Abstract
Type II secretion machinery is composed of 12 to 15 proteins for translocating extracellular proteins across the outer membrane. XpsL, XpsM, and XpsN are components of such machinery in the plant pathogen Xanthomonas campestris pv. campestris. All are bitopic cytoplasmic-membrane proteins, each with a large C-terminal periplasmic domain. They have been demonstrated to form a dissociable ternary complex. By analyzing the C-terminally truncated XpsN and PhoA fusions, we discovered that truncation of the C-terminal 103 residues produced a functional protein, albeit present below detectable levels. Furthermore, just the first 46 residues, encompassing the membrane-spanning sequence (residues 10 to 32), are sufficient to keep XpsL and XpsM at normal abundance. XpsN46(His6), synthesized in Escherichia coli, is able to associate in a membrane-mixing experiment with the XpsL-XpsM complex preassembled in X. campestris pv. campestris. The XpsN N-terminal 46 residues are apparently sufficient not only for maintaining XpsL and XpsM at normal levels but also for their stable association. The membrane-spanning sequence of XpsN was not replaceable by that of TetA. However, coimmunoprecipitation with XpsL and XpsM was observed for XpsN97::PhoA, but not XpsN46::PhoA. Only XpsN97::PhoA is dominant negative. Single alanine substitutions for three charged residues within the region between residues 47 and 97 made the protein nonfunctional. In addition, the R78A mutant XpsN protein was pulled down by XpsL-XpsM(His6) immobilized on an Ni-nitrilotriacetic acid column to a lesser extent than the wild-type XpsN. Therefore, in addition to the N-terminal 46 residues, the region between residues 47 and 97 of XpsN probably also plays an important role in interaction with XpsL-XpsM.
In the plant pathogen Xanthomonas campestris pv. campestris, several hydrolytic enzymes, including α-amylase, protease, pectate lyase, and cellulase, are secreted by the type II secretion machinery composed of a minimum of 12 Xps proteins (7). Among these components, the XpsN protein is a bitopic cytoplasmic membrane protein with a long C-terminal segment facing the periplasm. It exhibits an interactive relationship with two other cytoplasmic membrane proteins, XpsL and XpsM, as suggested by its requirement for an abundance of these two proteins (10). XpsL-XpsM complex formation has been demonstrated by immunoprecipitation data (10). In the absence of XpsN, the XpsL-XpsM complex tends to dissociate, probably leading to protein instability (23). Association of the XpsN protein with the XpsL-XpsM complex was confirmed by metal-chelating chromatography analysis, followed by immunoprecipitation (23). In addition, the XpsN protein interacts with the outer membrane protein XpsD, belonging to the secretin protein family (14), that serves as the secretion pore by forming a homomultimeric complex (3). Association between the two proteins was demonstrated by their coimmunoprecipitation and coelution on affinity chromatography (11). Thus, as a dissociable component of the XpsL-XpsM-XpsN complex located in the cytoplasmic membrane (23), the XpsN protein serves as an apparent intermediary between the ternary complex and the secretion pore located in the outer membrane.
Evidence implying similar interactive relationships for the PulC protein of Klebsiella oxytoca and the XcpPC protein of Pseudomonas aeruginosa is accumulating. A possible interactive relationship between PulC and the secretin PulD of K. oxytoca was implied by the observation that oligomerization of the former is dependent on the presence of the latter (15, 16). On the other hand, mutation in the pulC or pulM gene made the PulL protein sensitive to proteinase K treatment (16). Possible complex formation among PulC, PulL, and PulM was suggested by immunoprecipitation experiments using antibody against the PulM protein (16). Similarly, the abundance of the P. aeruginosa XcpPC protein was significantly reduced in mutants lacking a functional xcpYL or xcpZM gene (5), and it also became less abundant in an xcpQD mutant (1). Furthermore, XcpPC was demonstrated to modulate the stabilizing activity of XcpZM on XcpYL (18). These observations suggest that the PulC and XcpPC proteins are similar to the XpsN protein in interacting with analogous partners, the secretin in the outer membrane and two cytoplasmic membrane proteins. In spite of their low sequence similarity, these three proteins are alike in their molecular weights, subcellular locations, and topologies in the membrane. In addition, the pulN gene of K. oxytoca was clearly shown to be nonessential for type II secretion (16). In contrast, analogues of the pulC and xcpPC genes have so far not been identified in X. campestris pv. campestris or in its related species, Xylella fastidiosa (22) and X. campestris pv. citri (4). Accordingly, we propose here to change the category of the XpsN protein from its original assignment as GspN (11) to GspC.
To learn more about how the XpsN protein forms a complex with XpsL-XpsM and keeps the two proteins at normal abundance, we took the genetic approach by constructing various types of mutant xpsN genes and analyzing their interactive relationships with XpsL and XpsM. By making use of the random-insertion property of the transposon TnphoA/in (13), we obtained various C-terminally truncated XpsN proteins that were derived from XpsN::PhoA fusion proteins. Functional assay showed that the N-terminal 158 residues suffice for normal function. The N-terminal 46 residues have the capability to interact with XpsL and XpsM; however, this interaction appears weaker than that displayed by the N-terminal 97 residues. It is sensitive to structural perturbation caused by fusion at the C terminus with PhoA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phage.
