Abstract
To gain insight into the cell envelope of Escherichia coli grown under aerobic and anaerobic conditions, lipoproteins were examined by using functional genomics. The mRNA expression levels of each of these genes under three growth conditions—aerobic, anaerobic, and anaerobic with nitrate—were examined by using both Affymetrix GeneChip E. coli antisense genome arrays and real-time PCR (RT-PCR). Many genes showed significant changes in expression level. The RT-PCR results were in very good agreement with the microarray data. The results of this study represent the first insights into the possible roles of unknown lipoprotein genes and broaden our understanding of the composition of the cell envelope under different environmental conditions. Additionally, these data serve as a test set for the refinement of high-throughput bioinformatic and global gene expression methods.
Bacterial lipoproteins comprise a unique set of proteins modified at their amino-terminal cysteines by the addition of N-acyl and S-diacyl glyceryl groups (30). In Escherichia coli, this lipid serves to anchor these proteins to the inner or outer membrane so that they can function at the lipid aqueous interface. These proteins can be identified by the presence of a leader with a common consensus sequence (5). The leader is typically between 15 and 40 amino acid residues in length and has at least one arginine or lysine in the first seven residues. The leader is cleaved by signal peptidase II on the amino terminal side of the cysteine residue, which is then enzymatically modified (30).
The E. coli genome has previously been searched for potential lipoproteins. Various algorithms have been used for genome sequence analysis to identify potential lipoproteins, and these lipoproteins have been tabulated in databases on the World Wide Web (http://www.mrc-lmb.cam.ac.uk/genomes/dolop/, http://www.expasy.org/prosite, and http://www.projectcybercell.com); from these databases, we compiled a list of 96 lipoproteins. Fifty-six of these genes (58%) have completely unknown functions, a much higher fraction than that for the E. coli genome, in which approximately 25 to 30% of the genes have no known function. Thus, the examination of the expression of the lipoprotein genes under different growth conditions would be a beginning to understanding the function and importance of many of the unknown genes.
Other putative lipoproteins exist in E. coli but were not part of the gene expression study. First, the murein transglycosylase MltE (Blattner no. b1163) is not in any current lipoprotein database but has been experimentally shown to be a lipoprotein (17). Second, yifL (Blattner no. b3808.1) was originally not annotated in the E. coli genome sequencing project, but YifL now appears in the Prosite database (http://www.expasy.org/prosite) as a putative lipoprotein. Also, very small lipoproteins such as the entericidins (EcnA and EcnB) (3) were omitted from this study because they are below the Affymetrix cutoff for open reading frame (ORF) inclusion (150 bp).
In the present study, we used this set of protein genes to begin analyzing the global changes in gene expression during aerobic and anaerobic growth with a view to understanding the changes in the composition of the cell envelope. The expression of lipoprotein mRNAs in E. coli MG1655 incubated in glucose defined media (21) either aerobically with shaking in an Erlenmeyer flask or anaerobically in a sealed screw-cap tube, with 40 mM KNO3 being added to one set of anaerobic cultures as an alternative electron acceptor, was monitored. RNA was then isolated from the cells with a MasterPure RNA purification kit (Epicentre Technologies, Madison, Wis.), and cDNA synthesis and labeling was done as described in the Affymetrix GeneChip E. coli Antisense Genome Array Technical Manual (1). Affymetrix GeneChip antisense E. coli genome arrays were used to analyze the complete E. coli transcriptome. Each microarray contained 295,000 probes. Each identified ORF was covered by 15 probe pairs consisting of a perfect match and a 1-nucleotide mismatch pair. If the perfect match probe showed an intensity that was 200 U higher than that of the mismatch probe, the probe pair was considered to be present. An ORF was considered to be present with 95% confidence if neighboring probe pairs within an ORF were present.
