Abstract
Transcriptional analysis of a constitutively active mutant of the EvgA/EvgS two-component system of Escherichia coli resulted in enhanced expression of 13 PhoP/PhoQ-regulated genes, crcA, hemL, mgtA, ompT, phoP, phoQ, proP, rstA, rstB, slyB, ybjG, yrbL, and mgrB. This regulatory network between the two systems also occurred as a result of overproduction of the EvgA regulator; however, enhanced transcription of the phoPQ genes did not further activate expression of the PhoP/PhoQ-regulated genes. These results demonstrated signal transduction from the EvgA/EvgS system to the PhoP/PhoQ system in E. coli and also identified the genes that required the two systems for enhanced expression. This is one example of the intricate signal transduction networks that are posited to exist in E. coli.
The two-component signal transduction system is the major system in bacteria for sensing environmental stresses and transducing the information inside the cells for adaptation. This system is basically composed of a histidine kinase sensor residing in the inner membrane and a cognate response regulator in the cytoplasm. In Escherichia coli, 29 histidine kinase sensors, 32 response regulators, and one HPt (histidine-containing phosphotransmitter) domain have been found by analyses of the E. coli K-12 genome (23). Each sensor responds to individual environmental stresses to cope with the numerous environmental conditions that E. coli faces; however, 29 sensors are apparently insufficient, and it is posited that a complex regulatory network between the two-component systems exists.
Transcriptome analysis using the microarray technique is an extremely efficient method for analyzing regulatory networks. Oshima et al. (27) performed a global analysis of two-component regulatory system mutants of E. coli K-12 for a single growth condition (Luria-Bertani medium, aerobic, and mid-exponential phase) and proposed the existence of a network of functional interactions, such as cross talk and cascade signal transduction between the systems. In their study, some of the 36 two-component deletion mutants showed little or no change in their mRNA profiles, indicating that these two-component systems were not strongly operating under their culture condition.
One of these low- or nonoperating systems in mid-exponential-phase cells grown in a rich medium is the EvgA/EvgS system. The EvgA/EvgS system in E. coli is highly similar to the BvgA/BvgS system, which controls the expression of adhesins, toxins, and other virulence factors in Bordetella pertussis (2, 30, 31). Although the environmental signal to which the EvgS sensor responds is still unidentified, studies using a mutant with a constitutively active EvgS (evgS1 mutant [12]) or using overexpression of the response regulator EvgA demonstrated that the EvgA/EvgS system confers multidrug resistance to a drug-hypersusceptible strain that lacks constitutive multidrug efflux genes acrAB (16) and acid resistance to exponentially growing cells (5, 17, 18, 24-26). In our recent microarray analysis of the mutant with a constitutively active EvgS, we found that as many as 225 open reading frames showed significant increases in transcription and that 144 open reading frames showed significant decreases in transcription (http://www.nara.kindai.ac.jp/nogei/seiken/array2.html) (5). Comparison of this result with the systematic analysis of 36 two-component deletion mutants by Oshima et al. (27) revealed a surprisingly strong relationship between the EvgA/EvgS system and another two-component system, PhoP/PhoQ, which senses external Mg2+ concentration (8). Signal transduction between these two systems is investigated in this study.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli strains and plasmids used in this study are listed in Table 1. The phoP strains (KMP1 and KMP2001) and phoQ strains (KMQ1 and KMQ2001) were constructed with P1 transduction from donor strains WP3022 and WQ3007 (22) to recipient strains KMY1 (wild type) and KMY2001 (evgS1 mutant). Bacteria were grown at 37°C in Luria-Bertani (LB) medium (pH 7.5), containing 1% Bacto Tryptone (Difco), 0.5% Bacto Yeast Extract (Difco), and 1% NaCl. MgCl2 was added to a final concentration of 30 mM when necessary. For induction of the arabinose promoter of pA191, arabinose at a final concentration of 0.2% (wt/vol) was added to the culture at an optical density at 600 nm (OD600) of 0.2. Ampicillin, chloramphenicol, kanamycin, and tetracycline were used at 100, 25, 25, and 12.5 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference, construction, or source |
|---|---|---|
| E. coli strains | ||
| MC4100 | F− Δ(argF-lac)U169 araD139 rpsL150 ptsF25 fibB5301 rbsR | 3 |
| MK12 | MC4100 Δ(ara-leu)7696 | 12 |
| KMY1 | MK12 λRS45[φ(emrK′-lacZ)] | 12 |
| KMY2001 | KMY1 evgS1 | 12 |
| KMP1 | KMY1 phoP::cat | KMY1 × P1(WP3022)→Camra |
| KMP2001 | KMY2001 phoP::cat | KMY2001 × P1(WP3022)→Camr |
| KMQ1 | KMY1 phoQ::cat | KMY1 × P1(WQ3007)→Camr |
| KMQ2001 | KMY2001 phoQ::cat | KMY2001 × P1(WQ3007)→Camr |
| KMY204 | KMY1 evgA::cat | 12 |
| WP3022 | W3110 phoP::cat | 22 |
| WQ3007 | W3110 phoQ::cat | 22 |
| MG1601 | MC4100 mgtA::λplacMu55 | 11 |
| Plasmids | ||
| pMV191 | pBAD18 derivative carrying an EcoRI-AbaI fragment containing the tetR gene of pBR322 | 12 |
| pA191 | pMV191 derivative carrying a 0.7-kb fragment containing the evgA gene | 12 |
| pMW119 | Low-copy-number vector | Nippon Gene |
| pOH2001 | pMW119 derivative carrying a 5.2-kb fragment containing the evgA and evgS1 genes from KMY2001 | Laboratory stock |
| pOH2001D52A | pOH2001 derivative carrying an evgAD52A gene | This study |
| pHO119 | pMW119 derivative carrying a 3.6-kb fragment containing the phoPQ gene | 11 |
Camr, chloramphenicol resistance.
