prepared from E. coli DH5α, and 5 to 10 μg was used to transform H. pylori strains SD14, SD3, and SS1 on nonselective agar plates, as described previously (13). Following overnight incubation, the bacteria were scraped from the surface and inoculated onto plates containing 20 μg of chloramphenicol ml−1. Resistant colonies were subjected to Southern hybridization to confirm the disruption of the luxS gene. Also, oligomers were designed to a region upstream of the luxS gene and to the cat gene for PCR confirmation of the mutation in H. pylori.
In order to confirm the functional disruption of the luxS gene, the Vibrio harveyii luxN reporter strain BB170 (capable of sensing the LuxS autoinducer; kindly obtained from Bonnie Bassler) was used. The bioassay was performed as described previously (21), with the exception that the microtiter wells containing BB170 and H. pylori SD14 derivative supernatants were exposed to X-ray film and spots were quantified by densitometry. Essentially, SD14 and SD14luxS were grown in broth to a cell density of 6 × 108 ml−1, and an equal amount of cell-free culture supernatant was mixed with a stationary-phase culture of V. harveyii BB170 in a microtiter dish.
Construction of additional mutants of H. pylori.
The cagE mutant of H. pylori SD4 was previously described (15). The ppk, clpA, hpaA, and response regulator HP1365 mutations were constructed by PCR of the genes from H. pylori 26695, cloning into PCR-scriptSK (Stratagene), insertion of the modified cat gene, and transformation in H. pylori as described for the luxS mutant. Table 2 shows the gene identification numbers and the primers used to clone the genes, as well as the restriction sites used to insert the cat sequence for gene disruption. We were unable to obtain transformants of the clpB gene knockout in H. pylori SD14 with the cat gene in either orientation, suggesting that the mutation was lethal in this strain. Since this mutation was originally reported in strain N6, we attempted to transform H. pylori N6 with the inactivated clpB gene construct, made in E. coli and kindly provided by Elaine Allan (1). However, no mutants resulted from this transformation. In addition, mutation in the large clpP subunit gene was also lethal after a few passages on agar. We also tried to transform H. pylori SD14 with a mutated version of the folP gene, which is involved in anaerobic metabolism, but no mutants were obtained, suggesting that the gene is absolutely required for growth.
TABLE 2.
Primers and enzyme restriction sites used to construct H. pylori mutations by insertion of the modified cat gene
| Gene | H. pylori 26695 genea | Restriction site (bp)b | Sequence (5′-3′)c |
|---|---|---|---|
| clpA | HP0033 | StyI (32851) | F: GGACAAAGCCAAAGCGTTAG |
| R: CGGTATTCTTCAAAAGTGTCG | |||
| ppk | HP1010 | NsiI (1073285) | F: CCAAAAAATCCAAGCCCTAC |
| R: CGCTAAAAAAACTCACATCGG | |||
| hpaA | HP0410 | NheI (424060) | F: CATTACCTCTAAACCACCCAGGAACG |
| R: CAATCTGATGCTCTAACCGACTGAG | |||
| Response regulator | HP1365 | None; 141-bp deletiond | F: GCATCAACACAAACGAAGAGCC |
| R: ACCGCCTTACAGGATAACGCTAC | |||
| F: CGCTCAACAGAGAGCCTTTCATAA | |||
| R: TGAATTGCACCACATTGTGCC |
Gene numbers from the H. pylori 26695 sequence used to construct the mutations (45).
Restriction sites used to insert the cat gene. Numbers indicate the base pair number cited in the H. pylori 26695 sequence (45).
Primers used for PCR with genes from H. pylori strain 26695 for mutation and transformation into H. pylori. F, forward primer; R, reverse primer.
For mutation of response regulator HP1365, the two primer sets shown were used for PCR with 5′ end and 3′ end, respectively. The modified cat gene was inserted in the opposite orientation between the two PCR products.
RESULTS
Growth as a biofilm.