Using this cutoff, we were able to group the lipoproteins into four classes, as listed in Table 1. Twenty-one lipoprotein genes were not expressed (not present in the array analysis) under any of the selected conditions. Ten were present under one growth condition, 5 were present under two conditions, and 60 were present under all three conditions. Sixty-four of the lipoprotein genes were expressed at detectable levels during aerobic growth, the standard experimental growth condition for E. coli. Not surprisingly, lpp, the gene for the major structural outer membrane murein lipoprotein (25), has the highest expression level of all the genes. Other well-known lipoprotein genes highly expressed under aerobic growth conditions include pal, the gene for peptigoglycan-associated lipoprotein (19), and cyoA, which encodes a subunit of the cytochrome O terminal oxidase, the major terminal oxidase of the aerobic respiratory chain (7).
TABLE 1.
E. coli lipoprotein genes and their expression in microarray and RT-PCR analysesa
| Gene | Blattner no. | Analysis result for growth condition
|
|||||
|---|---|---|---|---|---|---|---|
| Aerobic
|
Anaerobic
|
Anaerobic + nitrate
|
|||||
| Microarrayb | RT-PCR | Microarrayb | RT-PCR | Microarrayb | RT-PCR | ||
| yaeF | b0193 | 2.85 A | 822.74 | 3.8 A | 904.07 | 5.2 M | 709.66 |
| yafL | b0227 | 13.9 A | 149.00 | 16.15 A | 129.12 | 8.95 A | 126.31 |
| yaiW | b0378 | 39.75 A | 1,094.73 | 39.75 A | 1,218.82 | 46.05 A | 1,153.40 |
| ybfP | b0689 | 4.35 A | 112.50 | 17.9 A | 207.54 | 11.75 A | 304.80 |
| ymcA | b0984 | 2.25 A | 370.83 | 6.05 A | 466.07 | 16.35 M | 972.86 |
| ycdR | b1023 | 0.3 A | 38.25 | 1.2 A | 77.86 | 0.7 A | 31.79 |
| ycjN | b1310 | 12.2 A | 23.84 | 13.1 A | 46.44 | 11.6 A | 47.93 |
| wza | b2062 | 8.65 A | 85.28 | 4.35 A | 59.48 | 4.55 A | 75.04 |
| yehR | b2123 | 1.85 A | 250.38 | 4.65 A | 325.93 | 4.75 A | 414.85 |
| yohG | b2138 | 8.85 A | 82.18 | 19 A | 128.07 | 13.7 A | 133.48 |
| yfbK | b2270 | 15.35 A | 19.51 | 11.6 A | 29.78 | 5.1 A | 39.77 |
| ypdI | b2376 | 0.45 A | 108.74 | 0.35 A | 238.47 | 0.55 A | 364.14 |
| yfgH | b2505 | 5.5 A | 256.39 | 4.1 A | 255.68 | 8.45 A | 223.25 |
| yfjS | b2636 | 3.9 A | 165.50 | 1.8 A | 112.20 | 0.9 A | 119.93 |
| yghG | b2971 | 1.65 A | 123.61 | 1.15 A | 82.76 | 1.1 A | 162.09 |
| yghH | b3014 | 12.6 A | 907.25 | 11.95 A | 1,012.54 | 9.05 A | 753.22 |
| acrE | b3265 | 0.7 A | 62.24 | 0.85 A | 58.61 | 1.35 A | 59.62 |
| yiaD | b3552 | 47.05 M | 952.14 | 24.5 A | 603.81 | 31.35 A | 680.38 |
| yiiG | b3896 | 3.8 A | 72.07 | 3.35 A | 107.60 | 4.8 A | 118.86 |
