Abstract
Fruits are an important evolutionary acquisition of angiosperms, which afford protection for seeds and ensure their optimal dispersal in the environment. Fruits can be divided into dry or fleshy. Dry fruits are the more ancient and provide for mechanical seed dispersal. In contrast, fleshy fruits develop soft tissues in which flavor compounds and pigments accumulate during the ripening process. These serve to attract animals that eat them and disseminate the indigestible seeds. Fruit maturation is accompanied by several striking cytological modifications. In particular, plastids undergo significant structural alterations, including the dedifferentiation of chloroplasts into chromoplasts. Chloroplast biogenesis, their remodeling in response to environmental constraints and their conversion into alternative plastid types are known to require communication between plastids and the nucleus in order to coordinate the expression of their respective genomes. In this review, we discuss the role of plastid modifications in the context of fruit maturation and ripening, and consider the possible involvement of organelle-nucleus crosstalk via retrograde (plastid to nucleus) and anterograde (nucleus to plastid) signaling in the process.
Keywords: tomato, fruit development, ripening, plastid, retrograde and anterograde signaling
FLOWERS ARE THE KEY FACTOR IN THE EVOLUTIONARY SUCCESS OF ANGIOSPERMS
Angiosperms are seed-producing vascular plants, in which the ovules – the precursors of the seeds – develop within the ovary. Developing seeds are enclosed inside the fruits, as also indicated by the term angiosperm, which derives from two Greek words: angeion, meaning “vessel” and sperma, meaning “seeds.” Estimates of the number of angiosperm species so far described range between 250,000–270,000 and 400,000 (Soltis et al., 2008; Magallón and Castillo, 2009), and they have established themselves in every type of terrestrial and aquatic (fresh and saltwater) habitat. In a letter to J. D. Hooker, written in July 1879 (Darwin and Seward, 1903), Charles Darwin referred to the sudden rise and rapid diversification of angiosperms as “an abominable mystery.” Researchers since have pointed to the innovative aspects of their mode of reproduction – their short reproductive cycles, flower formation, the development of closed carpels and the small size of the male and female gametophytes. The phenomenon of double fertilization, leading to formation of the diploid zygote, and the polyploid endosperm, is also thought to have contributed to the evolutionary success of the angiosperms (Haig and Westoby, 1989; Donoghue and Scheiner, 1992).
A dicot flower can be divided into four concentric but distinct whorls. The sepals of the first or outermost whorl form the calyx, while the corolla, consisting of the petals, lies in the second whorl; in the third whorl is the androecium and the gynoecium develops in the central (fourth) whorl. The female reproductive organ the gynoecium, include the carpels. Carpels are structures that are made up of an ovary and a stigma and contain one or more ovules. One or more carpels are combined into the pistil (ovary, style, stigma), forming the gynoecium as a whole. In the majority of flowering plants, fertilization is required to initiate the transition from ovule to seed, whereas the surrounding carpel(s) and, in some species, other floral organs differentiate into the fruit (Coombe, 1975). Furthermore, fruits represent a major evolutionary innovation, are essential for plant reproduction and adaptation, and greatly enhance the efficiency of seed dispersal. The ability to germinate and grow far away from the parent plant allows angiosperms to colonize new areas, reducing the risk of inbreeding and sibling competition (Willson and Traveset, 2000).
CLASSIFICATION OF ANGIOSPERM FRUITS
According to Brooks, fruits are “matured carpels with or without accessory structures and/or seeds” (Coombe, 1976). Nitsch, on the other hand, defined them as “the tissues which support the ovules and whose development is dependent upon the events occurring in these ovules.” Nitsch’s definition thus includes the “false” fruits, so called because extracarpellary tissues give rise to much of the fleshy tissue that bears or encloses the true fruits. Examples include pomes and strawberries, which form by the expansion and proliferation of the receptacle (Perkins Veazie, 1995; Velasco et al., 2010).
Based on their texture, fruits are mainly divided into two major groups: fleshy and dry. At maturity, dry fruits are characterized by dry pericarp (Simpson, 2011) and they can be further classified into dehiscent or indehiscent fruits. Dehiscent fruits open and release the mature seeds, while the indehiscent fruits do not disperse the seeds. It has been proposed that dry dehiscent fruits, found in all major clades of angiosperms, correspond to the ancestral type (Knapp, 2002), whereas the Arabidopsis silique with its specialized dehiscence zone may be a more recent evolutionary invention (Mühlhausen et al., 2013).
With regard to fleshy fruits Darwin writes “this (a fruit’s) beauty serves merely as a guide to birds and beasts in order that the fruit may be devoured and the manured seeds disseminated.” Darwin recognized that seeds protected by a fleshy fruit become more attractive for animals, which in turn play an essential role in their dispersal. The attractiveness and juiciness of fleshy fruits originate in the important cytological modifications which the parenchymal tissue undergoes during ripening – including chlorophyll degradation, accumulation of carotenoids and flavonoids, development of an aroma and flavors, and softening of the pulp (Willson and Whelan, 1990; Rodríguez et al., 2013).
Nevertheless, the specific biochemical programs that result in ripening phenomena vary among species, as highlighted by the fact that fleshy fruits can be further divided into two categories: climacteric and non-climacteric. The term “climacteric” was initially proposed to emphasize the dramatic increase in fruit respiration – marked by a burst of CO2 production (Biale, 1964). However, climacteric fruit ripening is actually stimulated by ethylene (Razali et al., 2013), although ethylene-dependent and -independent genes have been identified both in climacteric and non-climacteric fruits (Barry and Giovannoni, 2007). Intriguingly, recent data indicates that also the dry Arabidopsis silique shows a climacteric behavior as suggested by the patterns of ethylene production and respiration, and by its response to ethylene exogenous application (Kou et al., 2012).
DEVELOPMENT AND MATURATION OF THE TOMATO FRUIT
Among climacteric fleshy fruits, the tomato proved attractive to early inhabitants of the Americas, who initiated its domestication by selecting varieties with fruits larger than those of the wild ancestor Lycopersicon esculentum cv. cerasiforme (Tanksley, 2004; Peralta et al., 2006; Cong et al., 2008) – a process which has gone on up to the present day, as shown by the large collection of cultivars now in use, characterized by fruits with different sizes and shapes (Tanksley et al., 1996; Grandillo et al., 1999; Tanksley, 2004; Bai and Lindhout, 2007). Moreover, tomato fruits contribute more nutrients to the diet than any other fruit or vegetable, since they contain relatively large amounts of lycopene (Chalabi et al., 2004), vitamins C and A, potassium, folic acid and many other metabolites. Lycopene, for instance, has a strong antioxidant capacity because of its great ability to trap peroxyl radicals. Epidemiological studies recommend the consumption of foods containing high concentrations of lycopene, since it reduces the risk for certain types of cancer, including prostate cancer (Gann et al., 1999; Giovannucci et al., 2002; Jian et al., 2005).
From a botanical point of view, the tomato fruit is a berry, which can be bi- or multilocular (Figure 1). The septa of the carpels divide the ovary and the fruit into two or more locules. Seeds develop attached to the placenta, a parenchymatous tissue, which becomes gelatinous and fills the locular cavities during fruit development and maturation (Grierson and Kader, 1986; Ho and Hewitt, 1986; Bertin, 2005; Mintz-Oron et al., 2008).
FIGURE 1.
Different stages of tomato fruit development and anatomical details. (A) Tomato fruit development can be divided into different stages: IG, immature green; MG, mature green; BR, orange-breaker; and RR, red ripening stages are shown. (B) Transverse sections of fruits corresponding to the developmental stages shown in (A). p, pedicel; s, seed. Scale bar: 2 cm.
After fertilization, the ovary wall is transformed into the pericarp, which can be divided into three different structures: exocarp, mesocarp, and endocarp. The external exocarp consists of a cuticle layer that thickens as the fruit ages, and the skin, which includes an epidermal cell layer and three to four layers of a collenchymatous tissue, in which starch accumulates and few plastids are retained (Esau, 1953; Varga and Bruinsma, 1986; Joubès et al., 2000; Lemaire-Chamley et al., 2005; Mintz-Oron et al., 2008). The mesocarp, the intermediate layer, is a parenchymatous tissue formed by big cells with large vacuoles (Joubès et al., 2000; Lemaire-Chamley et al., 2005; Mintz-Oron et al., 2008). The cells of the mesocarp commonly undergo six to eight rounds of DNA duplication (endocycles) reaching ploidy levels of up to 512C (Bourdon et al., 2010) and are reminiscent of the palisade cells of leaves (Gillaspy et al., 1993) since they contain several chloroplasts, the organelle where photosynthesis occurs and produces up to 20% of fruit photosynthate, whereas the rest of photoassimilates are imported from source leaves (Hetherington et al., 1998). Nevertheless, the role of fruit photosynthesis in fruit metabolism and development is not fully understood. Early shading experiments (Tanaka et al., 1974) as well as the fruit-specific antisense inhibition of the chloroplastic Fructose 1,6-bisphosphatase (FBPase) indicated an important contribution of fruit photosynthesis to fruit yield, as shown by the reduction in weights of ripe fruits with reduced photosynthetic performance (Obiadalla-Ali et al., 2004). On the contrary, tomato lines exhibiting a fruit-specific reduction in the expression of glutamate 1-semialdehyde aminotransferase (GSA) and, as a consequence, lowered chlorophyll levels and photosynthetic activity, displayed almost no differences in fruit size and weight (Lytovchenko et al., 2011). However, these lines were characterized by a striking reduction in the rate of seed set as well as an altered seed morphology, which displayed a much reduced embryo-to-seed ratio, indicating that fruit photosynthesis is an important source of carbone assimilate for proper seed set and establishment under normal growth conditions.
Finally, the endocarp, the innermost structure, consists of a single cell layer adjacent to the locular region (Mintz-Oron et al., 2008; Xiao et al., 2009; de Jong et al., 2010).
Fertilization (stage 0) normally initiates the development of the tomato fruit, which proceeds through several major stages (Picken, 1984; Gillaspy et al., 1993; Figure 1). The first stage, immediately after fertilization, is characterized by rapid cell division, leading to a progressive increase in pericarp cell number. The end of this stage – around two weeks after pollination – is marked by a sharp fall in the rate of cell division, when the fruit is about 0.8–1 cm in diameter. During the second stage, fruit growth relies on cell expansion and leads to a significant increase in weight. Cell expansion coincides with endoreduplication (Bergervoet et al., 1996). By the end of this stage fruits have a diameter of around 2 cm. During the third phase, the fruit enters the mature green (MG) stage (Ho and Hewitt, 1986; Giovannoni, 2004; Czerednik et al., 2012) and attains its final size, which varies greatly among cultivars and is very susceptible to environmental influences (Chevalier, 2007).