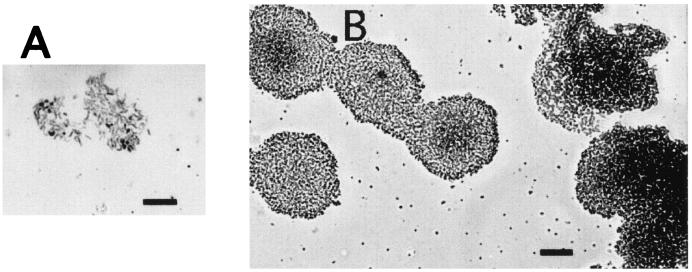
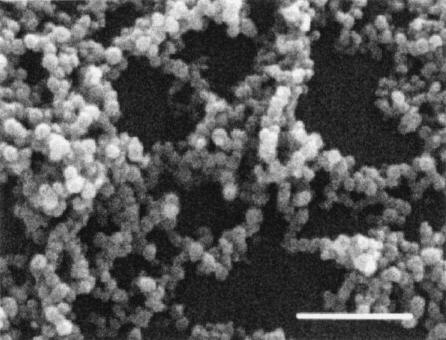
Figure 1 shows the progression of a typical biofilm of H. pylori SD14 at 3 to 4 days postinoculation. The bacteria adhered to the glass surface individually at the air-liquid interface and then divided to form circular microcolonies. The microcolonies grew to eventually merge together, forming a solid zone of bacteria at the air-liquid interface by 3 to 5 days, with or without gentle agitation. The three-dimensional architecture typical of other biofilms was observed for H. pylori in the SEM micrograph shown in Fig. 2. The bacteria (primarily coccoid in this micrograph) were stacked several layers thick, and numerous holes or pores could be seen extending to the base of the biofilm. As shown in the SEM micrograph, coccoid bacteria remained firmly attached. No obvious slime layer could be detected by SEM, but this could be due to the fixation of bacteria prior to processing for SEM (35). In our hands, H. pylori strains did not routinely form biofilms on plastic surfaces, such as the polystyrene used in microtiter trays.
FIG. 1.
Progression of biofilm formation by H. pylori SD14, as shown in light micrographs of fixed and carbol fuchsin-stained H. pylori SD14 cells at the air-liquid interface on borosilicate glass coverslips. (A) Single bacteria attached to glass at 3 days of stationary growth in broth. Attached bacteria began dividing in two dimensions to form a microcolony at 3 to 4 days. Bar, 10 μm. (B) Individual microcolonies merged into a dense biofilm at the air-liquid interface at >4 days of growth. Bar, 25 μm.
FIG. 2.
SEM micrograph of a 4-day-old biofilm of H. pylori SD14 grown as a stationary batch culture. Bar, 1 μm.
The cells of the actively growing biofilm (3 to 4 days old) were alive, as all of the bacteria fluoresced green (excluded propidium iodide) when exposed to the Live/Dead BacLight kit. At 5 days, more extensively colonized sections of the biofilm could be seen starting to peel off in sheets, and some dead bacteria could be seen (still adherent to the biofilm). Coccoid bacteria could be seen in the older biofilms, but most of these bacteria continued to fluoresce green with the BacLight stains.
Biofilm formation by multiple H. pylori strains.
All of the wild-type strains shown in Table 1 were able to form biofilms. This includes a gnotobiotic piglet-adapted strain (N6), mouse-adapted strains (SS1, SPM314, and SPM326), clinical isolates (SD3, SD4, SD7, and SD14), and laboratory strains (ATCC 26695 and 43504). The biofilms produced by all of these strains had similar three-dimensional architecture, observed by microscopy.
Effect of mucin on biofilm formation.
Mucin prevents H. pylori from binding to epithelial cells (12, 41, 46); therefore, we investigated its effect on adherence of H. pylori to a glass surface. As shown in Fig. 3, mucin significantly increased planktonic growth in a dose-dependent manner, while the numbers of biofilm bacteria were not appreciably altered, leading to a substantial decrease in the percent adherence. This was true using both the respiratory assay with strain N6 (Fig. 3A and B) and direct counts with strain SD4 (Fig. 3C and D).
FIG. 3.