Site-directed mutagenesis.
In order to create pOH2001D52A, a low-copy-number plasmid carrying the evgA gene with an Asp-to-Ala mutation at position 52, site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene), primers 5′-GATATCGTCATCATTGCTGTCGATATCCCC-3′ and 5′-GGGGATATCGACAGCAATGATGACGATATC-3′ (the mutation site is underlined), and pOH2001 carrying the evgA and evgS1 genes.
RNA preparation and S1 nuclease mapping.
Total RNA was extracted with hot phenol from a mid-log-phase culture (OD600 of about 0.8) at 60°C, as described by Aiba (1). Total RNA of EvgA-overexpressing strains was prepared from cells grown for an additional 30 min at 37°C after reaching an OD600 of about 0.8 for ample induction of EvgA from the arabinose promoter. S1 mapping was performed by the method of Kato et al. (12). Briefly, a 32P-end-labeled probe was prepared by PCR using the primers described by Minagawa et al. (22) and the primers listed in Table 2, E. coli MC4100 genomic DNA as the template, and ExTaq DNA polymerase (Takara). A mixture of 32P-end-labeled probe and 100 μg of total RNA was incubated for 10 min at 75°C, then gradually cooled to 37°C, and incubated overnight for hybridization. This was followed by S1 nuclease (Takara) digestion for 10 min at 37°C. Undigested RNA-probe DNA was extracted with phenol, precipitated with ethanol, and subjected to electrophoresis on a 6% (wt/vol) polyacrylamide sequencing gel. The radioactivity of the transcripts was measured by BAS1000 Mac (Fuji film) and analyzed by MacBAS2.2 (Fuji film).
TABLE 2.
PCR primers
| Gene | Primer | Primer sequence | GenBank accession no. | Nucleotide positions |
|---|---|---|---|---|
| crcA | crcAF | 5′-TGG TGT TAG CTG CGG CTG TG-3′ | AE000167 | 4147-4166 |
| crcAR | 5′-GGC TGT TGC CAG GTT TGT GC-3′ | AE000167 | 4631-4612 | |
| ybjG | ybjGF | 5′-GGC CCC AGC AGC CAT TGT AA-3′ | AE000186 | 5782-5763 |
| ybjGR | 5′-CCA GCA TAA CTT CCC GAC GC-3′ | AE000186 | 5271-5290 | |
| proP | proPF | 5′-GTG GTT TGC CTC TTC GAC CG-3′ | AE000483 | 4827-4846 |
| proPR | 5′-CCC AGT GAT GCT GCG GTA AT-3′ | AE000483 | 5398-5379 | |
| ompT | ompTF | 5′-GCG GCC CAC GAC TTA GAA GT-3′ | AE000161 | 8964-8945 |
| ompTR | 5′-GCA ATA GGG GTT GTC AGG AC-3′ | AE000161 | 8479-8498 |
PhoP detection.
Cells were grown to an OD600 of about 0.8 at 37°C in LB medium with constant shaking, collected by centrifugation, and resuspended in a solution of 25% sucrose and 40 mM Tris-HCl (pH 8.0). After treatment with 1 mM EDTA and 0.5 mg of lysozyme per ml at 0°C for 10 min, cells were lysed by adding 0.5% Brij 58. The Brij 58 lysate was supplemented with 0.01 M MgCl2 and 0.2 M KCl, digested at 37°C for 10 min with 20 μg of RNase A per ml in the presence of 1 mM phenylmethylsulfonyl fluoride, and sonicated. The supernatant after centrifugation for 30 min at 20,000 × g was used as the cell lysate. The protein concentration of cell lysates was determined by the Bradford method (protein assay kit; Bio-Rad). Equal amounts of protein (16 μg/lane) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were then transferred to a polyvinylidene difluoride membrane (Immobilon-P transfer membranes; Millipore), probed with anti-PhoP serum, and visualized by alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G, 5-bromo-4-chloro-3-indolylphosphate, and nitroblue tetrazolium (immunoblot kit; Bio-Rad).
β-Galactosidase assay.
Duplicate samples of cells grown to an OD600 of about 0.8 at 37°C in LB medium were subjected to β-galactosidase assay, and the results were expressed in Miller units (21). The data shown are means and standard deviations for three individual cultures.
RESULTS
DNA microarray-based analysis of the interaction between EvgA/EvgS and other two-component systems.