| yjbH | b4029 | 14.3 A | 81.04 | 14.2 A | 147.49 | 13.35 A | 212.67 |
| yjcP | b4080 | 3.25 A | 44.44 | 4.75 A | 81.48 | 3.85 A | 147.26 |
| yaaY | b0024 | 5.55 A | 416.62 | 10.5 M | 998.73 | 12.9 P | 1,417.36 |
| yajI | b0412 | 20.25 A | 300.04 | 28.65 P | 417.83 | 23.7 A | 357.06 |
| ybfN | b0682 | 15.6 M | 357.89 | 18.55 P | 576.64 | 14.95 M | 465.00 |
| yccZ | b0983 | 25.95 A | 341.56 | 23.15 M | 268.29 | 61.95 P | 672.07 |
| ymcC | b0986 | 15.1 M | 192.77 | 16 M | 267.76 | 47.05 P | 2,090.53 |
| csgG | b1037 | 2.8 A | 408.03 | 18.35 P | 1,182.28 | 12.6 A | 581.54 |
| yfiL | b2602 | 10.35 A | 495.61 | 11.3 A | 570.68 | 11.05 P | 368.30 |
| yghJ | b2974 | 26.6 P | 443.18 | 11.6 A | 218.09 | 9.9 A | 187.63 |
| yhiU | b3513 | 19.4 A | 80.34 | 109.6 P | 2,151.94 | 31.2 A | 295.74 |
| yjbF | b4027 | 27.8 A | 58.37 | 29.9 A | 54.12 | 25 P | 45.37 |
| yafT | b0217 | 15.35 P | 344.12 | 13.7 M | 207.16 | 18.6 P | 258.22 |
| ylcB | b0572 | 83.3 P | 2,207.72 | 11.75 P | 128.07 | 10.25 A | 64.35 |
| ybjP | b0865 | 23.3 P | 1,027.23 | 17.7 A | 986.19 | 58.35 P | 1,965.78 |
| yfhM | b2520 | 22 A | 490.08 | 65.3 P | 1,712.68 | 58.95 P | 795.13 |
| slp | b3506 | 5.6 A | 228.51 | 170.3 P | 10,280.10 | 41.25 P | 1,755.04 |
| cutF | b0192 | 49.65 P | 3,464.82 | 50 P | 3,315.12 | 68.95 P | 3,713.77 |
| yaeC | b0197 | 569.75 P | 5,357.99 | 673.1 P | 9,794.30 | 604.75 P | 9,094.88 |
| dniR | b0211 | 211 P | 14,770.97 | 332.75 P | 26,075.12 | 288.65 P | 18,758.77 |
| cyoA | b0432 | 1,508.05 P | 97,254.40 | 933.95 P | 64,118.31 | 975.1 P | 61,809.18 |
| yajG | b0434 | 467.95 P | 18,182.95 | 322.2 P | 11,028.77 | 286.75 P | 8,052.42 |
| ybaY | b0453 | 43.3 P | 10,698.22 | 52.85 P | 19,343.93 | 137.5 P | 8,323.34 |
| acrA | b0463 | 266 P | 21,274.95 | 262.05 P | 20,837.08 | 256.6 P | 10,104.47 |
| ybbC | b0498 | 12.75 P | 1,414.58 | 23 P | 2,338.44 | 36.15 P | 2,893.17 |
| fepG | b0589 | 68.8 P | 5,840.72 | 57.9 P | 4,170.60 | 70.8 P | 7,139.73 |
| rlpA | b0633 | 172.2 P | 8,577.58 | 173.4 P | 6,234.43 | 168.75 P | 3,118.40 |
| rlpB | b0641 | 336.65 P | 16,256.17 | 344.35 P | 18,380.11 | 280.8 P | 13,556.01 |
| ybgE | b0735 | 80.55 P | 4,025.33 | 625.2 P | 37,117.42 | 622.35 P | 26,673.41 |
| pal | b0741 | 1,088.75 P | 85,844.86 | 858.3 P | 72,334.53 | 737.95 P | 48,684.70 |
| ybhC | b0772 | 184.35 P | 4,837.29 | 152.35 P | 4,447.57 | 133.55 P | 3,426.66 |
| yliB | b0830 | 94.75 P | 4,305.95 | 239 P | 16,813.26 | 195.8 P | 8,175.31 |
| yceK | b1050 | 27 P | 2,825.53 | 32.9 P | 4,227.41 | 35.8 P | 5,551.83 |