Roughly 2 days after reaching the MG stage, the tomato fruit undergoes an extensive metabolic reorganization, which marks the beginning of the fruit ripening process (Ho and Hewitt, 1986). Two main phases can be distinguished, which are referred to as the breaking (BR) and the ripening (RR) stages. Conversion of chloroplasts into chromoplasts signals the BR phase, as indicated by the change in color to yellow-orange, owing to carotenoid accumulation, and concomitant chlorophyll degradation (Figure 1). Interestingly, proper ripening in tomato occurs only if fruits are harvested after having completed at least 40% of their normal growth: even exogenous application of ethylene fails to induce ripening in undeveloped locules.
At the end of the ripening process the abscission zone (AZ) is formed in the pedicel (Szymkowiak and Irish, 1999; Mao et al., 2000) to allow fruit to fall once mature. AZs differentiate at predetermined positions and contain a group of small cells lacking large vacuoles; in tomato, differentiation of the pedicel AZ is controlled by the MADS-box transcription factor JOINTLESS (Szymkowiak and Irish, 1999; Mao et al., 2000).
TOMATO: A MODEL SPECIES FOR FLESHY FRUIT STUDIES
Tomato is an ideal model plant for studying climacteric fruit ripening. Several tomato gene banks have been established and more than 75,000 accessions of tomato are maintained (Larry and Joanne, 2007; Minoia et al., 2010; Okabe et al., 2011; Saito et al., 2011). In addition, several mutants affected in fruit size, shape, development, and ripening have been isolated (Liu et al., 2002; Tanksley, 2004; Xiao et al., 2009; Rodríguez et al., 2011). Recently, the genome of Solanum lycopersicon cv. “Heinz 1706” (Tomato Genome Consortium, 2012; Aoki et al., 2013) has been fully sequenced and made publicly available. The predicted size of its diploid genome is approximately 900 megabases (Mb), distributed on 12 chromosomes, more than 75% of which is heterochromatin and largely devoid of genes. Around 33,000 genes have been predicted and some 5000 genes are preferentially expressed in fruits (Tikunov et al., 2013). With its short generation time, and the availability of a routine transformation technology, mapping populations, and microarrays of mapped DNA markers, tomato is a highly tractable experimental system. Several “omics” tools (transcriptomics, proteomics, and metabolomics) have been used to explore fruit formation and development (Alba et al., 2004; Fei et al., 2004; Fernie et al., 2004; Rose et al., 2004; Alba et al., 2005; Moore et al., 2005), leading to the genetic characterization of several important traits that have been selected during tomato domestication.
For instance several loci, named FRUIT WEIGHT (FW), have been recognized as key regulators of fruit mass (Grandillo et al., 1999; Paran and van der Knaap, 2007). Thus the FW2.2 allele increases FW by up to 30% and is found in commercial cultivars (Frary et al., 2000), whilst the small-fruited allele is present in wild tomato species. FW2.2 encodes a plasma membrane-localized protein that inhibits cell division; therefore low levels of FW2.2 mRNA promote cell cycling, leading to bigger fruits containing more and larger cells (Nesbitt and Tanksley, 2001).
Tomato fruit size is also influenced by locule number. Two loci, fasciated (f or fas) and locule number (lc), affect floral meristem size and organ/carpel number. FAS encodes a YABBY transcription factor, and it is down-regulated in the high-locule-number mutant (Barrero and Tanksley, 2004). The lc locus seems to correspond to two single nucleotide polymorphisms (SNPs) that map close to the tomato homolog of the WUSCHEL gene in Arabidopsis thaliana; however, no deregulation of WUS has been observed in low- or high-locule cultivars (Muños et al., 2011). In Arabidopsis the WUS protein is involved in stem cell maintenance, and its up-regulation leads to the formation of extra carpels (Carles et al., 2004).
GENETIC AND HORMONAL REGULATION OF FRUIT DEVELOPMENT: A TOMATO PERSPECTIVE
THE GENETICS OF FRUIT FORMATION
Fruit formation requires intimate exchange of developmental information between ovules and carpels. Signals that stimulate fruit development may be produced by pollen grains (O’Neill, 1997; O’Neill and Nadeau, 1997) and in ovules once fertilization has successfully occurred (Gillaspy et al., 1993), leading to alteration of the developmental fate of pistils from senescence to fruit set (Vercher et al., 1984; van Doorn and Woltering, 2008).
Since fruits are mature gynoecia, carpel patterning anticipates fruit architecture. Carpel identity is in turn controlled by the homeotic genes of class C, which includes all the members of the AGAMOUS sub-clade (AG; Dreni et al., 2011), named for the first member identified, in A. thaliana (Yanofsky et al., 1990; Becker and Theiβen, 2003). Several comparative studies indicate that the functions of members of the AG sub-clade are conserved from monocots to basal core eudicots (Bowman et al., 1989; Bradley et al., 1993; Pnueli et al., 1994; Mena et al., 1996; Davies et al., 1999; Pan et al., 2010; Yellina et al., 2010; Dreni et al., 2011).
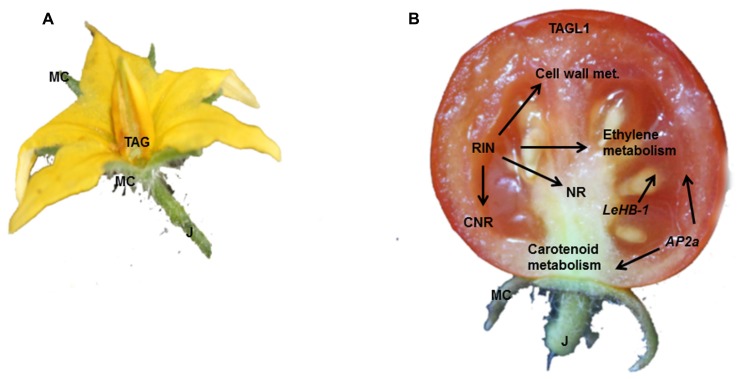
In tomato, as in snapdragon (Mizzotti et al., 2014), there are two AG-like genes (Figure 2), TAGL1 and TAG (TOMATO AG-LIKE 1 and TOMATO AG). Silencing of TAGL1 influences fruit ripening, without affecting floral organ specification (Vrebalov et al., 2009; Giménez et al., 2010; Pan et al., 2010). In particular, tagl1 fruits are characterized by a thinner pericarp, reduced firmness at the BR stage, and the maintenance of plastids in the collenchyma cells of the pericarp; consequently tagl1 fruits accumulate more chlorophyll and lutein than wild-type fruits (Itkin et al., 2009; Vrebalov et al., 2009; Table 1).
FIGURE 2.
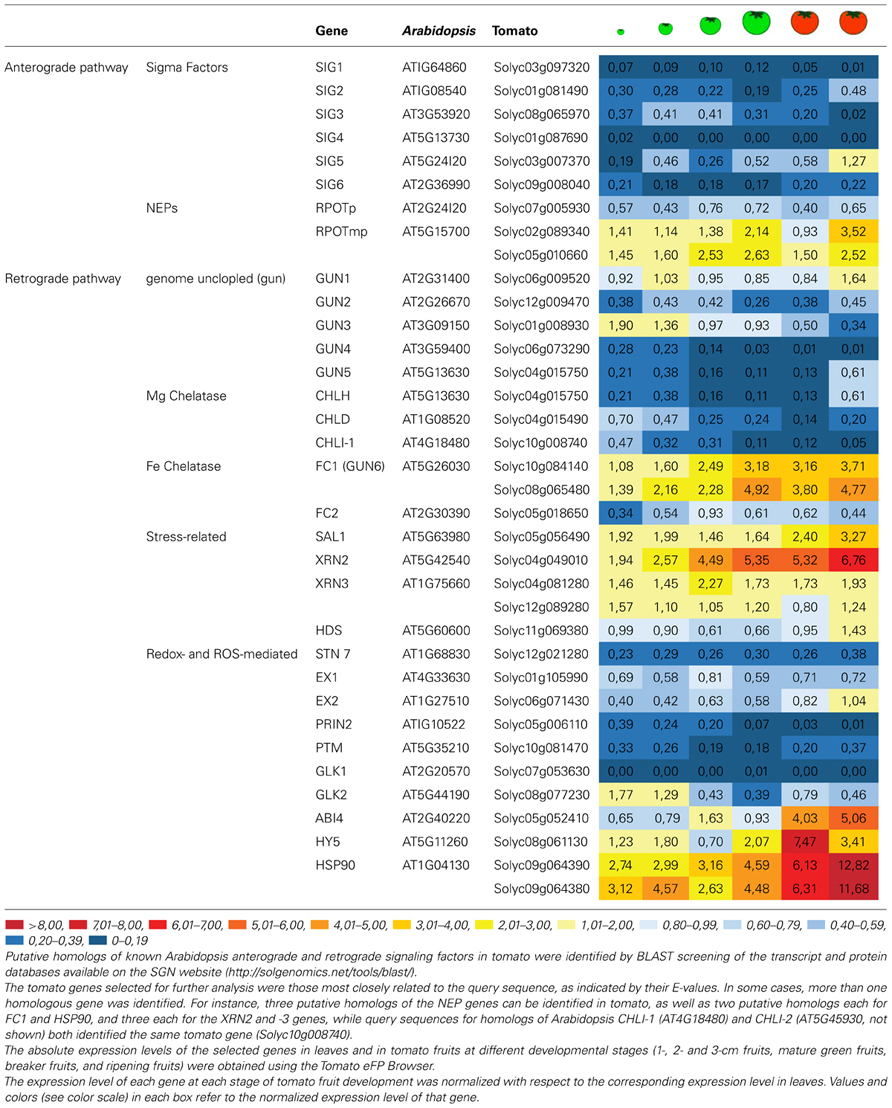
The key regulators involved in fruit commitment, formation and ripening. (A) The MADS-box transcription factors TAG controls pistil identity, whist J regulates the pedicel abscission zone differentiation. (B) TAGL1, RIN, AP2a, and LeHB-1 all impact the ethylene metabolism; although RIN controls also the transcription of genes involved in cell wall modification.