To search for two-component systems interacting with the EvgA/EvgS system, we first selected up-regulated genes of other two-component systems by picking genes that were down-regulated by deletion of each individual two-component system (http://ecoli.aist-nara.ac.jp/xp_analysis/2_components) (27). We thenmatched them with the up-regulated genes by the activation ofthe EvgS sensor (http://www.nara.kindai.ac.jp/nogei/seiken/array2.html) (5). As shown in Table 3, expression of 14 of 27 PhoP/PhoQ up-regulated genes was enhanced in the evgS1 mutant, clearly indicating interaction between the EvgA/EvgS and PhoP/PhoQ systems. The expression ratio of the 14 PhoP/PhoQ-regulated genes, which were up-regulated by the constitutively active evgS1 mutation, is shown in Table 4. Of these 14 genes, 9, namely, hemL, mgtA, phoP, phoQ, rstA, rstB, slyB, yrbL, and mgrB, were members of the PhoP/PhoQ-dependent Mg2+ stimulon reported by Minagawa et al. (22). These genes are directly regulated by PhoP via phosphorylation from the PhoQ sensor.
TABLE 3.
Interaction between EvgAS and other two-component systems
| TCSa | Ratiob |
|---|---|
| phoPQ | 14/27 |
| rssB | 3/23 |
| envZ ompR | 2/54 |
| basSR | 0/12 |
| narXL | 0/4 |
| narQ | 2/25 |
| yojN | 0/17 |
| hydHG | 0/13 |
| yfhA | 1/17 |
| rcsB | 0/11 |
| uhpAB | 0/3 |
| yfhK | 0/8 |
| narP | 0/9 |
| uvrY | 1/37 |
| ypdAB | 1/18 |
| citAB | 1/23 |
| yehUT | 0/7 |
| dcuSR | 2/29 |
| ntrBC | 1/8 |
| arcA | 0/38 |
| arcB | 3/112 |
| baeSR | 0/8 |
| cpxRA | 1/16 |
| creBC | 1/1 |
| kdpDE | 0/10 |
| phoBR | 0/8 |
| rstAB | 2/22 |
| torSR | 1/24 |
| yedWV | 0/7 |
| ygiXY | 0/6 |
| barA | 1/24 |
| cusRS | 1/7 |
| fimZ | 1/22 |
| cheABY | 1/28 |
| atoSC | 0/32 |
TCS, two-component system.
Ratio, number of up-regulated genes in the constitutively active evgS1 mutant among the genes up-regulated by the corresponding system to the number of up-regulated genes by the corresponding system (http://ecoli.aist-nara.ac.jp/xp_analysis/2_components) (27).
TABLE 4.
Up-regulation of PhoPQ regulated genes by constitutively active evgS1 mutation
| PhoPQ-regulated gene | Expression ratioa |
|---|---|
| copA | 10.0, 9.3 |
| crcA | 58.9, Hb |
| hemL | 3.5, 3.2 |
| mgtA | 14.7, 18.6 |
| ompT | 5.4, 7.0 |
| phoP | 7.2, 7.2 |
| phoQ | 4.5, 6.3 |
| proP | 13.3, 17.8 |
| rstA | 3.6, 5.4 |
| rstB | 5.0, 3.5 |
| slyB | 3.0, 3.6 |
| ybjG | 5.9, 5.3 |
| yrbL | 2.9, 2.5 |
| mgrB | 5.6, 2.5 |
Ratio of normalized signal intensities of Cy5-dUTP (strain KMY2001) against Cy3-dUTP (strain KMY1) from DNA microarray analysis. Two ratios are shown for each gene (http://www.nara.kindai.ac.jp/nogei/seiken/array2.html) (5).
H, signal not detected in strain KMY1 but signal was significantly detected in strain KMY2001.
Enhanced expression of PhoP regulons by EvgA/EvgS.
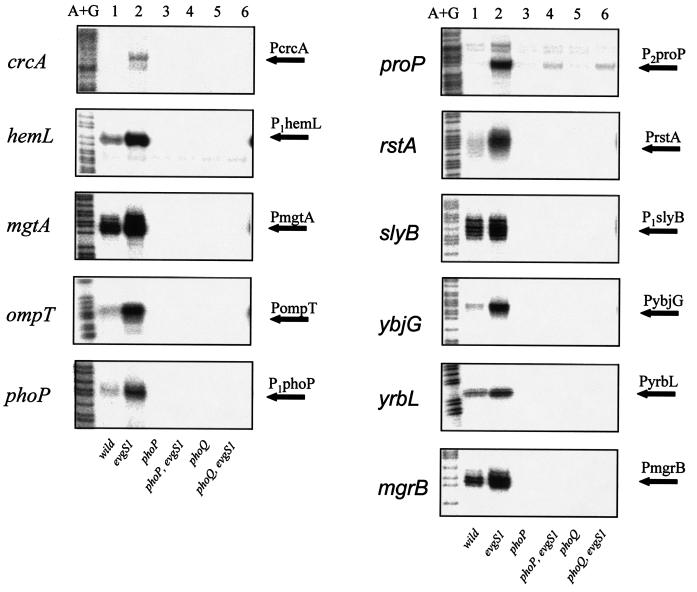
To further analyze the promoter regions of EvgA/EvgS-enhanced PhoP regulons, S1 mapping was performed for the 14 genes listed in Table 4. Enhanced expression of 13 genes in the evgS1 mutant (note that the phoP and phoQ genes and the rstA and rstB genes form operons) was confirmed (Fig. 1, lanes 2), validating our microarray data. Expression of these transcripts was dependent on PhoP/PhoQ, since deletion of phoP or phoQ resulted in a prominent decrease in transcription, even in the evgS1 mutants (Fig. 1, lanes 3 to 6). This decrease in transcription was complemented by pHO119, a plasmid carrying the phoPQ operon with its native promoter (data not shown). Thus, the enhancement of these promoters by EvgA/EvgS is clearly dependent on PhoP/PhoQ, indicating signal transduction from the EvgA/EvgS system to the PhoP/PhoQ system. As for the remaining copA gene, transcripts were searched for in a range of 1,000 bp upstream of the translational start codon, but no transcript was detected in our S1 mapping for unknown reasons.