| yceB | b1063 | 69.45 P | 585.53 | 101.15 P | 1,133.21 | 106.3 P | 481.83 |
| flgH | b1079 | 431.6 P | 22,619.34 | 274.5 P | 12,798.09 | 115.15 P | 5,517.09 |
| ycfL | b1104 | 90.75 P | 8,457.11 | 121.55 P | 11,679.68 | 160.85 P | 12,752.51 |
| ycfM | b1105 | 113.6 P | 3,414.83 | 185.75 P | 6,983.48 | 225.45 P | 9,351.10 |
| lolB | b1209 | 151.55 P | 7,612.79 | 205.95 P | 11,120.93 | 285.6 P | 13,613.65 |
| osmB | b1283 | 49.1 P | 1,493.47 | 132.65 P | 5,617.43 | 98.75 P | 3,191.80 |
| ynbE | b1382 | 16.25 P | 1,639.73 | 19.4 P | 1,998.16 | 20.3 P | 1,368.70 |
| ydcL | b1431 | 38.25 P | 1,839.59 | 31.85 P | 1,342.37 | 63.35 P | 2,357.52 |
| yddW | b1491 | 13.2 P | 327.56 | 23.5 P | 651.77 | 16.2 P | 372.78 |
| ydeK | b1510 | 34 P | 1,152.62 | 47.85 P | 3,366.87 | 47.3 P | 3,367.11 |
| slyB | b1641 | 1,177.4 P | 60,695.57 | 1,052.75 P | 61,458.84 | 1,110.9 P | 78,871.69 |
| lpp | b1677 | 2,142.5 P | 652,557.12 | 1,581.5 P | 488,494.78 | 1,501.85 P | 374,766.64 |
| nlpC | b1708 | 75 P | 2,709.71 | 78.35 P | 3,360.72 | 72.35 P | 3,052.40 |
| osmE | b1739 | 45.55 P | 3,368.38 | 63.3 P | 4,086.45 | 219.75 P | 14,538.54 |
| yoaF | b1793 | 48.6 P | 1,424.51 | 45.65 P | 1,424.32 | 47.3 P | 1,586.21 |
| yeaY | b1806 | 107.7 P | 4,334.04 | 86 P | 2,973.40 | 130.3 P | 2,282.15 |
| yebF | b1847 | 136.6 P | 3,104.08 | 132.2 P | 3,019.95 | 197.15 P | 5,182.22 |
| yecR | b1904 | 126.4 P | 14,835.41 | 25.95 P | 1,935.43 | 17.15 P | 940.23 |
| yedD | b1928 | 172.1 P | 12,536.98 | 171.3 P | 11,952.42 | 160.55 P | 19,869.35 |
| spr | b2175 | 587.4 P | 46,027.52 | 233.45 P | 17,688.27 | 220.05 P | 14,997.48 |
| rtn | b2176 | 56.55 P | 952.64 | 50.45 P | 874.74 | 48.65 P | 664.40 |
| yojL | b2214 | 46.1 P | 872.69 | 59.3 P | 1,142.29 | 61.25 P | 877.69 |
| vacJ | b2346 | 220.5 P | 3,966.98 | 167.4 P | 3,049.50 | 159.7 P | 2,716.95 |
| yfeY | b2432 | 96.85 P | 6,019.97 | 114.6 P | 6,788.17 | 245.15 P | 12,690.81 |
| nlpB | b2477 | 354 P | 11,015.25 | 426.5 P | 16,943.41 | 392.3 P | 10,879.40 |
| yfgL | b2512 | 313.4 P | 21,018.48 | 293.95 P | 23,681.22 | 328.35 P | 27,646.47 |
| yfiH | b2593 | 85.1 P | 4,998.32 | 95.3 P | 6,011.15 | 101.95 P | 5,639.94 |
| yfiO | b2595 | 332.25 P | 15,555.04 | 343.8 P | 17,027.70 | 376.75 P | 16,355.48 |
| yfiB | b2605 | 64.05 P | 3,947.83 | 77.95 P | 3,417.15 | 73.05 P | 4,036.95 |
| mltB | b2701 | 41.95 P | 560.42 | 65.95 P | 600.54 | 71.65 P | 656.93 |
| nlpD | b2742 | 1,018.55 P | 54,658.96 | 1,397.25 P | 93,860.15 | 1,480.25 P | 57,912.34 |
| ygdI | b2809 | 10.9 P | 593.84 | 15.5 P | 1,113.41 | 52.8 P | 2,160.82 |