Table 1.
Mutations associated with defects in fruit maturation and ripening.
| Tomato fruit mutants | |||
|---|---|---|---|
| Mutant | Phenotype/tissues affected | Reference | Gene product |
| tag | Flower meristem and inner whorl fate determination | Pnueli et al. (1994), Pan et al. (2010) | MADS-box transcription factor |
| tagl1 | Chlorophyll and carotenoid accumulation | Vrebalov et al. (2009), Giménez et al. (2010), Pan et al. (2010) | MADS-box transcription factor |
| Plastids present in the collenchyma of the exocarp | |||
| rin | Ripening delay | Vrebalov et al. (2002) | MADS-box transcription factor |
| mc | Sepal development | Vrebalov et al. (2009) | MADS-box transcription factor |
| j | Abscission zone formation | Szymkowiak and Irish (1999), Mao et al. (2000) | MADS-box transcription factor |
| Nr | Ripening delay | Lanahan et al. (1994), Yen et al. (1995) | Ethylene receptor |
| CNR | Ripening delay | Manning et al. (2006) | SBP transcription factor |
| AP2a | Regulation of carotenoid and chlorophyll metabolism | Chung et al. (2010), Karlova et al. (2011) | AP2 transcription factor |
| slarf7 (RNAi) | Parthenocarpy | de Jong et al. (2009) | Auxin-responsive factor 7 |
| gr | Ripening delay | Barry and Giovannoni (2006) | RTE-like proteins |
Recently, the semi-dominant insertion mutation Arlequin (Alq) has been mapped and found to correspond to an altered form of the TAGL1 gene (Giménez et al., 2010). In Alq plants, sepals are transformed into fruits which undergo a ripening process, like the true fruits originated by the pistils. Thus this mutant phenocopies transgenic lines that overexpress TAGL1 (Itkin et al., 2009; Vrebalov et al., 2009; Giménez et al., 2010).
TAG1, on the other hand, has been shown to be necessary for determination of stamens and carpels, as revealed by the effects of its down-regulation using antisense and RNAi approaches (Pnueli et al., 1994; Pan et al., 2010). Indeed, pistils are replaced by a reiteration of flowers in transgenic plants expressing TAG1 antisense (Pnueli et al., 1994) – just as in the Arabidopsis ag mutant (Yanofsky et al., 1990). In contrast, virtually complete silencing of TAG1 by RNAi does not affect pistil fate in this way instead, pistils develop into red fruits, indicating a loss of determinacy (Pan et al., 2010).
Besides AG genes, several other MADS-box transcription factors are involved in fruit formation and maturation (Figure 2). Vrebalov et al. (2002) showed that the classical mutation rin disrupts the function of RIN-MADS. RIN-MADS lies very close to another MADS-box gene, MACROCALYX (MC), which is also silenced in rin plants. However, antisense repression of RIN-MADS and MC confirmed that only RIN-MADS is necessary for tomato ripening (Table 1).
Several independent groups have described a plethora of direct targets for RIN-MADS (Ito et al., 2008; Fujisawa et al., 2011, 2013; Martel et al., 2011). Thus RIN-MADS binds to regulatory regions of several genes, whose products are involved in fruit metabolism and ripening, and transcriptionally regulates enzymes involved in cell wall (Polygalacturonase, PG; β-Galactosidase 4, TBG4; Endo-(1,4)-β-mannanase 4, LeMAN4; α-Expansin 1, LeEXP1), and carotenoid metabolism. RIN-MADS is also a master regulator of ethylene biosynthesis in developing fruits, acting via the control regions of the genes LeACS2 (1-aminocyclopropane-1-carboxylic acid synthase 2), LeACS4, LeACO1 (ACC oxidase 1). Moreover, RIN-MADS stimulates the transcription of Lipoxygenase (Lox), the product of which catalyzes the dioxygenation of 1,4 pentadiene cis-polyunsaturated fatty acids to their hydroperoxide derivatives (HPO), resulting in the production of volatile compounds that contribute to fruit flavor and aroma (Yilmaz, 2001).
RIN-MADS also binds and activates the promoter of NEVER RIPE (NR; Lanahan et al., 1994; Yen et al., 1995), which encodes an ethylene receptor protein. The loss of this protein/DNA interaction explains the delay in ripening seen in rin mutants (Klee and Tieman, 2002).
RIN-MADS also positively stimulates the transcription of colorless non-ripening (CNR), which codes for a SQUAMOSA-PROMOTER BINDING PROTEIN (Cardon et al., 1999; Manning et al., 2006) whose absence causes delay in fruit ripening and softening as a consequence of reduced ethylene production (Thompson et al., 1999). The interaction between CNR and RIN-MADS has been shown to be regulated by a complex mechanism. The CNR promoter is progressively demethylated during ripening, but in cnr mutants the promoter remains hypermethylated, which prevents RIN-MADS from binding to it (Zhong et al., 2013). Zhong et al. (2013) observed that the methylation states of several RIN-MADS targets change during ripening, indicating that progressive demethylation is necessary for RIN-MADS binding. Indeed, these authors showed that tens of thousands of sites in the tomato epigenome undergo modification during fruit development.
Transcriptomic studies suggest that many more transcription factors are involved in the regulation of ripening (Vriezen et al., 2008; Pascual et al., 2009) and recently members of the APETALA2 family have been shown to play a role in the process. For instance, the tomato APETALA2a gene (Karlova et al., 2011) participates in the control of fruit ripening by regulating genes involved in ethylene and auxin signaling, and in the differentiation of chloroplasts into chromoplasts. Down-regulation of AP2a in transgenic fruits is associated with the accumulation of β-carotene at the expense of lycopene (Chung et al., 2010). Ethylene metabolism is also controlled by the transcription factor Lycopersicum esculentum Homeobox-1 (LeHB-1), which binds to control regions of ACO1 (Lin et al., 2008).
HORMONES AND FRUIT DEVELOPMENT
Fruit development and maturation is tightly controlled by hormone homeostasis (Pandolfini, 2009). Indeed, several findings indicate that manipulation of hormone homeostasis is able to induce fruit development and ripening in the absence of fertilization – a phenomenon known as parthenocarpy.
Thus treatment of unpollinated flowers with auxins is sufficient to stimulate fruit growth in tomato and other horticultural plants, indicating that administration of the hormone can substitute for the signals provided by pollination and fertilization (Nitsch, 1952). Auxin homeostasis can be altered by manipulating its synthesis, perception or signaling. For example, AtARF8 (Arabidopsis thaliana auxin response factor 8) and tomato ARF7 have both been implicated in fruit initiation. atarf8 mutants develop parthenocarpic fruits, while tomato fruits that express the arf8-4 allele are seedless (Wang et al., 2005). Parthenocarpy can also be induced by silencing ARF7 in tomato via RNA interference (de Jong et al., 2009).
Besides auxins, gibberellins (GA) play an important role in coordinating fruit growth and seed development. Active GA induce fruit set in crop plants and in Arabidopsis (Gillaspy et al., 1993; Dorcey et al., 2009), in agreement with transcriptomic analyses showing that GA biosynthesis genes are highly expressed in pollinated ovaries (Lemaire-Chamley et al., 2005). Inhibition of GA production by paclobutrazol has negative effects on fruit growth and seed set in tomato (Serrani et al., 2007), while the transgenic tomato lines pat2 and pat3/4 show overexpression of GA biosynthetic genes in their parthenocarpic fruits (Rotino et al., 2005). This is consistent with the finding that silencing of DELLA genes (Hauvermale et al., 2012), which code for negative regulators of GA signaling, results in the development of parthenocarpic fruit in both tomato and Arabidopsis (Martì et al., 2007; Dorcey et al., 2009; Fuentes et al., 2012).
Tomato is a climacteric fruit and its ripening is dependent on an ethylene burst. Conversely, in several tomato mutants in which ripening is delayed (including rin, cnr and nr), ethylene production is compromised. Synthesis of ethylene depends on the action of two enzymes, ACC synthase (ACS) and ACC oxidase (ACO). ACS converts S-adenosylmethionine into 1-aminocyclopropane-1-carboxylate, which is subsequently transformed into ethylene by ACO. ACC synthases in tomato are encoded by a multigene family (Zarembinski and Theologis, 1994; Oetiker et al., 1997), but only LeACS2 and LeACS4 are up-regulated during climacteric fruit ripening (Olson et al., 1991; Barry et al., 1996, 2008; Baldwin et al., 2000; Alba et al., 2005; Barry and Giovannoni, 2006), and the down-regulation of LeACS2 and ACO delays ripening and the transgenic tomato fruits increase their shelf life (Xie et al., 2006).
NEVER RIPE is a semi-dominant mutation that affects one of the seven ethylene receptors (Lycopersicum esculentum ethylene receptor, LeETR1-7) present in the tomato genome. Of these seven genes, however, only LeETR4, LeETR6 and Nr (LeETR3) are strongly expressed during fruit ripening.
Green-ripe (Gr) is also a dominant non-ripening mutant (Barry and Giovannoni, 2006; Xie et al., 2013), whose phenotype is due to misexpression of the Gr gene in developing fruits and organs, where it is normally not active. GR codes for a homolog of the Arabidopsis RTE1 protein (Barry and Giovannoni, 2006), a factor that is able to bind to and modify ethylene receptors, although how it affects receptor function remains unclear.
THE CHLOROPLAST TO CHROMOPLAST TRANSITION AND NUCLEUS-PLASTID COMMUNICATION
The onset of fruit ripening and the consequent reprogramming of cellular metabolism is most strikingly reflected in the conversion of fully developed chloroplasts into chromoplasts, a type of plastid that accumulates massive amounts of colorful carotenoids to attract insects and mammals that facilitate the dispersal of the seeds contained in fleshy fruits (Egea et al., 2010).
The chloroplast to chromoplast transition involves various structural modifications, including changes in the density and size of the organelle (Rosso, 1968; Spurr and Harris, 1968; Harris and Spurr, 1969), breakdown of chlorophylls, disruption of the thylakoid membrane and the aggregation of carotenoids into crystals (Egea et al., 2011). Scanning confocal microscopy analyses indicate that at the MG stage of tomato development only chloroplasts are present, mainly located in the mesocarp cells.