Effect of mucin on biofilm production. Biofilm formation of H. pylori strains in the presence of 0 to 10% mucin was compared using the MTT respiratory assay (strain N6) (A and B) and direct counts (strain SD4) (C and D). Percent adherence (A and C) was calculated as described in the text for the two assays. In panels B and D, dark bars represent quantitation of biofilm bacteria and light bars indicate absolute values for planktonic bacteria. In both assays, note that while the percent adherence decreased with 1 and 10% mucin, the planktonic bacteria greatly outnumbered the biofilm bacteria (which remain relatively unchanged in number) in both assays. Error bars represent means ± standard deviations.
Effect of the luxS mutation on biofilm formation and coaggregation.
The luxS gene is the only known quorum-sensing gene present in the sequenced H. pylori genome. The luxS genes of H. pylori (21) strains SD14, SS1, and SD3 were disrupted by allelic exchange using the cat gene. The gene disruption was confirmed by both genetic analysis and bioassay. Southern blotting assays were performed using both the cat gene and the luxS gene on genomic DNA from strain SD14 and the isogenic luxS mutant. The blots showed an appropriate-sized band in the luxS mutant (data not shown), consistent with a cat cassette inserted in the luxS gene. The mutant was also confirmed by PCR using oligomers specific for the cat gene (3′) and a region upstream from the luxS gene (5′). The resulting fragment corresponded in size to the 5′ end of the luxS gene, suggesting that the cat cassette had indeed been inserted in the luxS gene. For confirmation by bioassay, we examined the ability of the H. pylori strains to stimulate the V. harveyii reporter strain BB170 to luminesce. The relative densities or light units for strain BB170 mixed with medium alone, H. pylori SD14, or H. pylori SD14luxS were 0.74, 1.17, and 0.73, respectively. Although the light output of the wild-type strain was less than those of rapid-growth bacteria such as salmonellae, the H. pylori luxS mutant induced light formation by V. harveyii less than wild type, confirming the mutation.
Although the luxS mutant had a similar growth rate to the wild type, we found that the mutant in both strains tested, with cat in either orientation, was capable of forming a mature biofilm microscopically similar to its isogenic parent. The bacteria were able to adhere, form microcolonies, and develop a three-dimensional architecture identical to the wild type. As planktonic bacteria, the mutant grew equally well in broth, self-aggregated (strain SD14luxS), and older cultures became coccoid, similarly to the wild type. In order to quantitate the amount of biofilm formed by the luxS mutant, it was compared to the wild type in the two assays described in Materials and Methods. As shown in Fig. 4A, the luxS mutant of strain SD3 was over threefold better at forming a biofilm than the wild-type parent strain in the respiratory assay. In addition, the same mutation in strain SD14 produced approximately twofold more biofilm than the wild type in the direct count assay, as indicated in Fig. 4B. We conclude that the lack of quorum sensing actually increased biofilm formation in two H. pylori clinical strains.
Effect of the cag type IV secretion system on biofilm formation.
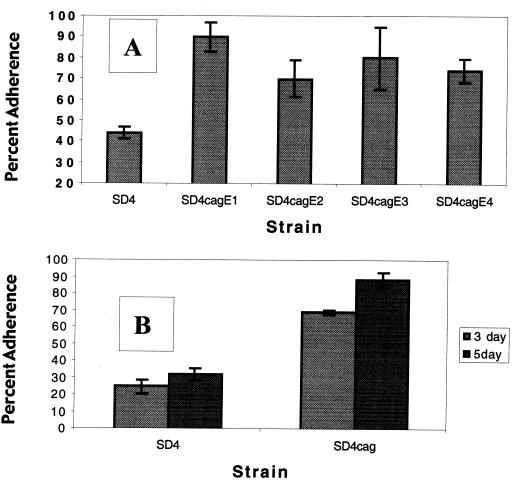
Inactivation of the cagE gene results in the inability to assemble the type IV secretion apparatus (reviewed in reference 39). We tested the effect of the cagE mutation on biofilm formation using the two assays described for the luxS mutant. Four separate isolates of the cagE mutant in strain SD4 were tested in the respiratory assay for their ability to form biofilms. As shown in Fig. 5A, all four isolates formed approximately twofold more biofilm than the wild-type strain. In addition, the cagE mutant consistently produced higher percentages of biofilm than wild-type SD4 in the direct count assay. This was confirmed in four independent experiments, with assays performed on 3-, 4-, and 5-day-old biofilms. In all cases, the cagE mutant produced between two- and fourfold more biofilm than the wild-type strain. A typical experiment is shown in Fig. 5B, which indicates that the cagE mutant of SD4 adhered to glass 2.8-fold more than the wild type in biofilms grown for either 3 or 5 days.