| mltA | b2813 | 64 P | 1,918.08 | 75.35 P | 2,154.47 | 91.3 P | 1,910.28 |
| ygdR | b2833 | 49.05 P | 1,914.44 | 50.15 P | 1,576.61 | 60.65 P | 2,155.33 |
| ygeR | b2865 | 42.3 P | 501.07 | 50.1 P | 573.73 | 48.2 P | 348.11 |
| mltC | b2963 | 126.6 P | 3,392.37 | 101.15 P | 2,259.74 | 99.35 P | 1,746.64 |
| yraP | b3150 | 119.05 P | 5,530.13 | 108.25 P | 6,152.70 | 174.3 P | 8,350.46 |
| nlpI | b3163 | 1,415.7 P | 197,056.23 | 1,460.15 P | 299,438.99 | 1,470.4 P | 281,225.58 |
| yhdV | b3267 | 28 P | 582.55 | 28.45 P | 815.84 | 21 P | 260.17 |
| yhfL | b3369 | 11.8 P | 338.64 | 11.45 P | 478.89 | 12.05 P | 406.85 |
| nlpA | b3661 | 62.9 P | 2,889.24 | 78 P | 4,949.62 | 94.3 P | 5,323.51 |
| blc | b4149 | 31.25 P | 1,599.72 | 22.8 P | 1,090.51 | 56.25 P | 1,189.99 |
| yjfO | b4189 | 26.9 P | 968.41 | 19.9 P | 963.99 | 25.45 P | 966.50 |
| fecD | b4288 | 15.85 P | 593.54 | 11.85 P | 392.53 | 12.3 P | 336.81 |
| rcsf | b0196 | 220.45 P | 232.75 P | 263.2 P | |||
| borD | b0557 | 98.2 P | 117.5 P | 62.85 P | |||
| ybjR | b0867 | 48.35 P | 49.85 P | 71.2 P | |||
| ycaL | b0909 | 15.1 P | 23.95 P | 23.2 P | |||
| mltE | b1193 | 106.5 P | 104.2 P | 89.6 P | |||
| hslJ | b1379 | 35.1 P | 75.15 P | 77 P | |||
| ynfC | b1585 | 25 P | 17 P | 19.5 P | |||
| smpA | b2617 | 199.45 P | 196.35 P | 257.5 P | |||
| yggG | b2936 | 47.3 P | 32.8 P | 34.25 P | |||
| yidQ | b3688 | 86.85 P | 164.35 P | 227 P | |||
| yidX | b3696 | 8.6 P | 6.4 P | 7.85 P | |||
| yjeI | b4144 | 228.5 P | 182.9 P | 216.4 P | |||
Genes are divided into four groups based on the microarray expression profile described in the text. RT-PCR measurements were made as described in the text, and data are expressed as the gene copy number in the presence of reverse transcriptase minus the gene copy number in the absence of reverse transcriptase and are normalized to 10 μg of mRNA. Each value is the average for two experiments.
mRNA status as determined by analysis with Affynaetrix Suite 5.0 software: P, present with 95% confidence; A, absent; M, marginal (present with 94% confidence).
We then used real-time PCR to help better quantify the expression levels for the lipoprotein genes. First, reverse transcription (RT) was carried out with the same total RNA samples used for the microarray analysis and random hexamer primer (Invitrogen, Burlington, Ontario, Canada). RT was performed with SuperScript II (Invitrogen) for reactions with RT (+RT reactions). Control reactions were also performed under the same conditions except that SuperScript II was omitted (−RT reactions). Both types of reactions were used in real-time PCRs.