During the breaker stage (BR), plastids begin to accumulate carotenoids, with the rate of accumulation of lycopene being three- to fourfold higher than that of chlorophyll decline (Trudel and Ozbun, 1970; Wu and Kubota, 2008; Egea et al., 2011).
From a structural point of view, the dedifferentiation of chloroplasts into chromoplasts begins with the breakdown of starch granules and the lysis of thylakoid membranes (Ljubesić et al., 1991; Egea et al., 2010). Concomitantly, new membranes are formed, which are derived from the plastid inner envelope and become sites of carotenoid accumulation and crystal formation (Simkin et al., 2007). During the transition plastoglobules and stromules increase in size and number (Harris and Spurr, 1969; Gray et al., 2001; Kwok and Hanson, 2004; Egea et al., 2010). Plastoglobules serve to sequester lipids and carotenoids (Klee and Giovannoni, 2011; Nogueira et al., 2013), whereas the stromules provide extra surface area for the import of novel plastid proteins (Kwok and Hanson, 2004).
The situation just described is typical for immature chromoplasts at early stages of differentiation. At the full ripening stage, the plastids in fruits are almost exclusively chromoplasts. Interestingly, using real-time recording of the transition occurring in the mesocarp tissues, Egea et al. (2011) were able to demonstrate that the transition from chloroplasts to chromoplasts occurs more synchronously within individual cells than between different cells of the fruit tissue. Moreover, since these authors found no evidence for de novo formation of plastids, they concluded that all chromoplasts originate from pre-existing chloroplasts, as previously suggested (Pyke and Howells, 2002; Waters et al., 2004; Egea et al., 2011).
Over 95% of the ~3000 proteins found in the chloroplast are encoded in the nuclear genome, translated in the cytoplasm and then imported into the organelle (Richly and Leister, 2004; Li and Chiu, 2010). Therefore, the transition from chloroplast to chromoplast must involve extensive exchange of information between the nucleus and the plastids, in order to regulate the plastid proteome and ensure that the organelle can meet the changing metabolic and energy demands of the cell (Chi et al., 2013). This notion is supported, for example, by the fact that mutation of the tomato lutescent2 locus (l2), encoding a chloroplast-targeted zinc metalloprotease, delays the onset of fruit ripening, which implies the existence of a chloroplast-derived signal that stimulates ripening (Barry et al., 2012).
Communication between plastids and the nucleus, and the nature of plastid-derived signals, have been widely studied in model organisms such as A. thaliana, and this has led to the identification of several key factors that are essential for chloroplast biogenesis (biogenic control) and adaptation to physiological and environmental conditions (operational control; for a review see Woodson and Chory, 2008; Chi et al., 2013).
Interestingly, the expression of these factors is maintained in Arabidopsis and tomato fruits at different developmental stages, suggesting a possible involvement of anterograde (nucleus-to-plastid) and retrograde (plastid-to-nucleus) signaling pathways in fruit maturation and ripening.
THE ANTEROGRADE PATHWAY
During anterograde regulation, nucleus-encoded transcriptional and post-transcriptional regulators convey information about cell type to the plastid, and nuclear genes direct the synthesis and delivery of proteins that are appropriate for the organelle’s development, division and differentiation into chloroplasts, amyloplasts, chromoplasts, and other plastid types (Leon et al., 1998; Raynaud et al., 2007). In general, nucleus-encoded post-transcriptional regulators, such as proteins of the tetratrico-peptide-repeat (TPR) and pentatrico-peptide-repeat (PPR) families (D’Andrea and Regan, 2003; Nakajima et al., 2012), bind to specific chloroplast mRNAs, and control their maturation and/or stabilization by acting as adaptors for enzymes of chloroplast RNA metabolism. Alternatively, they regulate protein synthesis initiation and/or elongation by recruiting the chloroplast translation machinery to specific mRNAs (Blatch and Lässle, 1999; Shikanai and Fujii, 2013). Through these processes, TPR, PPR, and other types of imported proteins mediate subtle regulatory changes, such as the assembly and abundance of specific protein complexes in response to developmental and environmental stimuli.
Conversely, large-scale developmental switches, such as the reprogramming that takes place during the chloroplast-to-chromoplast transition (Leon et al., 1998), lead to a general increase in transcription and in differential transcript accumulation. The plastome of higher plants is transcribed by two quite different transcription systems that originate from a cyanobacterial- and proteobacterial-like endosymbiont respectively (Maliga, 1998; Liere et al., 2011). The cyanobacterial ancestor of chloroplasts provided a eubacterial-type RNA polymerase (PEP) whose four-subunit core, comprising α, β, β′, and β′ ′ proteins, is encoded by the plastid genes rpoA, rpoB, rpoC1, and rpoC2. The PEP plays a prominent role in the expression of photosynthesis-related genes in leaf chloroplasts, but it is also present in dry seeds and is active during germination. The activity and specificity of PEP is regulated by nucleus-encoded sigma-like transcription factors (SIGs). In Arabidopsis six such sigma factors (SIG1-6) have been identified, and they appear to have distinct roles during embryonic photosynthesis (SIG5), seed maturation and germination (SIG3) and very early plant development (SIG2 and SIG6).
Two nuclear genes encode the plastid proteobacterial-like RNA polymerases (NEPs), named RPOTp and RPOTmp, the latter being targeted to and active in mitochondria also (Liere et al., 2011). NEPs are active in the expression of housekeeping genes in plastids, and they play an important role in the build-up of the plastid transcriptional and translational apparatus during stratification, germination and early seedling development.
Putative homologs of Arabidopsis anterograde signaling factors can be identified in tomato, using BLAST queries of transcript (cDNA ITAG release 2.31) and protein databases (ITAG release 2.31) available on the SGN website1. In addition, the expression patterns of the corresponding mRNAs in leaves and in tomato fruits at different times during maturation (1-cm fruit, 2-cm fruit and MG fruit, BR, and RR stages) can be assessed with the aid of the Tomato eFP browser2 (Table 2).
Table 2.
Relative expression levels of the putative homologs of Arabidopsis anterograde and retrograde factors in developing tomato fruit.
As expected, all putative homologs of sigma factors appear to be down-regulated in fruit with respect to leaves, confirming their predominant role in the PEP-mediated expression of photosynthesis-related genes. The only exception is represented by the SIG5 homolog (Solyc03g007370), which is expressed at slightly higher levels in ripening tomato fruits than in leaves.
Conversely, the three putative tomato homologs of plastid proteobacterial-like RNA polymerases (Solyc07g005930; Solyc02g089340; Solyc05g010660) display intriguing expression patterns in developing fruits. The closest homolog of RPOTp (Solyc07g005930) is down-regulated in fruit, while the other two (Solyc02g089340; Solyc05g010660), which are more similar to the RPTOPmp form found in both mitochondria and plastids, show higher expression levels relative to leaves in all the different fruit stages. In particular, their expression levels follow very similar patterns, with a first peak occurring at the MG fruit stage and a second at the ripening stage. These observations imply a very strict coordination of mitochondrial and plastid transcription activities during fruit formation and maturation.
THE RETROGRADE PATHWAY
The term retrograde signaling refers to the regulation of nuclear gene expression in response to the developmental stage and functional state of the plastids, including plastid differentiation (Enami et al., 2011). In the classical scenario, the retrograde signal is generated in the plastids, then exported, and traverses the cytosol to act in the nucleus. Several metabolites have been proposed to act as messengers during retrograde signaling. These include (1) tetrapyrroles (Mg-protoporphyrin IX or heme; Strand et al., 2003; Woodson et al., 2011); (2) 3-phosphoadenosine-5-phosphate (PAP; Estavillo et al., 2011); and (3) methylerythritolcyclodiphosphate (MEcPP; Xiao et al., 2012).
The involvement of tetrapyrroles in retrograde signaling in Arabidopsis was revealed by the identification of genome uncoupled (gun) mutants that, unlike wild type, continue to express photosynthesis-related nuclear genes including ribulose bisphosphate carboxylase small subunit (RBCS) and light harvesting complex of photosystem II (Lhcbs) even when chloroplasts have been photobleached by exposure to the herbicide norflurazon (Susek et al., 1993). In particular, GUN2, GUN3, and GUN6 are involved in the iron branch of tetrapyrrole biosynthesis leading to heme and phytochromobilin, and code for the enzymes heme oxygenase 1, phytochromobilin synthase, and Fe-chelatase 1, respectively. GUN4 and GUN5, on the other hand, operate in the magnesium branch that leads to chlorophylls, and form part of the Mg-chelatase enzymes together with CHLH, CHLD, CHLI-1, and CHLI-2 subunits (for a review, see Chi et al., 2013). However, the role of Mg-protoporphyrin IX (Mg-ProtoIX) as a plastid signal has been questioned, since its accumulation following norfluorazon treatment has not been observed in two independent studies (Mochizuki et al., 2008; Moulin et al., 2008). Consequently, it was suggested that either rapid changes in the flux through the tetrapyrrole pathway, or the accumulation of Mg-ProtoIX in a specific cellular compartment could be the origin of the plastid signal (Mochizuki et al., 2008; Moulin et al., 2008); however, these aspects deserve further investigations.
A novel role as a retrograde signaling messenger was recently assigned to PAP (Estavillo et al., 2011). PAP accumulates in the chloroplast under drought conditions or upon exposure to excess light, and functions as a mobile signal that alters nuclear RNA metabolism by inhibiting exoribonucleases (XRNs). Evidence for PAP-mediated chloroplast-to-nucleus communication came with the identification of the alx8 mutant, which exhibits constitutive up-regulation of genes normally induced by high-light stress. The alx8 phenotype is caused by a lesion in SAL1, a phosphatase that regulates PAP levels by dephosphorylating PAP to adenosine monophosphate (AMP).
Recently, a role as a retrograde signaling metabolite has been also reported for MEcPP, a precursor of isoprenoids produced by the plastidial methylerythritol phosphate (MEP) pathway (Xiao et al., 2012). This finding came from the observation that Arabidopsis plants showing constitutive expression of selected stress-responsive nuclear genes also accumulated high levels of MEcPP, as a consequence of a lesion in the enzyme HDS, which is responsible for the conversion of MEcPP to HMBPP in the plastid-specific, non-mevalonate MEP pathway.