Escherichia coli BL21(DE3) was a kind gift from F. W. Studier. E. coli CC191 and phage λ TnphoA/in were kindly provided by C. Manoil. X. campestris pv. campestris XC1701 was originally isolated as a spontaneous Rifr mutant from a wild-type isolate, XC17. XC17433 is a secretion mutant strain in which the entire xps gene cluster (xpsEFGHIJKLMND) was deleted. XC1709 is a chromosomal nonpolar mutant strain with in-frame deletion of the xpsN gene. All other bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strain | ||
| XL1-Blue | F′::Tn10 proA+B+ lacIq Δ(lacZ)M15/recA1 endA1 gyrA96(Nalr) thi hsdR17(rK+ mK+) supE44 relA1 lac | Stratagene |
| CC191 | F128 lac1q Δ(lacZ)M15 traD/Δ(ara-leu)7967 araD139 Δ(lac)X74 galE galK thi rpsE phoA20 rpoB argE(Am) recA1 | 13 |
| BL21(DE3) | hsdS gal (λcIts857ind1 Sam7 nin5 lacUV5-T7 gene 1) | 21 |
| Plasmid | ||
| Vector | ||
| pET32a | Apr; PT7-containing expression vector | Novagene |
| pSelect 1 | Tcr; phagemid vector; ColE1 replicon with an f1(−) ori | Promega |
| pBluescript SK(−) and KS(−) | Apr; f1(−) ori ColE1 replicon with multiple cloning sites in the order SacI-KpnI or KpnI-SacI, respectively | Stratagene |
| pCPP30 | Tra− Mob+ Tcr IncP replicon with multiple cloning sites downstream of Plac | D. W. Bauer |
| pBBR1MCS-5 | Tra− Mob+ Gmr; compatible with IncP, IncQ, IncW, ColE1- and P15a-based replicons, with multiple cloning sites downstream of Plac | 9 |
| Expression of wild-type and C-terminally truncated XpsN proteins | ||
| pNC2 | Wild-type XpsN expressed from Plac on pCPP30 | 11 |
| p46 | XpsNΔ47-261 expressed from Plac on pCPP30 | This study |
| p61 | XpsNΔ62-261 expressed from Plac on pCPP30 | This study |
| p97 | XpsNΔ98-261 expressed from Plac on pCPP30 | This study |
| p110 | XpsNΔ111-261 expressed from Plac on pCPP30 | This study |
| p117 | XpsNΔ118-261 expressed from Plac on pCPP30 | This study |
| p156 | XpsNΔ157-261 expressed from Plac on pCPP30 | This study |
| p158 | XpsNΔ159-261 expressed from Plac on pCPP30 | This study |
| p231 | XpsNΔ232-261 expressed from Plac on pCPP30 | This study |
| pHL1 | BamHI 0.3-kb PCR fragment containing the sequence encoding residues 1-97 of xpsN cloned in pET32a | This study |
| pHL2 | Derivative of pHL1 from which an XhoI fragment was deleted; encoding TrxA-XpsN97 with an N- and a C-terminal His6 tag | This study |
| pHL3 | Derivative of pHL2 from which an NdeI fragment was deleted; encoding the C-terminally His6-tagged XpsN97 and the XpsN N-terminal 97 residues expressed from PT7 on pET32a | This study |
| pHL6 | Derivative of pHL1 from which an NdeI fragment was deleted; encoding XpsN97 without a His6 tag; expressed from PT7 on pET32a | This study |
| pHM46 | DNA fragment amplified from pHL3, containing the sequence encoding residues 1-46 of xpsN cloned in pET32a as an XbaI-BamHI fragment | This study |
| pHM46H | DNA fragment amplified from pHL3 containing the sequence encoding residues 1-46 of xpsN cloned in pET32a as an XbaI-XhoI fragment in frame with a C-terminal His6 tag | This study |
| Expression of XpsN::PhoA fusion proteins | ||
| p46PhoA | XpsN46::PhoA expressed from Plac on pCPP30 | This study |
| p61PhoA | XpsN61::PhoA expressed from Plac on pCPP30 | This study |
| p97PhoA | XpsN97::PhoA expressed from Plac on pCPP30 | This study |
| Expression of XpsN N-terminal mutant proteins | ||
| pNΔ2 | XpsNΔ6-25 expressed from Plac on pCPP30 | This study |
| pAN2 | Amylase/XpsN expressed from Plac on pCPP30 | This study |
| pTNR2 | TetA/XpsN expressed from Plac on pCPP30 | This study |
| Expression of XpsN proteins with alanine substitutions | ||
| pCPPK64A | XpsNK64A expressed from Plac on pCPP30 | This study |
| pCPPE67A | XpsNE67A expressed from Plac on pCPP30 | This study |
| pCPPH71A | XpsNH71A expressed from Plac on pCPP30 | This study |
| pCPPR78A | XpsNR78A expressed from Plac on pCPP30 | This study |
| Coexpression of XpsL and XpsM | ||
| pCPP-LM | XpsL and XpsM coexpressed from Plac on pCPP30 | 10 |
| pRT40 | XpsL and XpsM(His6); XpsM with a C-terminal His6 tag, coexpressed from Plac on a broad-host-range vector, pBBR1MCS-5 | 23 |
| Plasmids used in mutagenesis | ||
| pND1 | 3.5-kb HindIII-EcoRI fragment containing xpsN and xpsD genes in pSelect 1 | This study |
| pND3m | 3.5-kb HindIII-EcoRI fragment containing mutated xpsN with newly created NcoI and NheI sites and the wild-type xpsD gene in pBluescript SK(−) | This study |
| pNC3 | 1.2-kb HindIII-BclI fragment carrying the xpsN gene subcloned in pBluescript SK(−) | This study |
| pHM116X | 1.2-kb HindIII-BclI fragment in pNC3 from which two BamHI sites were removed subcloned in pBluescript KS(−) | This study |
| pANX | 3.5-kb HindIII-EcoRI fragment containing amylase/xpsN and wild-type xpsD gene in pBluescript SK(−) | This study |
Construction of xpsN mutants mutated at the N terminus.
As a first step, two restriction sites, NcoI and NheI, were introduced into the xpsN gene through site-directed mutagenesis using the oligonucleotides pNNcoI and pNNheI, whose sequences are listed in Table 2. The NcoI site was introduced upstream of the 4th residue, and the NheI site was introduced at the 26th residue of the XpsN protein. The plasmid pND1, a derivative of pSelect 1 containing a 3.5-kb HindIII-EcoRI fragment that spans the xpsN and xpsD genes, was used as the template for mutagenesis. The derivative containing both the NcoI and NheI sites was subcloned into pBluescript SK(−) and named pND3m. Site-directed mutagenesis was conducted using the GeneEditor in vitro site-directed mutagenesis system (Promega), following the procedures suggested by the supplier.
TABLE 2.
Primer list
| Primer | Nucleotide sequence (5′→3′)a |
|---|---|
| M13 reverse | AGCGGATAACAATTTCACACAGGA |
| T7 | TAATACGACTCACTATAGGG |
| pNNcoI | TGCGCAGGCCGACCATGGCAAGGCGCATTGCT |
| pNNheI | GGCCACGGCTAGCACGCAGACCCA |
| pAmyFor | AAAACCATGGTGCACGCCACCTCTCGCCCCT |
| pAmyRev | AAAGGCTAGCACGATGACGTCGGCCTGTGCGCT |
| pTetAFor | ACCACCATGGTCAAATCTAACAATGCGCTCAT |
| pTetARev | AAAGGCTAGCACCGGCATAACCAAGCCTAT |
| pNxBam | AATCGGCCGGGTCCAGCGTCAGCGT |
| pNstop | GATCTCGAGTGATCACTCGA |
| pN46Rev | CGGGATCCTCACTCGAGCTGCACCAGGGCCGGGTCATC |
| pN64For | GAGCGGCTGGGGCCGTTCGAGGCCTACGCCGAGATCGCCGCGCAT |
| pN64Rev | ATGCGCGGCGATCTCGGCGTAGGCCTCGAACGGCCCCAGCCGCTC |
| pN67For | GGGCCGTTCGAGAAATACGCCGCGATCGCCGCGCATCCGGCGTTT |
| pN67Rev | AAACGCCGGATGCGCGGCGATCGCGGCGTATTTCTCGAACGGCCC |
| pN71For | AAATACGCCGAGATCGCCGCGGCTCCGGCGTTTGCCGAAGATCGC |
| pN71Rev | GCGATCTTCGGCAAACGCCGGAGCCGCGGCGATCTCGGCGTATTT |
| pN78For | CATCCGGCGTTTGCCGAAGACGCGTTGCCGCACCCGTTCTTCCTGTCC |
| pN78Rev | CAGGAAGAACGGGTGCGGCAACGCGTCTTCGGCAAACGCCGGATG |
| pN97For | CACTTGGGATCCCATATGCGCCTTGAAATGAT |
| pN97Rev | CACTGGGGATCCTCACTCGAGCACCGTGGAGGCGGCGCC |
Mutated bases are in boldface. The NcoI (CCATGG), NheI (GCTAGC), BamHI (GGATCC), and XhoI (CTCGAG) restriction sites are underlined, and the NdeI (CATATG) site is in italics.