FIG. 1.
S1 mapping of EvgA/EvgS-regulated PhoP regulons of E. coli strains. Lanes: A+G, Maxam-Gilbert sequencing ladder; 1, KMY1; 2, KMY2001 (evgS1); 3, KMP1 (phoP); 4, KMP2001 (phoP evgS1); 5, KMQ1 (phoQ); 6, KMQ2001 (phoQ evgS1). Transcripts with increased expression in the evgS1 mutants are indicated by arrows labeled with the gene promoter.
Promoters of the PhoP regulon activated by EvgA/EvgS.
All nine genes reported by Minagawa et al. (22) share a consensus PhoP binding sequence [consensus sequence of PhoP box, (T/G)GTTTAnnnnn(T/G)GTTTA (n is any nucleotide) (Fig. 2)] at their −35 region with a maximum mismatch of two nucleotides. Promoter regions of the four remaining genes, crcA, ybjG, proP, and ompT, are shown in Fig. 2 in comparison with several PhoP regulons. PhoP boxes with a mismatch of two and three nucleotides were found in the −35 regions of ybjG and crcA, respectively, suggesting that these two genes may be directly regulated by PhoP. On the other hand, PhoP boxes were not found in the promoter regions of proP and ompT, suggesting an indirect regulation of these genes by PhoP.
FIG. 2.
Promoter analysis of crcA, ybjG, proP, and ompT compared with other PhoP regulons. Transcription start sites (thin arrows labeled with the gene promoter) and direct repeats (PhoP box) (thick arrows) are indicated. The −10 region of each promoter and the −35 region of the ompT promoter are underlined. The −10 and −35 hexamers, start sites, and the direct repeats are indicated in capital letters.
Enhanced expression of the PhoP regulon by overproduced EvgA.
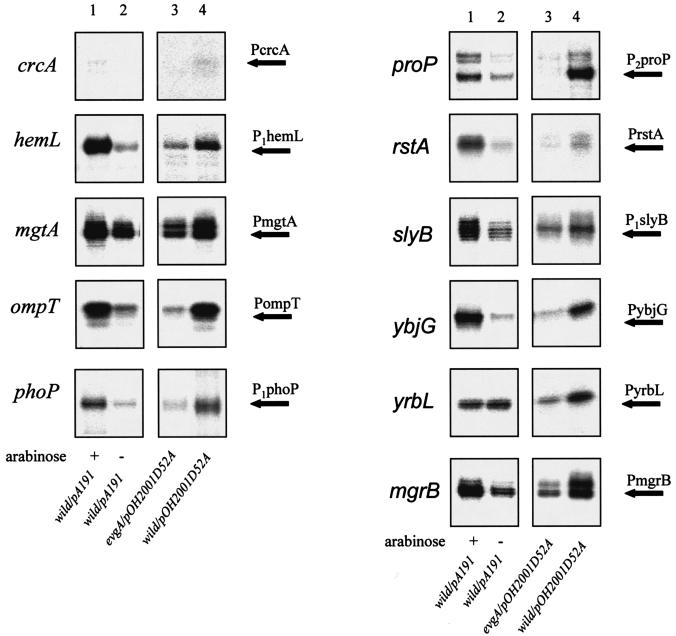
EvgA was overexpressed from an arabinose promoter in the wild-type strain, and the expression of EvgA/EvgS-enhanced PhoP regulons was determined. As shown in Fig. 3, lanes 1, overexpression of EvgA in a wild-type EvgS background also caused an increase in transcription from 10 of the 11 promoters under investigation. Furthermore, pOH2001D52A, which harbors evgAD52A (mutation in the phosphorylation site of evgA) and evgS1 genes and their native promoter was transformed into an evgA mutant and the wild-type strain (Fig. 3, lanes 3 and 4). Although introduction of the evgS1 gene did not enhance transcription of the PhoP-regulated genes in the absence of a wild-type evgA (Fig. 3, lanes 3), one copy of the wild-type evgA in the chromosome of the host strain resulted in enhanced transcription of the PhoP regulons (Fig. 3, lanes 4). These results clearly indicate that phosphotransfer from phospho-EvgS to EvgA is essential for the increased transcription of the EvgA/EvgS-enhanced PhoP regulons. They also show that cross talk from EvgS to PhoP is not the major pathway.
FIG. 3.