Changes in chloroplast homeostasis are also closely associated with changes in the redox state of the thylakoid electron transport chain (Baier and Dietz, 2005), particularly the redox state of the plastoquinone pool (PQ) and increases in the levels of reactive oxygen species (ROS), which also trigger retrograde signaling processes (Apel and Hirt, 2004; Asada, 2006). Components of the redox and ROS signaling circuits have been identified by genetic analysis in A. thaliana. They include STN7, a dual-function thylakoid protein kinase required for state transitions and photosynthetic acclimation (Bonardi et al., 2005; Pesaresi et al., 2009), Executor 1 (EX1) and Executor 2 (EX2; Lee et al., 2007) – both required for 1O2-dependent nuclear gene expression changes and stress responses – and PRIN2, which has been shown to be part of the plastid RNA polymerase (PEP) machinery (Kindgren et al., 2012a).
A further retrograde signaling pathway appears to originate from perturbation of plastid gene expression (PGE) both at the level of transcription and translation (Sullivan and Gray, 1999; Woodson et al., 2013). Arabidopsis mutants defective in SIG2 and SIG6 factors have been, indeed, shown to be the source of plastid retrograde signals (Woodson et al., 2013). Moreover, based on transcriptomic analyses, the transcription-, translation- and tetrapyrrole-mediated pathways seem to converge, within the chloroplast, at the level of the GUN1 protein (Koussevitzky et al., 2007; Woodson et al., 2013). Unlike the other GUN genes, GUN1 encodes a plastid-located PPR protein that is part of the transcriptionally active plastid chromosome (pTAC). However, the molecular details of GUN1 function remain elusive.
Generally speaking, the majority of tomato proteins that share homology with Arabidopsis retrograde signaling factors are encoded by genes that show reduced expression (with respect to leaves) in the fruits at the MG, breaker and ripening stages (Table 2). This is true of the tomato homologs of GUN2 (Solyc12g009470) and GUN3 (Solyc01g008930), and the subunits of the Mg-chelatase enzymes GUN4 (Solyc06g073290), GUN5 (Solyc04g015750), CHLD (Solyc04g015490), CHLI-1, and CHLI-2 (Solyc10g008740). In contrast, GUN6 transcripts (Solyc08g065480 and Solyc08g065480) encoding ferrochelatase 1 (FC1) accumulate to relatively high levels in all fruit stages, suggesting that FC1-dependent heme synthesis might play a key role as a source of messenger molecules to coordinate plastid and nucleus activities during fruit ripening. At all events, stress-related retrograde signals like PAP and MEcPP do not appear to have a major role in fruit formation, as shown by the leaf-like levels of HDS transcripts (Solyc11g069380) and the increased accumulation of both SAL1 (Solyc05g056490) and XRN2 (Solyc04g049010) and XRN3 (Solyc04g081280) mRNAs in all stages of fruit differentiation and maturation. Similarly, all factors involved in redox- and ROS-mediated retrograde signals are encoded by genes that are only weakly transcribed in fruits, such as STN7 (Solyc12g021280), EX1 (Solyc01g105990), EX2 (Solyc06g071430), and PRIN2 (Solyc05g006110), further supporting the inference that stress-related pathways are not involved in the chloroplast-chromoplast transition.
Once retrograde signals have been generated, they must be exported to the nucleus and interact with transcription factors to regulate gene expression. Hence, the discovery in Arabidopsis of a mechanism for the transduction of a retrograde signal in the nucleus represents a major breakthrough. The GUN1-dependent retrograde pathway has recently been shown to be mediated by N-PTM, an N-terminal fragment of the transcription factor PTM that is associated with the chloroplast envelope membrane. Once formed, N-PTM is translocated to the nucleus, where it activates the expression of ABI4, an AP2-type transcription factor reported to have a general role in plastid retrograde signaling (Sun et al., 2011). This pathway, however, does not seem to play a key role during fruit maturation and ripening, as indicated by the low accumulation of Solyc10g081470 transcripts, which code for the putative homolog of PTM in tomato, at all stages of fruit development.
GLK1 and GLK2 (Golden 2-like 1 and Golden 2-like 2) are MYB-GARP transcription factors that also act downstream of plastid retrograde signaling to regulate a large set of genes encoding photosynthetic thylakoid membrane proteins (Rossini et al., 2001). Two GLK genes are found in the tomato genome (GLK1, Solyc07g053630; GLK2, Solyc08g077230; Powell et al., 2012), and GLK2 accumulates during the earliest stages of fruit maturation, when new chloroplasts are needed to keep pace with cell division and expansion. Breeders have selected tomato varieties carrying light-green fruit before ripening, and Powell et al. (2012) have demonstrated recently that the light-green trait is due to the presence of a truncated version of GLK2/Solyc08g077230. These varieties produce fruits with a reduced sugar content, as a consequence of the reduced photosynthetic performance of the mesocarp cells. In agreement with that, overexpression of either GLK1 and GLK2 resulted in dark green tomato fruit with high chlorophyll and chloroplast levels in addition to more stacked thylakoid grana and elevated starch in the fruit (Nguyen et al., 2014).
Furthermore, the decrease in accumulation of GLK2/Solyc08g-077230 transcripts at later stages in fruit development agrees with the increased accumulation, at breaker and ripening stages, of ABI4 (Koussevitzky et al., 2007), HY5 and HSP90 genes (Kindgren et al., 2012b), which are known to inhibit photosynthesis-related gene expression. This indicates that they are part of the genetic program leading to the dismantling of the thylakoid membrane and its associated photosynthetic machinery.
CONCLUSION
In this survey we have explored the genetic and the hormonal regulation of fruit formation and development in tomato. Many players in the regulation of ripening have been identified, and their action clarified. However, the exchange of information between plastids and the nucleus has not been satisfactorily explored with regard to fruit ripening, despite the fact that the dedifferentiation of chloroplasts into chromoplasts is such a spectacular aspect of the whole process. Indeed comparative analyses reveal that several genes encoding protein involved in the retrograde and anterograde signaling undergo to transcriptional regulation and these waves can be associated to important developmental checkpoints. Indeed a better comprehension of these signaling pathways will provide new molecular tools to be used in breeding programs finalized to important applicative improvements, such as increase tomato fruit quality and tomato shelf-life.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Chiara Mizzotti and Simona Masiero are supported by the Cariplo Foundation (SEEFRUIT, Grant No. 2011-2257). We thank Paul Hardy for critical reading of the manuscript.
Footnotes
REFERENCES
- Alba R., Fei Z., Payton P., Liu Y., Moore S. L., Debbie P., et al. (2004). ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J. 39 697–714 10.1111/j.1365-313X.2004.02178.x [DOI] [PubMed] [Google Scholar]
- Alba R., Payton P., Fei Z., McQuinn R., Debbie P., Martin G. B., et al. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 2954–2965 10.1105/tpc.105.036053.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Ogata Y., Igarashi K., Yano K., Nagasaki H., Kaminuma E., et al. (2013). Functional genomics of tomato in a post-genome-sequencing phase. Breed. Sci. 63 14–20 10.1270/jsbbs.63.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Asada K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141 391–396 10.1104/pp.106.082040.Several [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Lindhout P. (2007). Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Ann. Bot. 100 1085–1094 10.1093/aob/mcm150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M., Dietz K.-J. (2005). Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J. Exp. Bot. 56 1449–1462 10.1093/jxb/eri161 [DOI] [PubMed] [Google Scholar]
- Baldwin E. A., Scott J. W., Shewmaker C. K., Schuch W. (2000). Flavor trivia and tomato aroma: biochemistry and possible mechanisms for control of important aroma components. HortScience 35 1013–1022 [Google Scholar]
- Barrero L. S., Tanksley S. D. (2004). Evaluating the genetic basis of multiple-locule fruit in a broad cross section of tomato cultivars. Theor. Appl. Genet. 109 669–679 10.1007/s00122-004-1676-y [DOI] [PubMed] [Google Scholar]
- Barry C. S., Aldridge G. M., Herzog G., Ma Q., McQuinn R. P., Hirschberg J., et al. (2012). Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiol. 159 1086–1098 10.1104/pp.112.197483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C. S., Blume B., Bouzayen M., Cooper W., Hamilton A. J., Grierson D. (1996). Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 9 525–535 10.1046/j.1365-313X.1996.09040525.x [DOI] [PubMed] [Google Scholar]
- Barry C. S., Giovannoni J. J. (2006). Ripening in the tomato green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 103 7923–7928 10.1073/pnas.0602319103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C. S., Giovannoni J. J. (2007). Ethylene and fruit ripening. J. Plant Growth Regul. 26 143–159 10.1007/s00344-007-9002-y [DOI] [Google Scholar]
- Barry C. S., McQuinn R. P., Chung M.-Y., Besuden A., Giovannoni J. J. (2008). Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 147 179–187 10.1104/pp.108.118430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Theißen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29 464–489 10.1016/S1055-7903(03)00207-0 [DOI] [PubMed] [Google Scholar]
- Bergervoet J., Verhoeven H., Glilissen L., Bino R. (1996). High amounts of nuclear DNA in tomato. Plant Sci. 116 141–145 10.1016/0168-9452(96)04383-X [DOI] [Google Scholar]
- Bertin N. (2005). Analysis of the tomato fruit growth response to temperature and plant fruit load in relation to cell division, cell expansion and DNA endoreduplication. Ann. Bot. 95 439–47 10.1093/aob/mci042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biale J. B. (1964). Growth, maturation, and senescence in fruits. Science 146 880–888 10.1126/science.146.3646.880 [DOI] [PubMed] [Google Scholar]
- Blatch G. L, Lässle M. (1999). The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. BioEssays 21 932–939 [DOI] [PubMed] [Google Scholar]
- Bonardi V., Pesaresi P., Becker T., Schleiff E., Wagner R., Pfannschmidt T., et al. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437 1179–1182 10.1038/nature04016 [DOI] [PubMed] [Google Scholar]