FIG. 5.
Effect of the cagE mutation on biofilm formation. (A) H. pylori SD4 and four different cagE isolates were grown as biofilms for 3 days and quantitated using the respiratory assay, as described in the text. (B) Strains SD4 and SD4cagE were grown as biofilms and assayed by direct counts, as described in Materials and Methods. Light bars represent results from biofilms grown for 3 days, while dark bars indicate values for 5-day-old biofilms. Error bars represent means of four experiments ± standard deviations.
Effect of other mutations on biofilm formation.
The clpA, ppk, hpaA, and response regulator HP1365 mutations were made in H. pylori SD14 and confirmed by PCR using primers outside of the inserted gene segments and within the cat cassette. All mutants were able to adhere to glass coverslips, and the resulting biofilms appeared similar to the wild type microscopically.
We tested the ability of a defined mutant deficient in terminal fucosylation of Lewis antigens (UA948 fucT) (37) to form a biofilm in our system. In H. pylori, the α(1,3/4)-fucosyltransferase is required for fucosylation of both type I (Lea) and type II (Lex) Lewis antigens; thus, mutation in the fucT gene eliminates all terminal fucosylated carbohydrates on the lipooligosaccharide (37). This mutant was capable of forming a dense biofilm at the air-liquid interface on glass coverslips, as observed microscopically.
Because none of these mutations showed significant differences in biofilm formation on glass coverslips, we did not submit them for further testing using the two quantitative assays.
DISCUSSION
The progression of biofilm formation by H. pylori mimics other biofilms described in the literature, beginning with individual bacteria adhering to the abiotic surface, expansion into colonies, and formation of a three-dimensional structure (Fig. 1 and 2). All strains tested were able to form biofilms on glass with overall similar biofilm structure (i.e., multiple layers of bacteria with channels for nutrient flow), including clinical isolates and laboratory strains, as well as mouse-adapted strains. H. pylori formed a biofilm only at the air-liquid interface, which is most likely indicative of its microaerobic, capnophilic character.
A number of global regulatory genes, controlling inducible genes such as those involved in virulence, cell-cell signaling, and responses to environmental signals, have been implicated in biofilm formation (reviewed in references 6 to 8, 19, 22, and 40). We investigated the importance of several genes involved in global regulation on biofilm formation by observing specific mutants in H. pylori. Because H. pylori has a paucity of potential global regulators (45), we made specific mutations in genes known to affect biofilm formation in other systems. Although the polyphosphokinase (Ppk) is required for biofilm formation in other gram-negative bacteria (32, 36), this does not appear to be the case for H. pylori. Likewise, although Clp protease mutations affect biofilm formation in Pseudomonas fluorescens (33), mutation in the clpA small subunit did not inhibit biofilms by H. pylori. However, it is possible that other Clp small subunits (ClpB or ClpX) affect this process. A number of studies have implicated two-component regulatory systems in controlling biofilm formation. The H. pylori 26695 genome has four homologous two-component sensor kinases and six genes encoding response regulators (45). Three of the response regulators are essential, and two are involved in flagellar and chemotaxis regulation (2). Therefore, we decided to inactivate the remaining response regulator, which exists at the beginning of an operon with its (presumably) cognate sensor (2). However, this mutant was able to form a biofilm essentially equivalent to that of the wild type, suggesting that the gene is not critical to biofilm formation. Again, this does not exclude the importance of other two-component systems in biofilms.