Effect of overproduction of EvgA and significance of the phosphorylation site of EvgA. S1 mapping of EvgA/EvgS-regulated PhoP regulons of E. coli strains was performed. Lanes 1, KMY1 (wild type) carrying plasmid pA191 grown in the presence of arabinose; 2, KMY1/pA191 grown without arabinose; 3, KMY204 (evgA) carrying pOH2001D52A (evgAD52A evgS1); 4, KMY1/pOH2001D52A (evgAD52A evgS1). Transcripts with increased expression in the evgS1 mutants are indicated by arrows labeled with the gene promoter.
Effect of enhanced transcriptional level of phoP on the expression of the PhoP regulon.
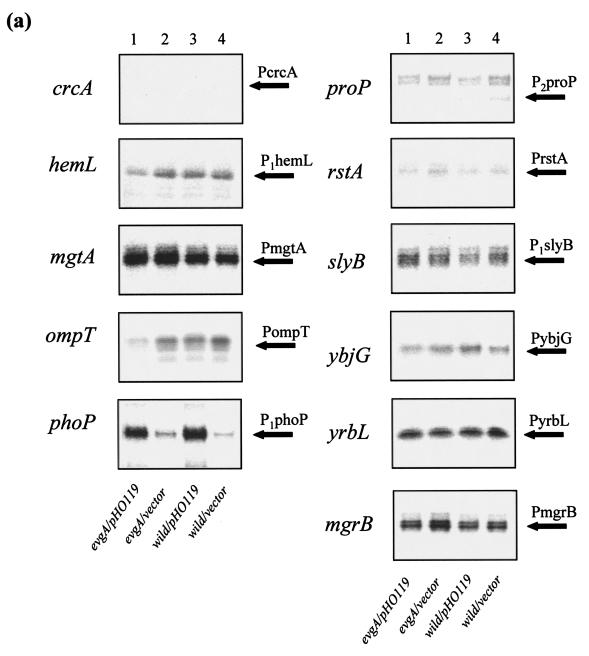
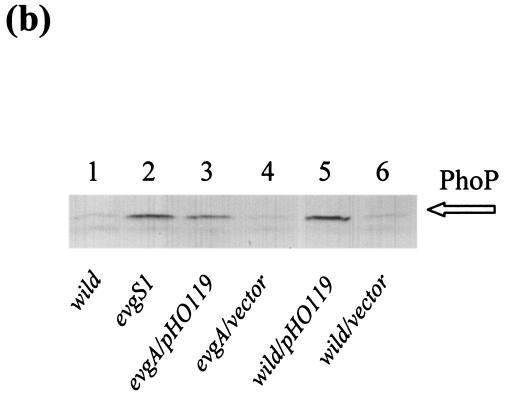
Transforming pHO119 to an evgA mutant and the wild-type strain caused an increase in phoP transcription to a level similar to that found in the evgS1 mutant (Fig. 4a, lanes 1 and 3). Immunoblotting with anti-PhoP serum confirmed that the increase in PhoP protein in the pHO119-transformed strains was also similar to the level found in the evgS1 mutant (Fig. 4b). However, the pHO119 transformants did not show increased expression of EvgA/EvgS-enhanced PhoP regulons, except for phoPQ, in both the evgA mutant and the wild-type strain (Fig. 4a, lanes 1 and 3). Thus, a simple increase in the level of PhoP was not involved in the signal transduction from EvgA/EvgS to PhoP/PhoQ.
FIG. 4.
(a) Effect of overproduction of PhoPQ. S1 mapping of EvgA/EvgS-regulated PhoP regulons of E. coli strains was performed. Lanes: 1, KMY204 (evgA)/pHO119; 2, KMY204/pMW119 (vector); 3, KMY1 (wild type)/pHO119; 4, KMY1/pMW119. Transcripts corresponding to those with increased expression in the evgS1 mutants are indicated by arrows labeled with the gene promoter. (b) Western blotting against anti-PhoP antibody. Lanes: 1, KMY1 (wild type); 2, KMY2001 (evgS1); 3, KMY204 (evgA)/pHO119; 4, KMY204/pMW119 (vector); 5, KMY1/pHO119; 6, KMY1/pMW119. The bands responding to anti-PhoP antibody are indicated by the open arrow to the right of the blot.
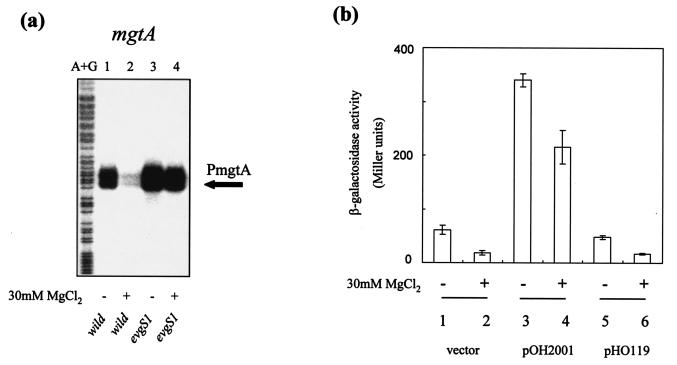
On the other hand, transcriptional activity of mgtA, a well-characterized member of the PhoP regulon (11, 36), was assayed in the presence of 30 mM MgCl2 (an inactivating condition for the PhoQ sensor [11]). Although transcription in the wild-type strain was repressed to 30% of that in the absence of MgCl2 (Fig. 5a, lanes 1 and 2) (ratio calculated from the radioactive intensity of the corresponding bands), transcription in the evgS1 mutant grown in the presence of 30 mM MgCl2 was still nearly twice that of the wild-type strain grown in the absence of MgCl2 (Fig. 5a, lanes 1 and 4).