- Bourdon M., Frangne N., Mathieu-Rivet E., Nafati M., Cheniclet C., Renaudin J.-P., et al. (2010). Endoreduplication and growth of fleshy fruits. Progr. Bot. 71 101–132 10.1007/978-3-642-02167-1_4 [DOI] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1 37–52 10.1105/tpc.1.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D., Carpenter R., Sommer H., Hartley N., Coen E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72 85–95 10.1016/0092-8674(93)90052-R [DOI] [PubMed] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237 91–104 10.1016/S0378-1119(99)00308-X [DOI] [PubMed] [Google Scholar]
- Carles C. C., Lertpiriyapong K., Reville K., Fletcher J. C. (2004). The ULTRAPETALA1 gene functions early in Arabidopsis development to restrict shoot apical meristem activity and acts through WUSCHEL to regulate floral meristem determinacy. Genetics 167 1893–1903 10.1534/genetics.104.028787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalabi N., Le Corre L., Maurizis J., Bignon Y., Bernard-Gallon D. (2004). The effects of lycopene on the proliferation of human breast cells and BRCA1 and BRCA2 gene expression. Eur. J. Cancer 40 1768–1775 10.1016/j.ejca.2004.03.028 [DOI] [PubMed] [Google Scholar]
- Chevalier C. (2007). “Cell cycle control and fruit development,” in Cell Cycle Control and Plant Development, Annual Plant Reviews Vol. 32 ed. Inzé D. (Oxford: Blackwell Publishing) 269–293 10.1002/9780470988923.ch12 [DOI] [Google Scholar]
- Chi W., Sun X., Zhang L. (2013). Intracellular signaling from plastid to nucleus. Annu. Rev. Plant Biol. 64 559–582 10.1146/annurev-arplant-050312-120147 [DOI] [PubMed] [Google Scholar]
- Chung M.-Y., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J.-D., et al. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64 936–947 10.1111/j.1365-313X.2010.04384.x [DOI] [PubMed] [Google Scholar]
- Cong B., Barrero L. S., Tanksley S. D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40 800–804 10.1038/ng.144 [DOI] [PubMed] [Google Scholar]
- Coombe B. G. (1975). Development and maturation of the grape berry. Aust. N. Z. Grapegrow. Winemak. 12 60–66 [Google Scholar]
- Coombe B. G. (1976). The development of fleshy fruits. Ann. Rev. Plant Physiol. 27 207–228 10.1146/annurev.pp.27.060176.001231 [DOI] [Google Scholar]
- Czerednik A., Busscher M., Bielen B. A. M., Wolters-Arts M., De Maagd R. A., Angenent G. C. (2012). Regulation of tomato fruit pericarp development by an interplay between CDKB and CDKA1 cell cycle genes. J. Exp. Bot. 63 2605–2617 10.1093/jxb/err451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea L. D., Regan L. (2003). TPR proteins: the versatile helix. Trends Biochem. Sci. 28 655–662 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- Darwin F., Seward A. (1903). More Letters of Charles Darwin. A Record of His Work in a Series of Hitherto Unpublished Letters, Vol. 2. London: John Murray; 10.5962/bhl.title.1477 [DOI] [Google Scholar]
- Davies B., Motte P., Keck E., Saedler H., Sommer H., Schwarz-Sommer Z. (1999). PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18 4023–4034 10.1093/emboj/18.14.4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong M., Wolters-Arts M., Feron R., Mariani C., Vriezen W. H. (2009). The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 57 160–170 10.1111/j.1365-313X.2008.03671.x [DOI] [PubMed] [Google Scholar]
- de Jong M., Wolters-Arts M., García-Martínez J. L., Mariani C., Vriezen W. H. (2010). The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J. Exp. Bot. 62 617–626 10.1093/jxb/erq293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M. J., Scheiner S. M. (1992). “The evolution of endosperm: a phylogenetic account,” in Ecology and Evolution of Plant Reproduction: New Approaches ed. Wyatt R. (New York: Chapman and Hall) 356–389 [Google Scholar]
- Dorcey E., Urbez C., Blázquez M. A., Carbonell J., Perez-Amador M. A. (2009). Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J. 58 318–332 10.1111/j.1365-313X.2008.03781.x [DOI] [PubMed] [Google Scholar]
- Dreni L., Pilatone A., Yun D., Erreni S., Pajoro A., Caporali E., et al. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23 2850–2863 10.1105/tpc.111.087007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea I., Barsan C., Bian W., Purgatto E., Latché A., Chervin C., et al. (2010). Chromoplast differentiation: current status and perspectives. Plant Cell Physiol. 51 1601–1611 10.1093/pcp/pcq136 [DOI] [PubMed] [Google Scholar]
- Egea I., Bian W., Barsan C., Jauneau A., Pech J.-C., Latché A., et al. (2011). Chloroplast to chromoplast transition in tomato fruit: spectral confocal microscopy analyses of carotenoids and chlorophylls in isolated plastids and time-lapse recording on intact live tissue. Ann. Bot. 108 291–297 10.1093/aob/mBR140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami K., Ozawa T., Motohashi N., Nakamura M., Tanaka K., Hanaoka M. (2011). Plastid-to-nucleus retrograde signals are essential for the expression of nuclear starch biosynthesis genes during amyloplast differentiation in tobacco BY-2 cultured cells. Plant Physiol. 157 518–530 10.1104/pp.111.178897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. (1953). Plant Anatomy. New York: Wiley; London: Chapman & Hall [Google Scholar]
- Estavillo G. M., Crisp P. A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23 3992–4012 10.1105/tpc.111.091033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Z., Tang X., Alba R. M., White J., Ronning C. M., Martin G. B., Tanksley S. D., et al. (2004). Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. Plant J. 40 47–59 10.1111/j.1365-313X.2004.02188.x [DOI] [PubMed] [Google Scholar]
- Fernie A. R., Trethewey R. N., Krotzky A. J., Willmitzer L. (2004). Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5 763–769 10.1038/nrm1451 [DOI] [PubMed] [Google Scholar]
- Frary A., Nesbitt T. C., Grandillo S., Knaap E., Cong B., Liu J., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289 85–88 10.1126/science.289.5476.85 [DOI] [PubMed] [Google Scholar]
- Fuentes S., Ljung K., Sorefan K., Alvey E., Harberd N. P, Østergaard L. (2012). Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell 24 3982–3996 10.1105/tpc.112.103192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Ito Y. (2011). Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol. 11:26 10.1186/1471-2229-11-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Shima Y., Ito Y. (2013). A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25 371–386 10.1105/tpc.112.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gann P. H., Ma J., Giovannucci E., Willett W., Sacks F. M., Hennekens C. H., et al. (1999). Lower prostate cancer risk in men with elevated plasma lycopene levels: results of a prospective analysis. Cancer Res. 59 1225–1230 [PubMed] [Google Scholar]
- Gillaspy G., Ben-David H., Gruissem W. (1993). Fruits: a developmental perspective. Plant Cell 5 1439–1451 10.1105/tpc.5.10.1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez E., Pineda B., Capel J., Antón M. T., Atarés A., Pérez-Martín F., et al. (2010). Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS ONE 5:e14427 10.1371/journal.pone.0014427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. J. (2004). Genetic regulation of fruit development and ripening. Plant Cell 16 170–180 10.1105/tpc.019158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci E., Rimm E. B., Liu Y., Stampfer M. J., Willett W. C. (2002). A prospective study of tomato products, lycopene, and prostate cancer risk. J. Natl. Cancer Inst. 94 391–398 10.1093/jnci/94.5.391 [DOI] [PubMed] [Google Scholar]
- Grandillo S., Ku H. M., Tanksley S. D. (1999). Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 99 978–987 10.1007/s001220051405 [DOI] [Google Scholar]
- Gray J. C., Sullivan J. A., Hibberd J. M., Hansen M. R. (2001). Stromules: mobile protrusions and interconnections between plastids. Plant Biol. 3:223–233 10.1055/s-2001-15204 [DOI] [Google Scholar]
- Grierson D., Kader A. (1986). “Fruit ripening and quality,” in The Tomato Crop: A Scientific Basis for Improvement eds Atherton J. G., Rudich J. (London: Chapman and Hall) 241–280 10.1007/978-94-009-3137-4_6 [DOI] [Google Scholar]
- Haig D., Westoby M. (1989). Selective forces in the emergence of the seed habit. Biol. J. Linn. Soc. 38 215–238 10.1111/j.1095-8312.1989.tb01576.x [DOI] [Google Scholar]
- Harris W., Spurr A. (1969). Chromoplasts of tomato fruits. II. The red tomato. Am. J. Bot. 56 380–389 10.2307/2440813 [DOI] [Google Scholar]
- Hauvermale A. L., Ariizumi T., Steber C. M. (2012). Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol. 160 83–92 10.1104/pp.112.200956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington S., Smillie R., Davies W. (1998). Photosynthetic activities of vegetative and fruiting tissues of tomato. J. Exp. Bot. 49 1173–1181 10.1093/jxb/49.324.1173 [DOI] [Google Scholar]
- Ho L., Hewitt J. (1986). “Fruit development,” in The Tomato Crop: A Scientific Basis for lmprovement eds Atherton J. G., Rudich J. (New York: Chapman and Hall) 201–239 10.1007/978-94-009-3137-4_5 [DOI] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60 1081–1095 10.1111/j.1365-313X.2009.04064.x [DOI] [PubMed] [Google Scholar]
- Ito Y., Kitagawa M., Ihashi N., Yabe K., Kimbara J., Yasuda J., et al. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55 212–223 10.1111/j.1365-313X.2008.03491.x [DOI] [PubMed] [Google Scholar]
- Jian L., Du C.-J., Lee A. H., Binns C. W. (2005). Do dietary lycopene and other carotenoids protect against prostate cancer? Int. J. Cancer 113 1010–1014 10.1002/ijc.20667 [DOI] [PubMed] [Google Scholar]
- Joubès J., Chevalier C., Dudits D., Heberle-Bors E., Inzé D., Umeda M., et al. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43 607–620 10.1023/A:1006470301554 [DOI] [PubMed] [Google Scholar]
- Karlova R., Rosin F. M., Busscher-Lange J., Parapunova V., Do P. T., Fernie A. R., et al. (2011). Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23 923–941 10.1105/tpc.110.081273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P., Kremnev D., Blanco N. E., De Dios Barajas López J., Fernández A. P., Tellgren-Roth C., et al. (2012a). The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 70 279–291 10.1111/j.1365-313X.2011.04865.x [DOI] [PubMed] [Google Scholar]
- Kindgren P., Norén L., López J. D. D. B., Shaikhali J., Strand A. (2012b). Interplay between Heat shock protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol. Plant 5 901–913 10.1093/mp/ssr112 [DOI] [PubMed] [Google Scholar]
- Klee H., Tieman D. (2002). The tomato ethylene receptor gene family: form and function. Physiol. Plant. 115 336–341 10.1034/j.1399-3054.2002.1150302.x [DOI] [PubMed] [Google Scholar]