The quantitation of biofilm formation by H. pylori is complicated by a number of factors. For example, H. pylori only forms a biofilm at the air-liquid interface and most strains do not form biofilms on plastic, similarly to other bacteria reported. In addition, H. pylori requires extended incubation (≥3 days of growth) in a microaerobic atmosphere for biofilm formation and readily converts to the viable but nonculturable (VBNC) form with extended incubation. Therefore, care had to be taken to choose quantitative assays that addressed these issues. The respiratory assay was chosen, since the ability to reduce tetrazolium salts remains high in VBNC H. pylori, at least under certain growth conditions (14). We also found that coccoid H. pylori cells from 7- to 10-day-old cultures respire at 40% of the rate of spiral forms (data not shown), a level far greater than that reported for protein synthesis or ATP content in H. pylori VBNC forms. In addition, quantitation of both planktonic and biofilm bacteria using tetrazolium salt reduction has been previously described for other difficult-to-culture bacteria (9, 35). However, because biofilm bacteria may not respire at the same rate as planktonic bacteria, we developed a second assay using direct counts of both planktonic and biofilm H. pylori. Direct counts are the preferred method for quantitation over colony counts, since biofilm formation required 3 to 5 days of incubation, which can produce many VBNC bacteria.
Using both of these assays, we found that mucin greatly accelerated planktonic growth over expansion of biofilm H. pylori. In the stomach, mucus is thought to form a protective layer to infection, since it is continually swept out by muscular contractions. In fact, H. pylori is chemoattracted to and binds human gastric mucin, the major component of mucus (reviewed in reference 44). Mucin is known to increase the growth of H. pylori in broth cultures (20). Our data complement these findings for planktonic cultures of H. pylori, showing that planktonic growth is enhanced with mucin addition. However, we also found that while planktonic growth was enhanced, mucin did not inhibit biofilm formation or affect the number of adherent bacteria, as shown in Fig. 3. Therefore, for the first time, our studies show the effect of mucin on biofilms produced by bacteria normally found in the gastrointestinal tract. These results were corroborated by the use of two quantitative assays and two different strains (N6, a mouse-adapted strain, and SD4, a clinical isolate). As a growth promoter, mucin may be used as a carbon source or may serve to modify the viscosity of the solution, but the growth-enhancing benefits of this supplement only pertained to planktonic bacteria. Therefore, we suggest that as H. pylori encounters mucin in the stomach, the selective growth of planktonic bacteria greatly accelerates, which explains the large number of free-swimming bacteria observed in the mucus layer.
Recently, the luxS gene in S. mutans was shown to be involved in biofilm formation (49); therefore, we tested the effect of LuxS on biofilms produced by H. pylori. Interestingly, a mutation in luxS actually increased monospecies biofilm formation by H. pylori in multiple experimental trials and with two separate strains. The mechanism for this enhancement of biofilm formation is likely to be complex; nevertheless, this represents the first phenotype associated with the luxS mutation in H. pylori.
Equally surprising was the effect of the cagE mutation on H. pylori biofilm formation. The Cag type IV secretion system forms a large membrane pore and contains many protein components. While a few components have been found to be secreted by this system, it is likely that other products are secreted through this apparatus. We found that an isogenic mutant lacking a functional cagE gene reproducibly adhered more strongly to glass surfaces than the wild type. Since this occurred with multiple isolates from the allelic replacement transformation, it is unlikely that some untoward effect from the cagE gene disruption (for example, mutation at another site) was responsible for these results. The reason for these findings remains obscure. The inability to secrete certain components could pertain to these findings, or perturbation of the membrane surface could result in nonspecific effects, such as alteration of membrane pH or hydrophobicity. It is possible that, since the cag pathogenicity island may not be required, or even beneficial, for survival outside the host, the additional energy cost of transcribing this operon hinders efficient biofilm formation. The ability to form a biofilm may be a remnant of a more primitive (premammalian) lifestyle for H. pylori. In addition, our data suggest that there may be an interplay between the luxS and cagE genes, although investigation of these interactions remains beyond the scope of this paper. Whatever the explanation for the effect of the cagE mutation on biofilm formation, our findings suggest a novel phenotype for the Cag type IV secretion system.
As mentioned in the introduction, H. pylori has been identified in multiple water systems throughout the world. We believe that the adherence of H. pylori to glass (43), stainless steel (26), cast iron (34), and clay represents a novel habitat and may allow growth in the environment at the air-liquid interface. For example, H. pylori may form biofilms on rocks containing silicates, iron, or other clay minerals at the air-liquid interface in surface wells. Our results indicate the relative importance of a number of genes to the production of biofilms by H. pylori and indicate the selective enhancement of planktonic growth in the presence of gastric mucin. We suggest that H. pylori exists primarily as a biofilm in the environment and, upon encountering gastric mucus, rapidly proliferates as planktonic bacteria in the stomach.