FIG. 5.
(a) Effect of high Mg2+ concentration. S1 mapping of the mgtA genes of E. coli strains was performed. Lanes: A+G, Maxam-Gilbert sequencing ladder; 1, KMY1 (wild type) without MgCl2; 2, KMY1 with 30 mM MgCl2; 3, KMY2001 (evgS1) without MgCl2; 4, KMY2001 with 30 mM MgCl2. (b) Effect of high Mg2+ concentration on the expression of mgtA in evgS1 strains and phoPQ-overexpressed strains by reporter assay. E. coli MG1601 carrying pMW119 (vector) (bars 1 and 2), pOH2001(bars 3 and 4), or pHO119 (bars 5 and 6) were grown in the absence (−) or presence (+) of 30 mM MgCl2. The data shown are means and standard deviations (error bars) for three individual cultures.
Furthermore, these results were confirmed by reporter assays (Fig. 5b). Plasmids pOH2001, carrying the evgA-evgS1 operon and its native promoter, and pHO119, carrying the phoPQ operon and its native promoter, were transformed into strain MG1601. The expression of the mgtA promoter was assayed by β-galactosidase activity. Introduction of evgS1 enhanced the mgtA expression over that of the control (Fig. 5b, bars 3 and 4), but increase in phoP expression did not enhance mgtA expression (Fig. 5b, bars 5 and 6).
DISCUSSION
Through a combination of microarray analysis and S1 mapping, we found evidence of signal transduction between EvgA/EvgS and PhoP/PhoQ systems. This signal transduction was observed to occur as a result of both EvgS activation and EvgA overproduction (Fig. 1 and 3). We also demonstrated that phospho-EvgA was essential to this signal transduction (Fig. 3). Although the level of phoPQ transcription increased as a result of activation of EvgS, transcription of the phoPQ genes from a heterologous promoter in a wild-type EvgS background did not promote expression of the PhoP regulons, except in the case of phoP (Fig. 4a).
One hypothesis presented to explain the mechanism underlying signal transduction between EvgA/EvgS and PhoP/PhoQ was that the phospho-EvgA directly regulated the PhoP/PhoQ-regulated genes in cooperation with PhoP. Masuda and Church (18) reported that EvgA binds to an 18-bp consensus sequence (5′-TTCPyTACA-3′ and its inverted repeat, 5′-TGTAPuGAA-3′, separated by two random bases [Py is a pyrimidine, and Pu is a purine]) for the direct regulation of the EvgA regulons. This 18-bp consensus sequence was not found in any of the PhoP/PhoQ-regulated genes enhanced by the EvgA/EvgS system. We also performed an in vitro transcription assay of the mgtA gene using DNA templates containing the promoter region of mgtA, RNA polymerase, acetyl phosphate, PhoP, and EvgA and found no additive effect of EvgA (data not shown). Thus, we were not able to find any evidence of a direct interaction between EvgA and the PhoP/PhoQ-regulated genes.
Another possibility was the indirect interaction of EvgA in cooperation with PhoP, where another transcriptional factor regulated by the EvgA/EvgS system directly interacted with the promoters of PhoP/PhoQ-regulated genes. If this were the case, a consensus sequence for the binding site of the unknown transcriptional factor should have been present in the 11 promoter regions of the PhoP/PhoQ-regulated genes. A search for such a consensus sequence was performed using Genespring software for positions +1 to −500 of each transcriptional start site of the 11 promoters but failed to find any such sequence. Therefore, direct and indirect interaction of EvgA with the 11 promoters in cooperation with PhoP also could not explain the signal transduction under investigation.
Taking all the aforementioned results together, the only possibility left was the increase in the level of phospho-PhoP by the EvgA/EvgS system. PhoQ of the PhoP/PhoQ system responds to extracellular Mg2+ levels (6, 7, 33). At a high concentration of Mg2+, its phosphatase activity is enhanced, resulting in a lower level of cellular phospho-PhoP and thus repression of PhoP regulon transcription (4). In the presence of 30 mM MgCl2, mgtA transcription in the wild-type strain was actually repressed to 30% of that in the absence of MgCl2 (Fig. 5). However, mgtA transcription in the evgS1 mutant grown in the presence of 30 mM MgCl2 was still nearly twice that of the wild-type strain grown without MgCl2 (Fig. 5). In other words, compared to the wild-type strain at a low Mg2+ concentration, a higher level of phospho-PhoP was present in the evgS1 mutant even at a high Mg2+ concentration, where the phosphatase activity of PhoQ should have been enhanced. The increase in the phospho-PhoP level may be due to inhibition of the phosphatase activity of PhoQ or to activation of the kinase activity of PhoQ by a mechanism requiring further investigation.