The gene encoding the Amy-XpsN chimera was constructed by replacing the NcoI-NheI fragment with a PCR fragment encoding the α-amylase signal peptide (residues 2 to 38). The DNA fragment was amplified using the primers pAmyFor and pAmyRev (Table 2). The PCR product was digested with NcoI and NheI and subsequently ligated with pND3m cut with the same enzymes. The sequencing results revealed that an extra NcoI site was unexpectedly created in the upstream region of the amyl-xpsN gene. To make the correction, the HindIII-NcoI fragment upstream of the amy-xpsN gene was replaced with a PCR fragment amplified from the wild-type xpsN gene using the primer pNNcoI and the M13 reverse primer (Table 2). The corrected end product, confirmed by sequencing data, was named pANX. In order to introduce the chimeric gene into X. campestris pv. campestris, a 1.2-kb HindIII-BclI fragment spanning the entire amy-xpsN gene was excised from pANX and ligated with the broad-host-range vector pCPP30 digested with HindIII and BamHI, resulting in the plasmid pAN2.
The gene encoding the TetA-XpsN chimera was constructed by replacing the NcoI-NheI fragment in the amyl-xpsN gene with a PCR fragment encoding the first membrane-spanning sequence of TetA (residues 2 to 26) and amplified with the primer pair pTetAFor and pTetARev (Table 2). The PCR products digested with NcoI and NheI were ligated with pANX digested with the same enzymes. Similarly, the chimeric gene was cut out as a 1.2-kb HindIII-BclI fragment and ligated into pCPP30 at HindIII and BamHI sites, resulting in the plasmid pTNR2.
The gene encoding XpsNΔ6-25 was constructed by deleting the NcoI-NheI fragment in the plasmid pANX. Following its cleavage with NcoI and NheI, both ends of the linearized plasmid were filled in with Klenow and ligated together. Again, the mutated gene was moved as a 1.2-kb HindIII-BclI fragment into pCPP30, generating the plasmid pNΔ2.
Construction of xpsN::phoA, xpsN::i31, and C-terminal deletion mutants of xpsN.
The xpsN mutants were constructed by a stepwise method modified from the procedures of Manoil and Bailey (13). Before conducting transposon mutagenesis, it is necessary to eliminate the two BamHI sites present on the plasmid pNC3, a derivative of pBluescript SK(−) containing a 1.2-kb fragment spanning the xpsN gene. To create a silent mutation at the BamHI site located in the xpsN gene, we performed site-directed mutagenesis using the oligonucleotide pNxBam (Table 2). To eliminate the BamHI site located downstream of the xpsN gene, the plasmid was linearized with BamHI, followed by filling in with Klenow and self-ligation. The derivative was then subcloned into pBluescript KS(−) and named pHM116X.
To generate the xpsN::phoA mutants, we transformed pHM116X into E. coli CC191 and transfected it with the λ phage carrying the transposon TnphoA/in. Plasmids isolated from PhoA-positive colonies were examined with restriction enzymes to determine the transposon insertion sites. Subsequently, each xpsN::phoA mutant was digested with BamHI and self-ligated to remove almost the entire transposon. The remaining 93 bases generate a coding sequence for 31 amino acid residues (called i31). The derivatives from such treatment were designated xpsN::i31. Fusion with PhoA is not permissive for secretion in any case.
To create the C-terminal deletion mutants of xpsN, we inserted a linker fragment containing a stop codon at the BamHI site that is left at the original transposon insertion site in each xpsN::i31 mutant. The linker was obtained by self-annealing of the oligomer pNstop (Table 2). Insertion of the linker introduced an in-frame TGA stop codon within i31, causing translation of the XpsN protein prematurely terminated at the original transposon insertion site.
All of the above-mentioned xpsN mutants subcloned in the broad-host-range vector pCPP30 were conjugated into the wild-type strain XC1701 or the xpsN mutant XC1709 for further analysis.
Construction of alanine substitution mutants of xpsN.
The four alanine substitution mutants, xpsNK64A, xpsNE67A, xpsNH71A, and xpsNR78A, were generated by PCR mutagenesis (6), using the plasmid pNC3 as a template and the paired complementary oligonucleotides pN64For/pN64Rev, pN67For/pN67Rev, pN71For/pN71Rev, and pN78For/pN78Rev, respectively. PCR was performed using Pfu Turbo DNA polymerase. The PCR products were digested with DpnI, followed by transformation into E. coli XL1-Blue. After being confirmed by DNA sequencing, the mutated genes were subcloned into pCPP30 at the HindIII and XbaI sites, giving rise to the plasmids pCPPK64A, pCPPE67A, pCPPH71A, and pCPPR78A encoding XpsNK64A, XpsNE67A, XpsNH71A, and XpsNR78A, respectively. The sequences of the oligonucleotides are listed in Table 2.
Overexpression of XpsN46/XpsN97 with or without a His6 tag.
PCR-amplified N-terminal fragments of XpsN were introduced downstream of the T7 promoter for overexpression in E. coli BL21(DE3). For amplification of the N-terminal 46 residues, the oligomers T7 and pN46Rev (Table 2) were used as the forward and reverse primer, respectively. For amplification of the N-terminal 97 residues, a forward primer, pN97For, and a reverse primer, pN97Rev (Table 2), were used. Both reverse primers were designed so that it would be possible for each amplified fragment to fuse with or without a C-terminal His6 tag. In the primer pN97For (Table 2), a BamHI site was present immediately upstream of the NdeI site, which provided the start codon for translating XpsN97. In the primer pN97Rev (Table 2), a TGA stop codon was introduced between the XhoI and BamHI sites. The PCR product was first digested with BamHI and inserted into the expression vector pET32a, giving rise to the plasmid pHL1. To remove the stop codon present in pHL1, it was digested with XhoI, followed by self-ligation, generating the plasmid pHL2, in which the xpsN97 gene is fused at its N terminus with the trxA gene. The trxA gene in pHL2 was removed by deleting an NdeI fragment, producing the plasmid pHL3, which expresses XpsN97(His6). Deletion of the NdeI fragment spanning trxA in pHL1, in which a stop codon is present downstream of xpsN97, generated the plasmid pHL6, which expresses XpsN97 without the His6 tag. The plasmid pHL3 was used as a template for amplifying the N-terminal 46 residues. The plasmid pHM46, encoding XpsN46 without the His6 tag, was obtained by cloning the amplified fragment digested with XbaI and BamHI into similarly digested pET32a. The plasmid pHM46H, encoding XpsN46(His6), was obtained by cloning the amplified fragment pretreated with XbaI and XhoI.
Overexpression of the XpsN46 or XpsN97 protein, with or without a His6 tag, was performed by growing the E. coli strains containing the respective plasmids to an optical density of 0.6 to 0.7 in Luria-Bertani medium (20) supplemented with ampicillin (at 50 μg/ml). Induction was carried out in the presence of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the cultures were incubated at 28 or 37°C for 3 h before being harvested. A lower temperature was employed so that XpsN46, with or without His6, and XpsN97(His6) could be produced as soluble proteins.
Assays for α-amylase secretion.