Acknowledgments
This work was supported by Public Health Service grant DK 53649 from the National Institute of Diabetes and Digestive and Kidney Diseases.
We thank Rino Rappuoli, Diane Taylor, Kathryn Eaton, Elaine Allen, Harry Mobley, and Bonnie Bassler for kindly providing strains and constructs.
REFERENCES
- 1.Allan, E., P. Mullany, and S. Tabaqchali. 1998. Construction and characterization of a Helicobacter pylori clpB mutant and role of the gene in the stress response. J. Bacteriol. 180:426-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunn, J. E. G., W. G. MacKay, J. E. Thomas, D. C. Reid, and L. T. Weaver. 2002. Detection of Helicobacter pylori DNA in drinking water biofilms: implications for transmission in early life. Lett. Appl. Microbiol. 34:450-454. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. P., D. Cirillo, M. F. Kagnoff, D. G. Guiney, and L. Eckmann. 1997. Coccoid and spiral Helicobacter pylori differ in their abilities to adhere to gastric epithelial cells and induce interleukin-8 secretion. Infect. Immun. 65:843-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, S. P., V. F. Kharitonov, and D. G. Guiney. 1999. Effect of nitric oxide on Helicobacter pylori morphology. J. Infect. Dis. 180:1713-1717. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 9.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 10.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enroth, H., and L. Engstrand. 1995. Immunomagnetic separation and PCR for detection of Helicobacter pylori in water and stool specimens. J. Clin. Microbiol. 33:2162-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falk, P., K. A. Roth, T. Borén, T. U. Westblom, J. I. Gordon, and S. Normark. 1993. An in vitro adherence assay reveals that Helicobacter pylori exhibits cell lineage-specific tropism in the human gastric epithelium. Proc. Natl. Acad. Sci. USA 90:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge, Z., and D. E. Taylor. 1997. H. pylori DNA transformation by natural competence and electroporation, p. 145-152. In C. L. Clayton and H. L. T. Mobley (ed.), Helicobacter pylori protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 14.Gribbon, L. T., and M. R. Barer. 1995. Oxidative metabolism in nonculturable Helicobacter pylori and Vibrio vulnificus cells studied by substrate-enhanced tetrazolium reduction and digital image processing. Appl. Environ. Microbiol. 61:3379-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guiney, D. G., P. Hasegawa, and S. P. Cole. 2003. Helicobacter pylori preferentially induces interleukin 12 (IL-12) rather than IL-6 or IL-10 in human dendritic cells. Infect. Immun. 71:4163-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegarty, J. P., M. T. Dowd, and K. H. Baker. 1999. Occurrence of Helicobacter pylori in surface water in the United States. J. Appl. Microbiol. 87:697-701. [DOI] [PubMed] [Google Scholar]
- 17.Hulten, K., H. Enroth, T. Nystrom, and L. Engstrand. 1998. Presence of Helicobacter species DNA in Swedish well water. J. Appl. Microbiol. 85:282-286. [DOI] [PubMed] [Google Scholar]
- 18.Hulten, K., S. W. Han, H. Enroth, P. D. Klein, A. R. Opekun, R. H. Gilman, D. G. Evans, L. Engstrand, D. Graham, and F. A. K. El-Zaatari. 1996. Helicobacter pylori in the drinking water in Peru. Gastroenterology 110:1031-1035. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, X., and M. P. Doyle. 2000. Growth supplements for Helicobacter pylori. J. Clin. Microbiol. 38:1984-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyce, E. A., B. L. Bassler, and A. Wright. 2000. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J. Bacteriol. 182:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landini, P., and A. J. B. Zehnder. 2002. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J. Bacteriol. 184:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, A., J. O'Rourke, M. Corazon de Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, Y., T. E. Redlinger, R. Avitia, A. Galindo, and K. Goodman. 2002. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl. Environ. Microbiol. 68:1436-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackay, W. G., L. T. Gribbon, M. R. Barer, and D. C. Reid. 1998. Biofilms in drinking water systems—a possible reservoir for Helicobacter pylori. Water Sci. Technol. 38:181-185. [DOI] [PubMed] [Google Scholar]