In Salmonella enterica serovar Typhimurium, a similar cascade signal transduction between the PhoP/PhoQ and PmrA/PmrB systems has been reported (9, 29). In signal transduction from PhoP/PhoQ to PmrA/PmrB, an increase in the phospho-PmrA levels by phospho-PhoP was suggested from the following two results: (i) transcription of the pmrAB genes from a heterologous promoter cannot restore expression of PmrA-regulated genes to strains lacking phoP or phoQ, and (ii) the pmrCAB operon is transcriptionally autoregulated (29). We also obtained similar results with signal transduction from EvgA/EvgS to PhoP/PhoQ in E. coli: transcription of the phoPQ genes from plasmid pHO119 could not restore expression of PhoP-regulated genes (Fig. 4a), and the phoPQ operon was transcriptionally autoregulated (Fig. 1). In Salmonella, a small protein, PmrD, was found to mediate activation of the PmrA/PmrB system by the PhoP/PhoQ system (14), and the PmrA/PmrB system negatively controls the expression of PmrD (13). However, it is not yet known how PmrD activates the PmrA/PmrB system. A similar signal transduction model between the EvgA/EvgS and PhoP/PhoQ systems may exist in E. coli and involve other factors. In addition to the PhoQ sensor sensing Mg2+ and Ca2+, PhoQ in E. coli was recently reported to be inactivated by acetate in a sensor domain-independent mechanism (15). Thus, there is a possibility that PhoQ in E. coli may sense stimulants other than Mg2+ and Ca2+, which may involve the EvgA/EvgS system.
The PhoP regulon candidate genes, crcA and ybjG, discovered in this study are genes detected only when the PhoP/PhoQ system is enhanced by EvgA/EvgS. These genes were not found in our earlier search (22) because the expression of these genes is very low without enhancement by the EvgA/EvgS system. The crcA gene is a homolog of pagP in Salmonella and encodes an enzyme located in the outer membrane. PagP is involved in cationic antimicrobial peptide resistance and is regulated by the PhoP/PhoQ system in Salmonella (10). It is interesting that PagP found in Bordetella is regulated by the BvgA/BvgS system (28), a virulence-related two-component system highly homologous to the E. coli EvgA/EvgS system (31).
Another gene regulated by EvgA/EvgS via PhoP/PhoQ is proP, which encodes an integral membrane transporter of proline, glycine betaine, and other osmoprotecting compounds. Two promoters, P1 and P2, have been reported for proP (20). EvgA/EvgS up-regulates transcription from the P2 promoter, which has been reported to be completely dependent on RpoS and coactivated by Fis and CRP (cyclic AMP receptor protein) by direct binding to the promoter (19, 34, 35). The absence of the PhoP box in the P2 promoter suggested that the regulation by phospho-PhoP was indirect. Transcript levels of fis and crp in the evgS1 mutant compared to the parent strain in our microarray were 0.44 and 0.37, respectively, for fis and 0.99 and 0.84, respectively, for crp (5), indicating that the changes in transcriptional levels of fis and crp were not related. It should be pointed out that the wild-type strain that did not overproduce EvgA showed enhanced transcription from the proP P2 promoter (Fig. 3, lane 2), whereas the wild-type strain in Fig. 1 did not (lane 1). This is probably due to the difference in the growth phase of the cells. Although the cultivation of the cells in Fig. 1 was stopped once the cells reached an OD600 of about 0.8, the cells in lanes 1 and 2 of Fig. 3 were grown for an additional 30 min after the cells reached an OD600 of about 0.8 for ample overproduction of EvgA. Thus, these cells were collected at an OD600 of 1.0 to 1.2, possibly in their late log phase, a stage when transcription from the P2 promoter is induced (34). We also performed S1 mapping of proP with rpoS mutants and found that the EvgA/EvgS-dependent up-regulation of the P2 promoter was completely dependent on RpoS (data not shown).
The difference in growth phase may also explain why transcription of yrbL was enhanced without the overproduction of EvgA (Fig. 3, lane 2). Furthermore, in our previous study (5), the EvgA/EvgS-dependent up-regulation of transcription from the P2 promoter of tolC, which encodes an outer membrane channel protein, markedly decreased in the absence of phoP or phoQ, indicating that regulation of tolC also involved both EvgA/EvgS and PhoP/PhoQ systems.
Then, why should the EvgA/EvgS system enhance the expression of the PhoP regulons? The PhoP/PhoQ system regulates various genes involved in functions such as modification of the components in the bacterial cell envelope besides those known for the adaptation to Mg2+-limiting environments (8). We confirmed that deletion of phoP or phoQ in E. coli decreased the MICs of cholate and deoxycholate (data not shown), as has been reported for Salmonella (32). In the natural environment, there may be a situation where E. coli requires the activation of the PhoP/PhoQ system at high Mg2+ concentrations. We believe that this situation may overlap with the unknown signal for the EvgS sensor.
In conclusion, the present study demonstrated a novel interaction between the EvgA/EvgS and PhoP/PhoQ systems in E. coli and also identified genes that required the two systems for enhanced expression. This is one example of the intricate signal transduction networks posited to exist in E. coli.
Acknowledgments
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, CREST of JST (Japan Science and Technology), the Sasakawa research grant from the Japan Science Society, and the Hayashi Memorial Foundation for Female Natural Scientists.