The starch plate assay for α-amylase secretion was conducted as described before (8). Semiquantitative secretion activity was determined by measuring the secreted α-amylase activity of a liquid culture. The bacteria were grown in a semisynthetic XOL medium (7) supplemented with 0.4% maltose to an A600 of 1 before the supernatant was collected. The α-amylase activity was assayed by incubating aliquots of the supernatant with equal volumes of the starch solution, in which 1.5% soluble starch was made in 10 mM Tris-HCl, pH 7.0, in a final volume of 0.2 ml at 28°C for 30 min. At the end of incubation, 0.8 ml of a stop solution (prepared by mixing 0.5 M acetic acid and 0.5 M HCl at a 5:1 ratio) was added. The amount of starch remaining in the enzyme assay mixture was subsequently determined by mixing 0.1 ml of the assay mixture with 0.1 ml of an iodine solution (0.4% potassium iodine, 0.125% iodine). The A600 was measured with the solution at fivefold dilution (A1). To determine the total amount of starch (A2), similar steps were followed, except that the stop solution was added before the starch solution. The percentage of starch digested by the enzyme was calculated by subtracting A1 from A2 and dividing by A2.
Immunoprecipitation.
The procedures of Lee et al. (10) for immunoprecipitation were followed. The anti-PhoA serum, which was obtained by immunizing rabbits with the E. coli PhoA protein, purchased from Sigma, was used at a 1:45 dilution.
Membrane-mixing experiment.
A membrane-mixing experiment was conducted between X. campestris pv. campestris XC17433(pCPPLM) and E. coli BL21(DE3), which expresses XpsN46 or XpsN97 with or without the His6 tag, by following the procedures of Tsai et al. (23). In brief, each bacterial strain, harvested from separately grown cultures at A600s of 1.0 to 1.3, was resuspended in 20 mM phosphate buffer, pH 7.5. In order to facilitate the generation of mixed membrane vesicles, the two concentrated (40×) cell suspensions were mixed before breakage by passing them through a French press. In addition, to provide the opportunity for membrane proteins to interact with each other in the mixed membrane vesicles, the membrane vesicles collected as a pellet from ultracentrifugation of the broken cells were resuspended and incubated at 4°C for 3 h with gentle shaking. Subsequently, the membrane proteins were extracted with 2% (vol/vol) Triton X-100 for 2 h at 4°C. The membrane protein extract collected as the supernatant from a second ultracentrifugation was then separated on a 1 ml Ni-nitrilotriacetic acid (NTA) affinity column.
Pull-down experiment.
The pull-down experiment was performed following the procedures of Tsai et al. (23). Briefly, the XpsL-XpsM(His6) complex, prepared as a Triton X-100 extract from XC17433(pRT40), was immobilized on Ni-NTA resin via the XpsM C-terminal His6 tag. After it was thoroughly washed with a buffer containing 20 mM imidazole, the resin was divided evenly into three fractions. Each was incubated with an equivalent amount of the Triton X-100 extract of membrane proteins from XC17433 that carry the plasmid pCPPE67A, pCPPR78A, or pNC2. After being washed, the bound proteins were eluted with a buffer containing 250 mM imidazole and analyzed for XpsL, XpsM, and XpsN by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by immunoblotting.
RESULTS
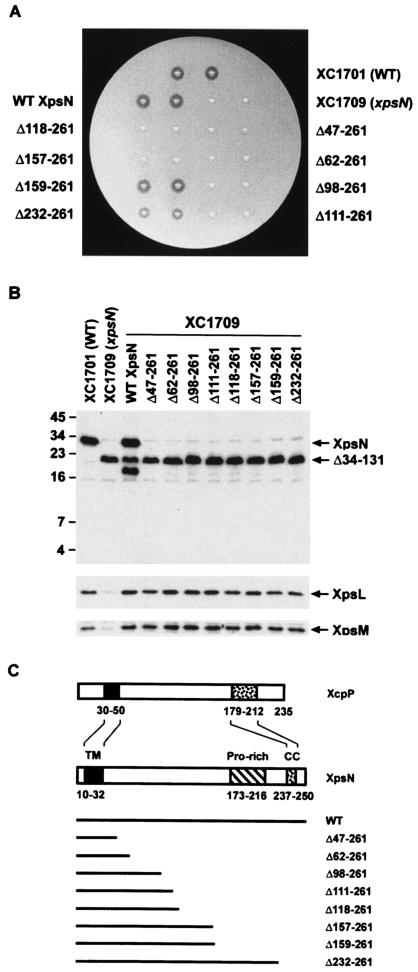
The N-terminal 158 amino acid residues of XpsN are sufficient for its function.
In order to define the functional domains of the XpsN protein, we constructed a series of C-terminal deletion mutants. These mutants were cloned in a broad-host-range vector, pCPP30, conjugated into the xpsN mutant XC1709, and analyzed for the ability to secrete α-amylase on starch plates. All but two were unable to complement XC1709. One mutant, XpsNΔ159-261, produced a clear zone on a starch plate with a size similar to that produced by the wild-type strain XC1701, as well as the xpsN mutant XC1709 complemented with the wild-type XpsN (Fig. 1A). This mutant also secreted other extracellular proteins, including protease, pectate lyase, and cellulase, in addition to α-amylase (data not shown). These results suggest that the N-terminal 158 amino acid residues of XpsN are sufficient for its function in type II secretion in X. campestris pv. campestris. Interestingly, deletion of two more residues made the protein XpsNΔ157-261 nonfunctional, suggesting that the XpsNΔ159-261 protein probably possesses the minimal requirements for a functional XpsN. Similarly, the C-terminal approximately one-third of the Erwinia chrysanthemi OutC protein is dispensable for normal secretion of at least three extracellular enzymes (2).
FIG. 1.
(A) Starch plate assay of XpsN C-terminal deletion mutants. The xpsN mutant strain XC1709 containing plasmids, each encoding a C-terminally truncated XpsN protein, was assayed for extracellular α-amylase secretion on a starch plate. After overnight incubation at 28°C, the clear zone surrounding the colony was scored as positive secretion. Wild-type (WT) XC1701 and the xpsN mutant XC1709 (xpsN) were included as positive and negative controls, respectively. WT XpsN, XC1709 complemented with the wild-type XpsN. (B) Immunoblot analysis of total lysates for abundances of XpsL, XpsM, and XpsN. XC1709 containing plasmids, each encoding a C-terminally truncated XpsN protein, was analyzed by Western blotting with anti-XpsN antibody (top), anti-XpsL antibody (middle), or anti-XpsM antibody (bottom). The molecular mass standards (in kilodaltons) are shown on the left. (C) Schematic diagram of deletion mutants, aligned with the full-length XpsN and the XcpP protein of E. chrysanthemi. The positions of the TM region, the proline-rich region (Pro-rich), and the predicted coiled-coil (CC) are marked with residue numbers indicating positions in the respective protein.
The other mutant, XpsNΔ232-261, exhibited a slightly smaller clear zone on a starch plate, indicating that it is only partially functional. To confirm the difference observed between the two mutants on starch plates, we measured α-amylase activity in the extracellular fraction from the liquid culture. While the α-amylase secretion activity exerted by the xpsN mutant XC1709 was null, the activity of the XpsNΔ232-261 mutant was ∼27% of that of the wild-type XpsN. Consistent with the results from the starch plate assay, the secretion activity of XpsNΔ159-261 was 94% of that of the wild type.