- Klee H. J., Giovannoni J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45 41–59 10.1146/annurev-genet-110410-132507 [DOI] [PubMed] [Google Scholar]
- Knapp S. (2002). Tobacco to tomatoes: a phylogenetic perspective on fruit diversity in the Solanaceae. J. Exp. Bot. 53 2001–2022 10.1093/jxb/erf068 [DOI] [PubMed] [Google Scholar]
- Kou X., Watkins C., Gan S. (2012). Arabidopsis AtNAP regulates fruit senescence. J. Exp. Bot. 63 6139–6147 10.1093/jxb/err313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T. C., Hong F., Sachetto-Martins G., Surpin M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316 715–719 10.1126/science.1140516 [DOI] [PubMed] [Google Scholar]
- Kwok E. Y., Hanson M. R. (2004). Stromules and the dynamic nature of plastid morphology. J. Microsc. 214 124–37 10.1111/j.0022-2720.2004.01317.x [DOI] [PubMed] [Google Scholar]
- Lanahan M. B., Yen H. C., Giovannoni J. J., Klee H. J. (1994). The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6 521–530 10.1105/tpc.6.4.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larry R., Joanne L. (2007). “Genetic resources of tomato (Lycopersicon esculentum Mill) and wild relatives,” in Tomato Vol. 2 Genetic Improvement of Solanaceous Crops eds Razdan M. K., Mattoo A. K. (New Hampshire, USA: Science Publishers) 25–75 [Google Scholar]
- Lee K. P., Kim C., Landgraf F., Apel K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104 10270–10275 10.1073/pnas.0702061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley M., Petit J., Garcia V., Just D., Baldet P., Fagard M., et al. (2005). Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol. 139 750–769 10.1104/pp.105.063719.plex [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon P., Arroyo A., Mackenzie S. (1998). Nuclear control of plastid and mitochondrial development in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 453–480 10.1146/annurev.arplant.49.1.453 [DOI] [PubMed] [Google Scholar]
- Li H., Chiu C.-C. (2010). Protein transport into chloroplasts. Annu. Rev. Plant Biol. 61 157–180 10.1146/annurev-arplant-042809-112222 [DOI] [PubMed] [Google Scholar]
- Liere K., Weihe A, Börner T. (2011). The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J. Plant Physiol. 168 1345–1360 10.1016/j.jplph.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Lin Z., Hong Y., Yin M., Li C., Zhang K., Grierson D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55 301–310 10.1111/j.1365-313X.2008.03505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Van Eck J., Cong B., Tanksley S. D. (2002). A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 99 13302–13306 10.1073/pnas.162485999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubesić N., Wrischer M, Devidé Z. (1991). Chromoplasts – the last stages in plastid development. Int. J. Dev. Biol. 35 251–258 [PubMed] [Google Scholar]
- Lytovchenko A., Eickmeier I., Pons C., Osorio S., Szecowka M., Lehmberg K., et al. (2011). Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol. 157 1650–1663 10.1104/pp.111.186874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S., Castillo A. (2009). Angiosperm diversification through time. Am. J. Bot. 96 349–365 10.3732/ajb.0800060 [DOI] [PubMed] [Google Scholar]
- Maliga P. (1998). Two plastid RNA polymerases of higher plants: an evolving story. Trends Plant Sci. 3 17–19 10.1016/S1360-1385(97)87981-87982 [DOI] [Google Scholar]
- Manning K., Tor M., Poole M., Hong Y., Thompson A., King G., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SPB-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38 949–952 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- Mao L., Begum D., Chuang H., Budiman M., Szymkowiak E., Irish E., et al. (2000). JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406 910–913 10.1038/35022611 [DOI] [PubMed] [Google Scholar]
- Martel C., Vrebalov J., Tafelmeyer P., Giovannoni J. J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157 1568–79 10.1104/pp.111.181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martìl C., Orzaez D., Ellul P., Moreno V., Carbonell J., Granell A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52 865–876 10.1111/j.1365-313X.2007.03282.x [DOI] [PubMed] [Google Scholar]
- Mena M., Ambrose B., Meeley R. B., Briggs S. P., Yanofsky M. F., Schmidt R. J. (1996). Diversification of C-function activity in maize flower development. Science 274 1537–1540 10.1126/science.274.5292.1537 [DOI] [PubMed] [Google Scholar]
- Minoia S., Petrozza A., D’Onofrio O., Piron F., Mosca G., Sozio G., et al. (2010). A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 3:69 10.1186/1756-0500-3-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S., Mandel T., Rogachev I., Feldberg L., Lotan O., Yativ M., et al. (2008). Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 147 823–851 10.1104/pp.108.116004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzotti C., Galliani B. M., Masiero S. (2014). The backstage of the ABC model: the Antirrhinum majus contribution. Plant Biosyst. 148 176–186 10.1080/11263504.2013.877531 [DOI] [Google Scholar]
- Mochizuki N., Tanaka R., Tanaka A., Masuda T., Nagatani A. (2008). The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105 15184–15189 10.1073/pnas.0803245105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S., Payton P., Wright M., Tanksley S., Giovannoni J. (2005). Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae. J. Exp. Bot. 56 2885–2895 10.1093/jxb/eri283 [DOI] [PubMed] [Google Scholar]
- Moulin M., McCormac A. C., Terry M. J., Smith A. G. (2008). Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. U.S.A. 105 15178–15183 10.1073/pnas.0803054105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhausen A., Lenser T., Mummenhoff K, Theißen G. (2013). Evidence that an evolutionary transition from dehiscent to indehiscent fruits in Lepidium (Brassicaceae) was caused by a change in the control of valve margin identity genes. Plant J. 73 824–835 10.1111/tpj.12079 [DOI] [PubMed] [Google Scholar]
- Muños S., Ranc N., Botton E., Bérard A., Rolland S., Duffé P., et al. (2011). Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156 2244–2254 10.1104/pp.111.173997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y., Zenda S., Minashi K., Yano T., Tahara M., Doi T., et al. (2012). Non-surgical approach to small cell carcinoma of the esophagus: does this rare disease have the same tumor behavior as SCLC? Int. J. Clin. Oncol. 17 610–615 10.1007/s10147-011-0332-1 [DOI] [PubMed] [Google Scholar]
- Nesbitt T. C., Tanksley S. D. (2001). fw2.2 directly affects the size of developing tomato fruit, with secondary effects on fruit number and photosynthate distribution. Plant Physiol. 127 575–583 10.1104/pp.010087.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C. V., Vrebalov J. T., Gapper N. E., Zheng Y., Zhong S., Fei Z., et al. (2014). Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell. 10.1105/tpc.113.118794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch J. (1952). Plant hormones in the development of fruit. Q. Rev. Biol. 27 33–57 10.1086/398643 [DOI] [PubMed] [Google Scholar]
- Nogueira M., Mora L., Enfissi E. M., Bramley P. M., Fraser P. D. (2013). Subchromoplast sequestration of carotenoids affects regulatory mechanisms in tomato lines expressing different carotenoid gene combinations. Plant Cell 25 4560–4579 10.1105/tpc.113.116210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obiadalla-Ali H., Fernie A. R., Lytovchenko A., Kossmann J., Lloyd J. R. (2004). Inhibition of chloroplastic fructose 1,6-bisphosphatase in tomato fruits leads to decreased fruit size, but only small changes in carbohydrate metabolism. Planta 219 533–540 10.1007/s00425-004-1257-y [DOI] [PubMed] [Google Scholar]
- Oetiker J. H., Olson D. C., Shiu O. Y., Yang S. F. (1997). Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum). Plant Mol. Biol. 34 275–286 10.1023/A:1005800511372 [DOI] [PubMed] [Google Scholar]
- Okabe Y., Asamizu E., Saito T., Matsukura C., Ariizumi T., Brès C., et al. (2011). Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 52 1994–2005 10.1093/pcp/pBR134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson D. C., White J. A., Edelman L., Harkins R. N., Kende H. (1991). Differential expression of two genes for 1-aminocyclopropane-1-carboxylate synthase in tomato fruits. Proc. Natl. Acad. Sci. U.S.A. 88 5340–5344 10.1073/pnas.88.12.5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill S. (1997). Pollination regulation of flower development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 58 47–72 10.1146/annurev.arplant.48.1.547 [DOI] [PubMed] [Google Scholar]
- O’Neill S., Nadeau J. (1997). Postpollination flower development. Hortic. Rev. 19 1–58 10.1002/9780470650622.ch1 [DOI] [Google Scholar]
- Pan I. L., McQuinn R., Giovannoni J. J., Irish V. F. (2010). Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 61 1795–1806 10.1093/jxb/erq046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfini T. (2009). Seedless fruit production by hormonal regulation of fruit set. Nutrients 1 168–177 10.3390/nu1020168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran I, van der Knaap E. (2007). Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J. Exp. Bot. 58 3841–3852 10.1093/jxb/erm257 [DOI] [PubMed] [Google Scholar]
- Pascual L., Blanca J. M., Cañizares J., Nuez F. (2009). Transcriptomic analysis of tomato carpel development reveals alterations in ethylene and gibberellin synthesis during pat3/pat4 parthenocarpic fruit set. BMC Plant Biol. 9:67 10.1186/1471-2229-9-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta I., Knapp S., Spooner D. (2006). Nomenclature for wild and cultivated tomatoes. Tomato Genet. Coop. Rep. 56 6–12 [Google Scholar]
- Perkins Veazie P. (1995). Growth and ripening of strawberry fruit. Hortic. Rev. 17 267–297 10.1002/9780470650585.ch8 [DOI] [Google Scholar]
- Pesaresi P., Hertle A., Pribil M., Kleine T., Wagner R., Strissel H., et al. (2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21 2402–2423 10.1105/tpc.108.064964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picken A. (1984). A review of pollination and fruit set in the tomato (Lycopersicun esculentum Mill). J. Hortic. Sci. 59 1–13 [Google Scholar]
- Pnueli L., Hareven D., Broday L., Hurwitz C., Lifschitz E. (1994). The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6 175–186 10.1105/tpc.6.2.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell A. L., Nguyen C. V., Hill T., Cheng K. L., Figueroa-Balderas R., Aktas H., et al. (2012). Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336 1711–1715 10.1126/science.1222218 [DOI] [PubMed] [Google Scholar]