Primers for real-time PCR were designed with Primer Express 2.0 software from Applied Biosystems (ABI) (Foster City, Calif.). Forward and reverse primer pairs were designed for the 5′ and 3′ regions of each gene and purchased from Sigma Genosys (Oakville, Ontario, Canada). Real-time PCRs were carried out for each primer set with both the +RT and −RT reactions for each growth condition. The reaction buffer contained, in part, 1× ROX glycine conjugate of 5-carboxy-X-rhodamine, with succinimidyl ester as the inert-passive reference dye, and SYBR Green I. The reaction mixtures were aliquoted into 384-well ABI reaction plates. The plates were then placed in an ABI Prism 7900HT RT-PCR machine under the following conditions: stage 1 consisted of 95°C for 45 s; stage 2 consisted of 40 cycles of 95°C for 15 s, followed by 60°C for 1 min; stage 3 consisted of 95°C for 15 s; stage 4 consisted of 60°C for 15 s; and for stage 5, the temperature was ramped to 95°C for 5 s. The RT-PCR data were analyzed with SDS 2.0 software (ABI). Each +RT-versus-−RT reaction set was compared against a standard curve generated for each primer set by using E. coli linear DNA as a standard. A cycle threshold value was chosen that gave a linear regression value greater than 0.996 for each primer set standard curve. The calculated quantity values for each +RT or −RT reaction were standardized within each individual primer set-generated standard curve.
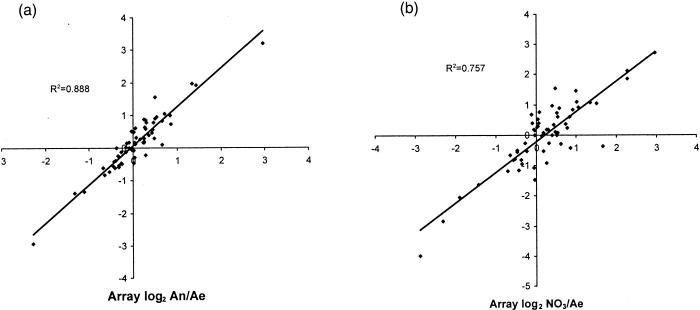
The RT-PCR data correlate well with the microarray data in that highly expressed genes found in the microarray study also give high RT-PCR signals. However, the RT-PCR results are much more accurate and sensitive and give a wider dynamic range of numbers. Signal intensity ratios for anaerobic-versus-aerobic and anaerobic-plus-nitrate-versus-aerobic data sets were calculated for both microarray (“present” values only) and RT-PCR data and are compared in a scatter plot in Fig. 1. The anaerobic/aerobic ratios had very good correlation between RT-PCR and microarray data, with an R2 value of 0.888; the nitrate/aerobic ratios had a slightly lower correlation (R2 = 0.757).
FIG. 1.
Scatter plot comparative analysis of microarray and RT-PCR data. The signal intensities of 61 genes which were designated “present” under all three growth conditions in the microarray data set were compared to the signal intensities generated by RT-PCR. Each datum point represents the log2 ratio of the signal intensity determined for one growth condition to the signal intensity determined for another growth condition, as determined by both RT-PCR and microarray analyses. (a) Ratios for anaerobically (An) versus aerobically (Ae) grown cells; (b) ratios for anaerobic-plus-nitrate (NO3) versus aerobic cells.