To examine whether the XpsN protein was detectable in the nonfunctional deletion mutants, we performed immunoblot analysis using antibody against the XpsN protein. To our surprise, none of the truncated XpsN protein was detected (Fig. 1B), suggesting that the C-terminally truncated forms of XpsN are subject to degradation. Alternatively, truncation at the C terminus may have removed major epitopes of XpsN. A protein band with an apparent molecular mass of ∼22 kDa was clearly visible in all samples with the genetic background of XC1709, in which the chromosomal xpsN gene was deleted in frame at its 5′ region. The signal at the same position in XC1701 is much weaker, indicating that the 22-kDa protein probably represents the truncated XpsNΔ34-131 protein encoded by the mutated chromosomal xpsN gene in XC1709. Probably because of its richness in proline, the presumed truncated protein migrated more slowly than expected. For the same reason, the full-length XpsN, whose predicted molecular mass is 29 kDa, migrated as if its apparent molecular mass were 36 kDa. In the xpsN mutant XC1709 complemented with the wild-type XpsN, an extra band migrating slightly faster than the 22-kDa protein was visible. It might be a degradation product from the full-length XpsN protein encoded by the plasmid-borne gene. Two other protein bands present in all samples are probably anti-XpsN antibody-cross-reactive materials.
The first 46 residues, encompassing an XpsN-specific membrane-anchoring sequence, are adequate to keep the XpsL and XpsM proteins at normal abundance.
Previous studies showed that knockout of the chromosomal xpsN gene caused reduction of the XpsL and XpsM protein levels (10), indicating that the wild-type XpsN is required to keep XpsL and XpsM stable. To examine which of the XpsN deletion mutants could restore these two proteins to normal levels, we performed immunoblot analysis using an antibody against each of them. The results clearly showed that both XpsL and XpsM were present at near-normal levels in all mutants (Fig. 1B, bottom two blots), indicating that the first 46 amino acid residues of XpsN alone are sufficient to keep XpsL and XpsM stable.
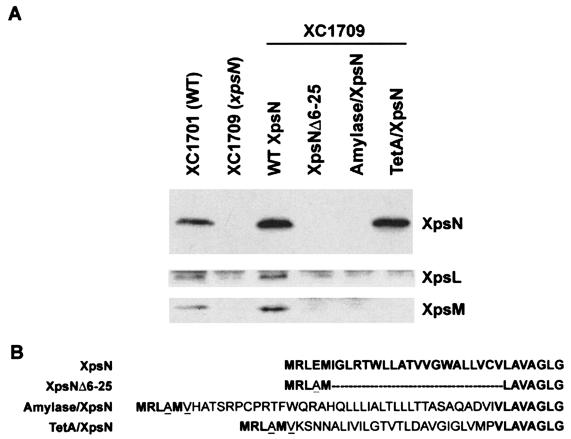
A stretch of hydrophobic residues constituting the membrane-anchoring sequence of XpsN is located between residues 10 and 32. This membrane-anchoring sequence makes up a major portion of the smallest truncated XpsNΔ47-261 protein that restores normal XpsL and XpsM protein levels. To find the significance of the XpsN membrane-anchoring sequence in keeping the XpsL and XpsM proteins at normal levels, we constructed three full-length XpsN mutants that were mutated in the N-terminal hydrophobic region. They included a deletion mutant with an incomplete transmembrane region (XpsNΔ6-25) and two replacement mutants, whose hydrophobic regions were replaced with the signal peptide of α-amylase or with the first membrane-spanning sequence of the TetA protein (Fig. 2B). Immunoblot analysis revealed that only the TetA-XpsN protein was detectable (Fig. 2A). This implies that the XpsN protein needs to be anchored in the membrane in order to keep itself stable. Despite its presence at a normal level, the TetA-XpsN protein is not functional (data not shown), nor was it able to make XpsL and XpsM detectable (Fig. 2A, bottom two blots). These results indicate that the mere presence of a membrane-anchoring sequence is not adequate for XpsN to be functional or for XpsL and XpsM to be maintained at normal abundance. Probably the specific hydrophobic sequence of the XpsN protein is required.
FIG. 2.
(A) Immunoblot analysis of XpsN N-terminal mutants. Total cell lysates were separated on SDS-PAGE, followed by immunoblotting with anti-XpsN antibody (top), anti-XpsL antibody (middle), and anti-XpsM antibody (bottom). Wild-type (WT) XpsN, XC1709 supplemented with the plasmid pNC2 encoding WT XpsN. XpsNΔ6-25, Amylase/XpsN, and TetA/XpsN represent XC1709 containing plasmids encoding the XpsNΔ6-25 deletion mutant and the amylase/XpsN and TetA/XpsN fusion proteins, respectively. (B) Alignment of N-terminal sequence of XpsN and its derivatives. The amino acids in boldface are derived from XpsN; those in normal type are derived from the N terminus of α-amylase or TetA, as indicated. The dashes represent deletions at the N terminus of XpsN. The underlined amino acids were introduced as a consequence of cloning.
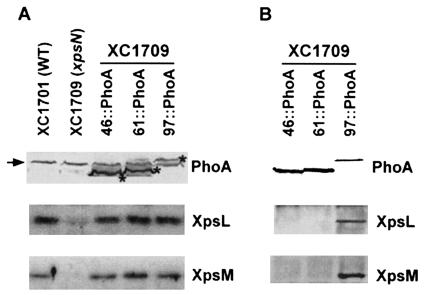
The N-terminal 97 residues of XpsN are required for its coimmunoprecipitation with XpsL and XpsM.
Unlike the truncated XpsN proteins, fusions at the C termini of some mutants with the E. coli alkaline phosphatase (PhoA) made them detectable with antibody against the PhoA protein (Fig. 3A). Similar to the truncated proteins, all fusion proteins remained capable of restoring normal levels of XpsL and XpsM (Fig. 3A and data not shown). Wild-type XpsN was demonstrated to form a ternary complex with XpsL and XpsM (23). Detection of the XpsN::PhoA fusion proteins with PhoA antibody permitted us to perform immunoprecipitation experiments so that the minimal requirement for XpsN sequence responsible for ternary-complex formation could be determined. Coimmunoprecipitation with XpsL and XpsM was observed only with XpsN97::PhoA and not with XpsN46::PhoA or XpsN61::PhoA (Fig. 3B). Possibly, physicochemical properties of the PhoA fusions prevented the last two from coimmunoprecipitating with XpsL-XpsM.
FIG. 3.
(A) Immunoblot analysis of XpsN::PhoA fusion proteins. Total cell lysates of X. campestris pv. campestris were analyzed by separating them on SDS-PAGE, followed by Western blotting using anti-PhoA antibody (top), anti-XpsL antibody (middle), or anti-XpsM antibody (bottom). Each XpsN::PhoA fusion protein is designated by the position, in amino acid residues, at which PhoA is fused. The asterisks mark the protein bands of the fusion proteins. The extra protein band (indicated by an arrow) appearing in all samples is probably a cross-reactive material detected by the anti-PhoA antibody. (B) Coimmunoprecipitation of XpsN::PhoA fusion proteins with XpsL and XpsM. Membrane vesicles prepared from XC1709 that expresses different XpsN::PhoA proteins were extracted with 2% Triton X-100, followed by precipitation with anti-PhoA antibody. The immunoblots were detected with anti-PhoA antibody (top), anti-XpsL antibody (middle), and anti-XpsM antibody (bottom).