REFERENCES
- 1.Aiba, H. 1983. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor of its own gene. Cell 32:141-149. [DOI] [PubMed] [Google Scholar]
- 2.Arico, B., V. Scarlato, D. M. Monack, S. Falkow, and R. Rappuoli. 1991. Structural and genetic analysis of the bvg locus in Bordetella species. Mol. Microbiol. 5:2481-2491. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban, M. J. 1976. Transposition and fusion of lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 4.Castelli, M. E., E. García Véscovi, and F. C. Soncini. 2000. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J. Biol. Chem. 275:22948-22954. [DOI] [PubMed] [Google Scholar]
- 5.Eguchi, Y., T. Oshima, H. Mori, R. Aono, K. Yamamoto, A. Ishihama, and R. Utsumi. 2003. Transcription regulation of the drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819-2828. [DOI] [PubMed] [Google Scholar]
- 6.García Véscovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 7.García Véscovi, E., Y. M. Ayala, E. DiCera, and E. A. Groisman. 1997. Characterization of the bacterial sensor protein PhoQ. Evidence for distinct binding sites for Mg2+ and Ca2+. J. Biol. Chem. 272:1440-1443. [DOI] [PubMed] [Google Scholar]
- 8.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, L., K. B. Lim, J. S. Gunn, B. Bainbridge, R. P. Darvean, M. Hackett, and S. I. Miller. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250-253. [DOI] [PubMed] [Google Scholar]
- 11.Kato, A., H. Tanabe, and R. Utsumi. 1999. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, A., H. Ohnishi, K. Yamamoto, E. Furuta, H. Tanabe, and R. Utsumi. 2000. Transcription of emrKY is regulated by the EvgA-EvgS two-component system in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 64:1203-1209. [DOI] [PubMed] [Google Scholar]
- 13.Kato, A., T. Latifi, and E. A. Groisman. 2003. Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc. Natl. Acad. Sci. USA 100:4706-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kox, L. F. F., M. M. S. M. Wösten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesley, J. A., and C. D. Waldburger. 2003. Repression of Escherichia coli PhoP-PhoQ signaling by acetate reveals a regulatory role for acetyl coenzyme A. J. Bacteriol. 185:2563-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda, N., and G. M. Church. 2002. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184:6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 19.McLeod, S. M., J. Xu, and R. C. Johnson. 2000. Coactivation of the RpoS-dependent proP P2 promoter by Fis and cyclic AMP receptor protein. J. Bacteriol. 182:4180-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellies, J., A. Wise, and M. Villarejo. 1995. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J. Bacteriol. 177:144-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Minagawa, S., H. Ogasawara, A. Kato, K. Yamamoto, Y. Eguchi, T. Oshima, H. Mori, A. Ishihama, and R. Utsumi. 2003. Identification and molecular characterization of the Mg2+-stimulon of Escherichia coli. J. Bacteriol. 185:3696-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4:161-168. [DOI] [PubMed] [Google Scholar]
- 24.Nishino, K., and A. Yamaguchi. 2001. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol. 183:1455-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishino, K., and A. Yamaguchi. 2002. EvgA of the two-component signal transduction system modulates production of the YhiUV multidrug transporter in Escherichia coli. J. Bacteriol. 184:2319-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishino, K., Y. Inazumi, and A. Yamaguchi. 2003. Global analysis of genes regulated by EvgA of the two-component regulatory system in Escherichia coli. J. Bacteriol. 185:2667-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshima, T., H. Aiba, Y. Masuda, S. Kanaya, M. Sugiura, B. L. Wanner, H. Mori, and T. Mizuno. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol. Microbiol. 46:281-291. [DOI] [PubMed] [Google Scholar]
- 28.Preston, A., E. Maxim, E. Toland, E. J. Pishko, E. T. Harvill, M. Caroff, and D. J. Maskell. 2003. Bordetella bronchiseptica PagP is a Bvg-regulated lipid A palmitoyl transferase that is required for persistent colonization of the mouse respiratory tract. Mol. Microbiol. 48:725-736. [DOI] [PubMed] [Google Scholar]
- 29.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stibitz, S., and M.-S. Yang. 1991. Subcellular localization and immunological detection of proteins encoded by the vir locus of Bordetella pertussis. J. Bacteriol. 173:4288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Utsumi, R., S. Katayama, M. Taniguchi, T. Horie, M. Ikeda, S. Igaki, H. Nakagawa, A. Miwa, H. Tanabe, and M. Noda. 1994. Newly identified genes involved in the signal transduction of Escherichia coli K-12. Gene 140:73-77. [DOI] [PubMed] [Google Scholar]
- 32.van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waldburger, C. D., and R. T. Sauer. 1996. Signal detection by the PhoQ sensor-transmitter. J. Biol. Chem. 271:26630-26636. [DOI] [PubMed] [Google Scholar]
- 34.Xu, J., and R. C. Johnson. 1995. Fis activates the RpoS-dependent stationary-phase expression of proP in Escherichia coli. J. Bacteriol. 177:5222-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, J., and R. C. Johnson. 1997. Activation of RpoS-dependent proP P2 transcription by the Fis protein in vitro. J. Mol. Biol. 270:346-359. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto, K., H. Ogasawara, N. Fujita, R. Utsumi, and A. Ishihama. 2002. Novel mode of transcription regulation of divergently overlapping promoters by PhoP, the regulator of two-component system sensing external magnesium availability. Mol. Microbiol. 45:423-438. [DOI] [PubMed] [Google Scholar]