A further examination of gene expression patterns based on the RT-PCR data was then undertaken in order to gain some insight into possible functions of unknown lipoproteins. Growth under anaerobic conditions results in significant changes in the expression of many of the lipoprotein genes relative to aerobic expression. The expression of key structural protein genes such as lpp and pal remained fairly constant under these conditions, which was expected given their “housekeeping” role. However, transcripts of 14 genes are induced twofold or more under anaerobic growth. The gene with the strongest anaerobic induction is slp (Table 1). This gene is known to be induced under conditions of starvation or in the stationary phase (2), so perhaps it is not surprising that it is also induced during slow anaerobic growth. Other genes with strong anaerobic induction include ybgE, which appears to be cotranscribed with the cydAB cytochrome D terminal oxidase (also strongly induced anaerobically [data not shown]), and osmB, a lipoprotein gene which is also induced by high osmotic strength and in the stationary phase (15, 16). Only five genes had twofold or greater reductions of signal intensity under anaerobic conditions relative to that under aerobic conditions. These genes included cusC/ibeB, which is induced by high concentrations of copper ions (20) and appears to be important for virulence and invasion across the blood-brain barrier for other E. coli strains (12, 13), and spr, which encodes a putative penicillin binding protein (11).
The addition of nitrate to an anaerobic culture as an alternative electron acceptor also influences the expression of several of the lipoprotein genes. It is known that the addition of nitrate to anaerobic cultures regulates the expression of many genes, especially those involved in alternative electron transport pathways (26). When grown anaerobically with nitrate, the RT-PCR signals for 13 genes decreased twofold or more and those for 4 genes increased twofold or more relative to those under anaerobic conditions without nitrate. Anaerobically induced genes such as slp and yhiU are repressed with the addition of nitrate. Two of the four lipoproteins induced by the addition of nitrate to an anaerobic culture, albeit with low overall signals, are ymcA and ymcC, which form part of a putative ymcCBA operon. Another gene induced by nitrate is osmE, a putative lipoprotein gene which is induced by high osmotic strength (10).
Microarrays have fast become a commonly used tool to examine global expression profiles for many bacterial species (see reference 6 for a recent review). Many of these microarray studies have gone one step further by selection of a subset of genes to determine expression by RT-PCR data, which are then compared to the microarray data (4, 8, 9, 18, 22-24, 27-29). However, in these cases, the genes studied by RT-PCR are chosen as a subset of genes of interest that were originally identified by the microarray assay. The approach presented here is different; the gene set to be studied by RT-PCR was not chosen on the basis of the microarray results; instead, it was chosen based on known or predicted functions of the gene products. Even with this unbiased approach to selecting genes for RT-PCR analysis, the correlation between the RT-PCR data and the microarray data is very good. The more accurate and quantitative RT-PCR data were not used just for comparative purposes, however. These data were then used to identify potentially significant unknown lipoprotein genes with either high gene expression levels or significant changes in gene expression depending on growth conditions. This study has produced the first real data reported for these unknown genes and may lead to more effective investigation of these genes in the future. With the usefulness of this approach assured, it is now time to further study the other unknown lipoprotein genes showing either strong or varied expression levels by other means.
ADDENDUM
As this paper was under review, other potential lipoproteins in E. coli came to our attention, especially those predicted in a recent related paper (14). These genes—rcsF (Blattner no. b0196), borD (b0557), ybjR (b0867), ycaL (b0909), hslJ (b1379), ynfC (b1585), smpA (b2617), yggG (b2936), yidQ (b3688), yidX (b3696), and yjeI (b4144)—are included in the microarray results, but RT-PCR data were not generated.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council and by Project CyberCell.
J.H.W. is a Canada Research Chair on Membrane Biochemistry. We thank Philip Winter for data analysis.
REFERENCES
- 1.Affymetrix, Inc. 2002, posting date. GeneChip® E. coli antisense genome array technical manual. [Online.] Affymetrix, Inc., Santa Clara, Calif. http://www.affymetrix.com/support/technical/manuals.affx.