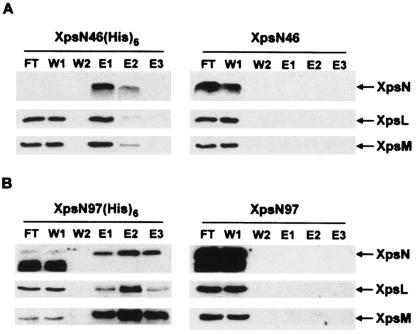
The N-terminal 46 residues of XpsN are adequate for in vitro assembly with the XpsL-XpsM complex.
In order to eliminate possible effects that might have been caused by PhoA, we overproduced XpsN46 in E. coli and examined its interaction with XpsL-XpsM by performing a membrane-mixing experiment. The same experiment had been employed previously to demonstrate that individually expressed wild-type XpsN could assemble in vitro with a preformed XpsL-XpsM complex (23). Because the XpsL and XpsM proteins produced from plasmid-encoded genes are present in greater abundance than those produced from chromosomally encoded genes, stable association of the two in the absence of XpsN was probably made possible by their abundance. Although the protein expression level was not as high as anticipated, the truncated XpsN protein produced in E. coli could be detected with antibody against XpsN. XpsN97 was included in a parallel experiment, since complex formation between the XpsN97::PhoA fusion protein and XpsL and XpsM was demonstrated with coimmunoprecipitation data.
The E. coli strain BL21(DE3) that expressed the truncated XpsN proteins, with or without His6, was mixed before cell breakage with the X. campestris pv. campestris strain XC17433(pCPP-LM), which expressed no other Xps proteins but XpsL and XpsM. Membrane vesicles prepared from the mixed cultures were incubated for 3 h at 4°C before Triton X-100 extraction of the membrane proteins, as described previously for membrane mixing between two X. campestris pv. campestris strains (23). Coelution of XpsL and XpsM with XpsN46 or with XpsN97 from nickel affinity columns was clearly observed only when the truncated XpsN protein was labeled with a His6 tag (Fig. 4A and B, left), but not in its absence (Fig. 4A and B, right). These results suggest that the truncated protein XpsN46 probably suffices to assemble with the XpsL-XpsM complex. A fast-migrating band appearing in the flowthrough and the first wash fractions indicates that both XpsN97(His6) and XpsN97 are subject to degradation and that the products seemed to have lost their C termini.
FIG. 4.
Membrane mixing between truncated XpsN and preassembled XpsL-XpsM, followed by nickel affinity chromatography. X. campestris pv. campestris XC17433(pCPP-LM), which expresses XpsL and XpsM, and E. coli BL21(DE3) containing a plasmid that expresses XpsN46 with or without a C-terminal His6 tag (A) or XpsN97 with or without a C-terminal His6 tag (B) were mixed before cell breakage. Membrane proteins extracted from the mixed membrane vesicles with 2% Triton X-100 were loaded on an Ni-NTA column. After being washed, the bound proteins were eluted with buffer containing 500 mM imidazole. Each fraction, collected at 1 ml/fraction, was concentrated with acetone and analyzed on SDS-PAGE, followed by immunoblotting with anti-XpsN antibody (top), anti-XpsL antibody (middle), or anti-XpsM antibody (bottom). FT, flowthrough; W1 and W2, first and last fractions, respectively, collected from washing; E1 to E3, eluted fractions concentrated 30-fold.
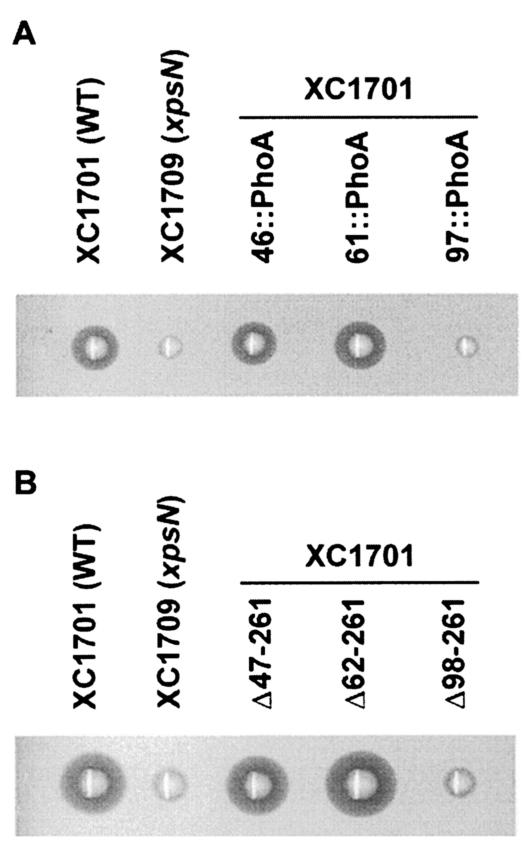
XpsN97, but not XpsN46, causes secretion interference in wild-type strain XC1701.
Could any of the truncated XpsN proteins exert a dominant-negative effect, presumably by forming complexes with XpsL and XpsM, in vivo? We examined whether their coexpression with the wild-type XpsN interferes with secretion. The results of the starch plate assay (Fig. 5A) indicate that only the XpsN97::PhoA fusion causes interference with the secretion of α-amylase, which correlates with their ability to coprecipitate with XpsL and XpsM. All three XpsN::PhoA fusion proteins are present in similar amounts, as shown by immunoblotting with anti-PhoA antibody (Fig. 3). It appears unlikely that the observed difference in interference is due to different amounts of protein. Moreover, despite the invisibility of all truncated proteins on the immunoblot (Fig. 1B), introduction of the plasmid encoding the truncated XpsNΔ98-261 protein in XC1701 clearly interfered with secretion (Fig. 5B). In contrast, the other two truncated proteins with shorter XpsN N termini did not interfere.
FIG. 5.
Starch plate assay for secretion interference in XC1701 by XpsN::PhoA fusion proteins (A) and C-terminally truncated XpsN proteins (B). Wild-type (WT) XC1701 and the xpsN mutant XC1709 (xpsN) were included as positive and negative controls, respectively.
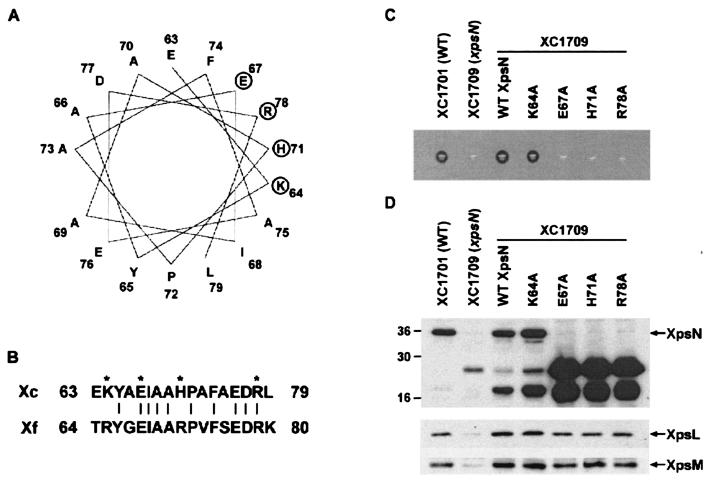
Mutations within a predicted α-helix between residues 63 and 79 of XpsN abolish its normal function.