- Pyke K., Howells C. (2002). Plastid and stromule morphogenesis in tomato. Ann. Bot. 90 559–566 10.1093/aob/mcf235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Loiselay C., Wostrikoff K., Kuras R., Girard-Bascou J., Wollman F.-A., et al. (2007). Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast. Proc. Natl. Acad. Sci. U.S.A. 104 9093–9098 10.1073/pnas.0703162104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razali M., Ali Z. M., Othman R. (2013). Effects of 1-methylcyclopropene on activities of ethylene biosynthesis and cell wall degrading enzyme during ripening of papaya “Sekaki”. J. Trop. Agric. Food Sci. 41 1–13 [Google Scholar]
- Richly E., Leister D. (2004). An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329 11–16 10.1016/j.gene.2004.01.008 [DOI] [PubMed] [Google Scholar]
- Rodríguez A., Alquézar B, Peña L. (2013). Fruit aromas in mature fleshy fruits as signals of readiness for predation and seed dispersal. New Phytol. 197 36–48 10.1111/j.1469-8137.2012.04382.x [DOI] [PubMed] [Google Scholar]
- Rodríguez G. R., Muños S., Anderson C., Sim S.-C., Michel A., Causse M., et al. (2011). Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 156 275–285 10.1104/pp.110.167577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. K. C., Bashir S., Giovannoni J. J., Jahn M. M., Saravanan R. S. (2004). Tackling the plant proteome: practical approaches, hurdles and experimental tools. Plant J. 39 715–733 10.1111/j.1365-313X.2004.02182.x [DOI] [PubMed] [Google Scholar]
- Rossini L., Cribb L., Martin D. J., Langdale J. A. (2001). The maize golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 13 1231–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso S. (1968). The ultrastructure of chromoplast development in red tomatoes. J. Ultrastruct. Res. 25 307–322 10.1016/S0022-5320(68)80076-0 [DOI] [PubMed] [Google Scholar]
- Rotino G. L., Acciarri N., Sabatini E., Mennella G., Lo Scalzo R., Maestrelli A., et al. (2005). Open field trial of genetically modified parthenocarpic tomato: seedlessness and fruit quality. BMC Biotechnol. 5:32 10.1186/1472-6750-5-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Ariizumi T., Okabe Y., Asamizu E., Hiwasa-Tanase K., Fukuda N., et al. (2011). TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 52 283–296 10.1093/pcp/pBR004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrani J. C., Sanjuán R., Ruiz-Rivero O., Fos M, García-Martínez J. L. (2007). Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 145 246–257 10.1104/pp.107.098335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T., Fujii S. (2013). Function of PPR proteins in plastid gene expression. RNA Biol. 10 1446–1456 10.4161/rna.25207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A. J., Gaffé J., Alcaraz J.-P., Carde J.-P., Bramley P. M., Fraser P. D., et al. (2007). Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 68 1545–1556 10.1016/j.phytochem.2007.03.014 [DOI] [PubMed] [Google Scholar]
- Simpson M. G. (2011). Plant Systematics, 2nd Edn London: Elsevier-Academic Press [Google Scholar]
- Soltis D., Bell C., Kim S., Soltis P. (2008). Origin and early evolution of the angiosperms. Ann. N. Y. Acad. Sci. 1133 3–25 10.1196/annals.1438.005 [DOI] [PubMed] [Google Scholar]
- Spurr A., Harris W. (1968). Ultrastructure of chloroplasts and chromoplasts in Capsicum annuum. I. Thylakoid membrane changes during fruit ripening. Am. J. Bot. 55 1210–1224 10.2307/2440743 [DOI] [Google Scholar]
- Strand A., Asami T., Alonso J., Ecker J. R., Chory J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421 79–83 10.1038/nature01250.1 [DOI] [PubMed] [Google Scholar]
- Sullivan J., Gray J. (1999). Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Feng P., Xu X., Guo H., Ma J., Chi W., et al. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2 477 10.1038/ncomms1486 [DOI] [PubMed] [Google Scholar]
- Susek R. E., Ausubel F. M., Chory J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74 787–799 10.1016/0092-8674(93)90459-4 [DOI] [PubMed] [Google Scholar]
- Szymkowiak E. J., Irish E. E. (1999). Interactions between jointless and wild-type tomato tissues during development of the pedicel abscission zone and the inflorescence meristem. Plant Cell 11 159–175 10.1105/tpc.11.2.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Fujita K., Kikuchi K. (1974). Nutrio-physiological studies on the tomato plant. I. Outline of growth and nutrient absorption. Soil Sci. Plant Nutr. 20 57–68 10.1080/00380768.1974.10433228 [DOI] [Google Scholar]
- Tanksley S. D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16 181–189 10.1105/tpc.018119.S182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S. D., Grandillo S., Fulton T. M., Zamir D., Eshed Y., Petiard V., et al. (1996). Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl. Genet. 92 213–224 10.1007/BF00223378 [DOI] [PubMed] [Google Scholar]
- Thompson A., Tor M., Barry C., Vrebalov J., Orfila C., Jarvis M., et al. (1999). Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 120 383–390 10.1104/pp.120.2.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y. M., Molthoff J., De Vos R. C. H., Beekwilder J., Van Houwelingen A., Van der Hooft J. J. J., et al. (2013). NON-smoky glycosyltransferase1 prevents the release of smoky aroma from tomato fruit. Plant Cell 25 3067–3078 10.1105/tpc.113.114231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485 635–641 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudel M., Ozbun J. (1970). Relationship between chlorophylls and carotenoids of ripening tomato fruit as influenced by potassium nutrition. J. Exp. Bot. 21 881–886 10.1093/jxb/21.4.881 [DOI] [Google Scholar]
- van Doorn W. G., Woltering E. J. (2008). Physiology and molecular biology of petal senescence. J. Exp. Bot. 59 453–480 10.1093/jxb/erm356 [DOI] [PubMed] [Google Scholar]
- Varga A., Bruinsma J. (1986). “Tomato,” in CRC Handbook of Fruit Set and Development ed. Monselise S. P. (Boca Raton, FL: CRC Press) 461–491 [Google Scholar]
- Velasco R., Zharkikh A., Affourtit J., Dhingra A., Cestaro A., Kalyanaraman A., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42 833–839 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Vercher Y., Molowny A., López C., García-Martínez J., Carbonell J. (1984). Structural changes in the ovary of Pisum sativum L. induced by pollination and gibberellic acid. Plant Sci. Lett. 36 87–91 10.1016/0304-4211(84)90240-2 [DOI] [Google Scholar]
- Vrebalov J., Pan I. L., Arroyo A. J. M., McQuinn R., Chung M., Poole M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21 3041–3062 10.1105/tpc.109.066936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., et al. (2002). A MADS-box gene necessary for ripening at the tomato ripening-inhibitor (rin) locus. Science 296 343–346 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- Vriezen W. H., Feron R., Maretto F., Keijman J., Mariani C. (2008). Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 177 60–76 10.1111/j.1469-8137.2007.02254.x [DOI] [PubMed] [Google Scholar]
- Wang H., Jones B., Li Z., Frasse P., Delalande C., Regad F., et al. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17 2676–2692 10.1105/tpc.105.033415.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T., Fray R. G., Pyke K. A. (2004). Stromule formation is dependent upon plastid size, plastid differentiation status and the density of plastids within the cell. Plant J. 39 655–667 10.1111/j.1365-313X.2004.02164.x [DOI] [PubMed] [Google Scholar]
- Willson M. F., Traveset A. (2000). “The ecology of seed dispersal,” in Seeds: The ecology of regeneration in plant communities 2nd Edn ed. Fenner M. (Wallingford: CAB International) 85–110 10.1079/9780851994321.0085 [DOI] [Google Scholar]
- Willson M. F., Whelan C. J. (1990). The evolution of fruit color in fleshy-fruited plants. Am. Nat. 136 790–809 10.1086/285132 [DOI] [Google Scholar]
- Woodson J. D., Chory J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9 383–395 10.1038/nrg2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J. D., Perez-Ruiz J. M., Chory J. (2011). Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21 897–903 10.1016/j.cub.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J. D., Perez-Ruiz J. M., Schmitz R. J., Ecker J. R., Chory J. (2013). Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 73 1–13 10.1111/tpj.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Kubota C. (2008). Effects of high electrical conductivity of nutrient solution and its application timing on lycopene, chlorophyll and sugar concentrations of hydroponic tomatoes during ripening. Sci. Hortic. 116 122–129 10.1016/j.scienta.2007.11.014 [DOI] [Google Scholar]
- Xiao H., Radovich C., Welty N., Hsu J., Li D., Meulia T., et al. (2009). Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol. 9:49 10.1186/1471-2229-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Savchenko T., Baidoo E. E. K., Chehab W. E., Hayden D. M., Tolstikov V., et al. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149 1525–1535 10.1016/j.cell.2012.04.038 [DOI] [PubMed] [Google Scholar]
- Xie F., Liu Q., Wen C.-K. (2006). Receptor signal output mediated by the ETR1 N terminus is primarily subfamily I receptor dependent. Plant Physiol. 142 492–508 10.1104/pp.106.082628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhu B., Luo Y., Chen X., Zhang H. (2013). Preparation of polyclonal antibody and expression analysis of Gr in tomato. Adv. Technol. Teach. 163 239–247 10.1007/978-3-642-29458-7 [DOI] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 35–39 10.1038/346035a0 [DOI] [PubMed] [Google Scholar]
- Yellina A. L., Orashakova S., Lange S., Erdmann R., Leebens-Mack J., Becker A. (2010). Floral homeotic C function genes repress specific B function genes in the carpel whorl of the basal eudicot California poppy (Eschscholzia californica). EvoDevo 1 13 10.1186/2041-9139-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H., Lee S., Tanksley S. D., Lanahan M. B., Klee H. J., Giovannoni J. J. (1995). The tomato Never-ripe locus regulates ethylene-inducible gene expression and is linked to a homolog of the Arabidopsis ETR1 gene. Plant Physiol. 1343–1353 10.1104/pp.107.4.1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz E. (2001). Oxylipin pathway in the biosynthesis of fresh tomato volatiles. Turk. J. Biol. 25 351–360 [Google Scholar]
- Zarembinski T. I., Theologis A. (1994). Ethylene biosynthesis and action: a case of conservation. Plant Mol. Biol. 26 1579–1597 10.1007/BF00016491 [DOI] [PubMed] [Google Scholar]
- Zhong S., Fei Z., Chen Y., Zheng Y., Huang M., Vrebalov J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31 154–159 10.1038/nbt.2462 [DOI] [PubMed] [Google Scholar]