- 27.Marchetti, M., B. Arico, D. Burroni, N. Figura, R. Rappuoli, and P. Ghiara. 1995. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science 267:1655-1658. [DOI] [PubMed] [Google Scholar]
- 28.Marshall, B. J., and C. S. Goodwin. 1987. Revised nomenclature of Campylobacter pyloridis. Int. J. Syst. Bacteriol. 37:68. [Google Scholar]
- 29.Mazari-Hiriart, M., Y. Lopez-Vidal, and J. J. Calva. 2001. Helicobacter pylori in water systems for human use in Mexico City. Water Sci. Technol. 43:93-98. [PubMed] [Google Scholar]
- 30.Mazari-Hiriart, M., Y. Lopez-Vidal, G. Castillo-Rojas, S. Ponce de Leon, and A. Cravioto. 2001. Helicobacter pylori and other enteric bacteria in freshwater environments in Mexico City. Arch. Med. Res. 32:458-467. [DOI] [PubMed] [Google Scholar]
- 31.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, and W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa, N., C.-M. Tzeng, C. D. Fraley, and A. Kornberg. 2000. Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J. Bacteriol. 182:6687-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 34.Park, S. R., W. G. Mackay, and D. C. Reid. 2001. Helicobacter sp. recovered from drinking water biofilm sampled from a water distribution system. Water Res. 35:1624-1626. [DOI] [PubMed] [Google Scholar]
- 35.Ramage, G., K. Vande Walle, B. L. Wickes, and J. L. López-Ribot. 2001. Biofilm formation by Candida dubliniensis. J. Clin. Microbiol. 39:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasko, D. A., G. Wang, M. M. Palcic, and D. E. Taylor. 2000. Cloning and characterization of the α(1,3/4) fucosyltransferase of Helicobacter pylori. J. Biol. Chem. 275:4988-4994. [DOI] [PubMed] [Google Scholar]
- 38.Sasaki, K., Y. Tajiri, M. Sata, Y. Fujii, F. Matsubara, M. Zhao, S. Shimuzu, A. Toyonaga, and K. Tanikawa. 1999. Helicobacter pylori in the natural environment. Scand. J. Infect. Dis. 31:275-279. [DOI] [PubMed] [Google Scholar]
- 39.Selbach, M., S. Moese, T. F. Meyer, and S. Backert. 2002. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependcent and VirD4-CagA-independent mechanisms. Infect. Immun. 70:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro, J. A. 1998. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52:81-104. [DOI] [PubMed] [Google Scholar]
- 41.Simon, P. M., P. L. Goode, A. Mobasseri, and D. Zopf. 1997. Inhibition of Helicobacter pylori binding to gasterointestinal epithelial cells by sialic acid-containing oligosaccharides. Infect. Immun. 65:750-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stark, R. M., G. J. Gerwig, R. S. Pitman, L. F. Potts, N. A. Williams, J. Greenman, I. P. Weinzweig, T. R. Hirst, and M. R. Millar. 1999. Biofilm formation by Helicobacter pylori. Lett. Appl. Microbiol. 28:121-126. [DOI] [PubMed] [Google Scholar]
- 44.Testerman, T. L., D. J. McGee, and H. L. T. Mobley. 2001. Adherence and colonization, p. 381-417. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 45.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchem, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 46.Tzouvelekis, L. S., A. F. Mentis, A. M. Makris, C. Spiliadis, C. Blackwell, and D. M. Weir. 1991. In vitro binding of Helicobacter pylori to human gastric mucin. Infect. Immun. 59:4252-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Doorn, N. E. M., F. Namavar, M. Sparrius, J. Stoof, E. P. Van Rees, L.-J. Van Doorn, and C. M. J. E. Vandenbroucke-Grauls. 1999. Helicobacter pylori-associated gastritis in mice is host and strain specific. Infect. Immun. 67:3040-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]