The results from the interference experiments suggest that XpsN97 exhibits protein-protein interactions not executed by XpsN46. Within the extended region in XpsN97, an α-helix structure is predicted between residues 63 and 79. Similar helical structure could be predicted in the equivalent positions in most other GspC proteins. Helical-wheel analysis revealed a cluster of four charged residues, K64, E67, H71, and R78, next to one another (Fig. 6A). These four residues are well conserved in the X. fastidiosa XpsN protein (Fig. 6B). To examine the significance of the region between residues 47 and 97 of XpsN for its complex formation with XpsL-XpsM, we chose to replace each of these four charged residues with alanine, which has a high propensity for the α-helix, and analyzed its ability to complement the xpsN mutant XC1709. Unlike the substitution at K64, the other three substitutions, at E67, H71, and R78, were not functional (Fig. 6C). Examination of protein abundance by immunoblotting revealed that the three nonfunctional mutants appeared to be highly degraded (Fig. 6D, top). The full-length XpsN was no longer detectable. Instead, two strong signals with apparent molecular masses of 25 and 18 kDa appeared. These results suggested that alanine substitution at E67, H71, or R78 significantly influenced XpsN protein stability. Curiously, in these mutants, XpsL and XpsM could be detected, albeit at slightly lower abundance than those produced in XC1709 complemented with the wild-type XpsN but higher than those detected in XC1709 (Fig. 6D, middle and bottom). Equal loading of all samples was confirmed by Coomassie blue staining (data not shown).
FIG. 6.
(A) Helical-wheel plot of the predicted α-helix located between residues 63 and 79 of XpsN. The circles mark the four charged residues K64, E67, H71, and R78 that were individually replaced with alanine. (B) Alignment of the X. campestris pv. campestris (Xc) XpsN residues 63 to 79 with the X. fastidiosa (Xf) XpsN residues 64 to 80. The asterisks mark the mutated residues; the vertical lines connect identical residues. (C) Starch plate assay for complementation of XpsN mutant proteins with alanine substituted. Wild-type (WT) XC1701 and the mutant XC1709 (xpsN) were included as positive and negative controls, respectively. WT XpsN, wild-type XpsN protein encoded by pNC2. (D) Immunoblot analysis of XpsL, XpsM, and XpsN proteins in XC1709 that expresses the wild-type XpsN (WT XpsN) or XpsN mutants with alanine substituted. The wild-type strain XC1701 and the xpsN mutant XC1709 were included as positive and negative controls, respectively. Numbers on the left are molecular mass standards, in kilodaltons.
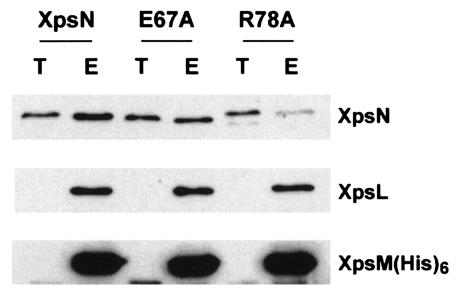
Alanine substitution at the 78th residue of XpsN reduced its ability to associate with XpsL-XpsM complex.
Since we were not able to detect the full-length mutant XpsN proteins when attempting complementation of XC1709, we could not perform immunoprecipitation to determine whether substitutions at these positions affect the ability of XpsN to associate with other Xps protein components. Interestingly, by introducing the mutated xpsN gene into XC17433, whose entire xps gene cluster had been deleted, we observed that two mutated XpsN proteins, E67A and R78A, were detected at levels similar to that of the wild-type XpsN (data not shown). We therefore performed a His6 pull-down experiment on them. XpsL-XpsM(His6) immobilized on Ni-NTA resin, after thorough washing, was employed to pull down membrane extract of the wild-type XpsN protein (23). For comparison, the wild-type XpsN encoded by pNC2 was included in a parallel experiment. Nearly equal amounts of XpsN proteins were mixed with equal amounts of XpsL-XpsM(His6) immobilized on resin (Fig. 7, lanes T). The amount of R78A detected in the eluted fraction was significantly reduced relative to that in the wild-type XpsN (Fig. 7), although the difference between the mutant E67A and the wild-type protein was not so obvious. This result suggests that the region between residues 47 and 97 is also important in the interaction between XpsN and the XpsL-XpsM complex. For unknown reasons, the R78A mutant protein migrated slightly behind the wild-type XpsN protein.
FIG. 7.
Pull-down assay of XpsN proteins with alanine substitutions, with XpsL-XpsM(His6) immobilized on Ni-NTA resin. The XpsL-XpsM(His6) complex prepared as Triton X-100 extract from XC17433(pRT40) was immobilized on Ni-NTA resin. After a thorough wash, the resin was mixed with the Triton X-100 extract of the membrane proteins prepared from XC17433 supplemented with plasmid that expresses the wild-type XpsN(pNC2), mutated XpsNE67A(pCPPE67A), or XpsNR78A(pCPPR78A). After being washed, the bound proteins were eluted with buffer containing 250 mM imidazole. T, total Triton X-100 extract of membrane proteins; E, bound proteins eluted from the resin at five times the concentration of T. For immunoblotting, anti-XpsN antibody (top), anti-XpsL antibody (middle), and anti-XpsM antibody (bottom) were used.
DISCUSSION
Complex formation between XpsN and XpsL-XpsM was originally implied by their mutual dependence for protein abundance (10). The prediction was later substantiated by the observation that XpsN indeed forms a complex with XpsL and XpsM (23). Furthermore, the XpsL-XpsM complex has a tendency to dissociate in the absence of XpsN (23). As a consequence, the XpsL and XpsM proteins become unstable. By analyzing different XpsN mutants for the ability to keep XpsL and XpsM at normal abundance and to form complexes with them, we demonstrated in this study that the N-terminal 46 residues by themselves possess both properties exhibited by the full-length XpsN. Perhaps specific residues in the respective transmembrane (TM) sequences are responsible for stable association of XpsN46(His6) with XpsL-XpsM. Alternatively, their association might be reinforced by interaction between regions that are immediately C terminal to the respective TM sequence. A region encompassing 35 residues (designated TM/HR [highly conserved region]) located immediately C terminal of the TM sequence in P. aeruginosa XcpPC was demonstrated to be species specific (5). Replacement of the TM/HR sequences by a corresponding region from E. chrysanthemi OutC caused XcpPC to be defective in stabilizing XcpYL in the presence of a defective XcpZM, implying its involvement in stabilizing the XcpYL-XcpZM complex.