- 2.Alexander, D. M., and A. C. St John. 1994. Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol. Microbiol. 11:1059-1071. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, R. E., B. K. Leskiw, R. S. Hodges, C. M. Kay, and J. H. Weiner. 1998. The entericidin locus of Escherichia coli and its implications for programmed bacterial cell death. J. Mol. Biol. 280:583-596. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V., and H. C. Wu. 1993. Lipoproteins, structure, function, biosynthesis and model for protein export, p. 319-342. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier Science, Amsterdam, The Netherlands. [Google Scholar]
- 6.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 7.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 8.Graham, M. R., L. M. Smoot, C. A. Migliaccio, K. Virtaneva, D. E. Sturdevant, S. F. Porcella, M. J. Federle, G. J. Adams, J. R. Scott, and J. M. Musser. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc. Natl. Acad. Sci. USA 99:13855-13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guckenberger, M., S. Kurz, C. Aepinus, S. Theiss, S. Haller, T. Leimbach, U. Panzner, J. Weber, H. Paul, A. Unkmeir, M. Frosch, and G. Dietrich. 2002. Analysis of the heat shock response of Neisseria meningitidis with cDNA- and oligonucleotide-based DNA microarrays. J. Bacteriol. 184:2546-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez, C., S. Gordia, and S. Bonnassie. 1995. Characterization of the osmotically inducible gene osmE of Escherichia coli K-12. Mol. Microbiol. 16:553-563. [DOI] [PubMed] [Google Scholar]
- 11.Hara, H., N. Abe, M. Nakakouji, Y. Nishimura, and K. Horiuchi. 1996. Overproduction of penicillin-binding protein 7 suppresses thermosensitive growth defect at low osmolarity due to an spr mutation of Escherichia coli. Microb. Drug Resist. 2:63-72. [DOI] [PubMed] [Google Scholar]
- 12.Huang, S. H., Y. H. Chen, Q. Fu, M. Stins, Y. Wang, C. Wass, and K. S. Kim. 1999. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect. Immun. 67:2103-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, S. H., and A. Y. Jong. 2001. Cellular mechanisms of microbial proteins contributing to invasion of the blood-brain barrier. Cell. Microbiol. 3:277-287. [DOI] [PubMed] [Google Scholar]
- 14.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung, J. U., C. Gutierrez, F. Martin, M. Ardourel, and M. Villarejo. 1990. Transcription of osmB, a gene encoding an Escherichia coli lipoprotein, is regulated by dual signals. Osmotic stress and stationary phase. J. Biol. Chem. 265:10574-10581. [PubMed] [Google Scholar]
- 16.Jung, J. U., C. Gutierrez, and M. R. Villarejo. 1989. Sequence of an osmotically inducible lipoprotein gene. J. Bacteriol. 171:511-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraft, A. R., M. F. Templin, and J. V. Holtje. 1998. Membrane-bound lytic endotransglycosylase in Escherichia coli. J. Bacteriol. 180:3441-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, J. M., S. Zhang, S. Saha, S. Santa Anna, C. Jiang, and J. Perkins. 2001. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 183:7371-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno, T. 1981. A novel peptidoglycan-associated lipoprotein (PAL) found in the outer membrane of Proteus mirabilis and other gram-negative bacteria. J. Biochem. (Tokyo) 89:1039-1049. [PubMed] [Google Scholar]
- 20.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182:5864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paustian, M. L., B. J. May, and V. Kapur. 2002. Transcriptional response of Pasteurella multocida to nutrient limitation. J. Bacteriol. 184:3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 24.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki, H., Y. Nishimura, S. Yasuda, A. Nishimura, M. Yamada, and Y. Hirota. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 167:1-9. [DOI] [PubMed] [Google Scholar]
- 26.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 27.Voyich, J. M., D. E. Sturdevant, K. R. Braughton, S. D. Kobayashi, B. Lei, K. Virtaneva, D. W. Dorward, J. M. Musser, and F. R. DeLeo. 2003. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. USA 100:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei, Y., J. M. Lee, D. R. Smulski, and R. A. LaRossa. 2001. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J. Bacteriol. 183:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, J. W., R. Ramamurthy, S. Porwollik, M. McClelland, T. Hammond, P. Allen, C. M. Ott, D. L. Pierson, and C. A. Nickerson. 2002. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc. Natl. Acad. Sci. USA 99:13807-13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, H. C. 1996. Biosynthesis of lipoproteins, p. 1005-1014. In F. C. Neidhardt, R. Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.