The function of the region between residues 47 and 97 was further addressed because XpsN97 (either as a PhoA fusion or in a truncated form), but not XpsN46, interferes with secretion in a dominant-negative fashion. Substitution of alanine for arginine at the 78th residue in full-length XpsN caused the protein to lose function and weakened its ability to associate with the XpsL-XpsM complex. In addition, the levels of XpsL and XpsM were also reduced, more so for the former than the latter. These results suggest that, in its full-length form, residues 47 to 97 of XpsN may also be crucial for its association with XpsL-XpsM and for keeping the proteins stable. However, other possible functions for the 47-to-97 region of XpsN, for instance, involvement in its interaction with other unexamined Xps members, cannot be ruled out. The degraded form of XpsN (E67A-H71A-R78A) accumulated to great abundance only in XC1709 but was negligible in XC17433 (data not shown). This phenomenon suggests that the stability of the mutated XpsN is favored by excluding other Xps components. It would be interesting to identify which Xps members are involved.
The truncated protein XpsNΔ159-261 complemented the secretion defect in the xpsN mutant XC1709, as well as the wild-type XpsN protein. Likewise, truncation of the C-terminal 61 residues of the P. aeruginosa XcpPC protein did not eliminate its normal function (1). On the contrary, this truncated XcpPC protein appeared to be even more effective than the wild-type protein, as if the C-terminal 61 residues are slightly inhibitory. Similarly, the truncated protein XpsNΔ232-261 complemented XC1709 only partially, implying that the region spanning residues 159 to 231 in XpsN may be inhibitory. However, the presence of the very C-terminal 30 residues (232 to 261), within which a coiled-coil motif is predicted (Fig. 1C), appears to suppress the inhibitory effect caused by including the region between residues 159 and 231. In contrast to the C-terminally truncated form, deletion of the coiled-coil sequence alone from XcpPC made it stop secreting (1). However, the coiled-coil sequence in XcpPC could be replaced with a similarly located PDZ motif of the E. chrysanthemi OutC protein (5), which was demonstrated to be involved in recognition of specific secreted substrates (2). To determine what role the coiled-coil motif predicted at the C terminus of XpsN may play, further studies are needed.
Since XpsNΔ159-261 is fully functional, what purpose could the C-terminal region missing from this protein serve in the secretion process? Perhaps it forms a stable but dispensable domain in XpsN and confers stability on its physically linked N-terminal part. Located in this region is a unique proline-rich sequence (Fig. 1C). As predicted for the formation of unordered structure, this region could serve as a linker between two separable domains. All XpsN proteins with their C termini truncated were not detectable on immunoblotting. This implies significance of the C-terminal region in keeping the XpsN protein stable. In agreement with this, the XpsNΔ34-131 protein encoded by the deleted xpsN gene of the XC1709 chromosome, which contains the C-terminal region, was detected as a stable protein of 22 kDa (Fig. 1B). Alternatively, the C-terminal region of XpsN could be required only for specific recognition of some untested exoproteins, as was proposed for the PDZ domain of E. chrysanthemi OutC. Bouley et al. (2) demonstrated that the PDZ domain is required for the secretion of certain exoproteins, but not others. The likelihood that XpsNΔ159-261 gained its function by forming a complex with XpsNΔ34-131, encoded by the chromosomal gene of XC1709, appears slim, since the slightly shorter protein XpsNΔ157-261 was not functional in the same genetic background (Fig. 1A).
Unlike the C-terminally truncated functional XcpPC protein (1), the XpsNΔ159-261 protein was not detectable on immunoblotting. Since its fusion at the C terminus with PhoA did not permit its detection with either anti-XpsN or anti-PhoA antibody (data not shown), it seems reasonable to assume that this truncated form, with or without PhoA at its C terminus, is subject to degradation, resulting in low abundance. The secretion function exerted by subdetectable levels of the XpsNΔ159-261 protein implies that XpsN could be a component that functions “catalytically” in the secretion machinery. It is conceivable that XpsN could act in a catalytic way by momentarily interacting with the secretin XpsD in the outer membrane. Such a transient interaction could serve to trigger opening of the secretion pore, like the pI protein triggering opening of the secretion pore constituted from the secretin pIV protein in filamentous-phage assembly (19). Consistent with such a prediction, the OutC and OutD proteins have been proposed to play the role of gatekeepers as a pair in the Erwinia type II secretion machinery (12). Alternatively, XpsN could interact with XpsD repeatedly as an energy transducer for pore opening, similar to the TonB protein function in iron uptake across the outer membrane (17). XpsN is prone to dissociating from the XpsL-XpsM-XpsN ternary complex in vitro (23). However, whether it acts catalytically in vivo together with XpsL-XpsM or as an entity by itself requires further study.
Acknowledgments
This work was supported by grant E-91-B-FA05-2-4 from the Ministry of Education and grant NSC90-2311-B-005-020 from the National Science Council of the Republic of China.
We thank Jiun-Li Chang for performing the semiquantitative secretion assay and Nei-Li Chan for reading the manuscript and providing helpful suggestions. We are also grateful to all reviewers for their inspiring comments.
REFERENCES
- 1.Bleves, S., M. Gerard-Vincent, A. Lazdunski, and A. Filloux. 1999. Structure-function analysis of XcpP, a component involved in general secretory pathway-dependent protein secretion in Pseudomonas aeruginosa. J. Bacteriol. 181:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouley, J., G. Condemine, and V. E. Shevchik. 2001. The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II out pathway of Erwinia chrysanthemi. J. Mol. Biol. 308:205-219. [DOI] [PubMed] [Google Scholar]
- 3.Chen, L. Y., D. Y. Chen, J. Miaw, and N. T. Hu. 1996. XpsD, an outer membrane protein required for protein secretion by Xanthomonas campestris pv. campestris, forms a multimer. J. Biol. Chem. 271:2703-2708. [DOI] [PubMed] [Google Scholar]
- 4.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, et al. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 5.Gerard-Vincent, M., V. Robert, G. Ball, S. Bleves, G. P. Michel, A. Lazdunski, and A. Filloux. 2002. Identification of XcpP domains that confer functionality and specificity to the Pseudomonas aeruginosa type II secretion apparatus. Mol. Microbiol. 44:1651-1665. [DOI] [PubMed] [Google Scholar]
- 6.Hemsley, A., N. Arnheim, M. D. Toney, G. Cortopassi, and D. J. Galas. 1989. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 17:6545-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu, N. T., M. N. Hung, S. J. Chiou, F. Tang, D. C. Chiang, H. Y. Huang, and C. Y. Wu. 1992. Cloning and characterization of a gene required for the secretion of extracellular enzymes across the outer membrane by Xanthomonas campestris pv. campestris. J. Bacteriol. 174:2679-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, N. T., M. N. Hung, C. T. Liao, and M. H. Lin. 1995. Subcellular location of XpsD, a protein required for extracellular protein secretion by Xanthomonas campestris pv. campestris. Microbiology 141:1395-1406. [DOI] [PubMed] [Google Scholar]
- 9.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 10.Lee, H. M., S. W. Tyan, W. M. Leu, L. Y. Chen, D. C. Chen, and N. T. Hu. 2001. Involvement of the XpsN protein in formation of the XpsL-XpsM complex in Xanthomonas campestris pv. campestris type II secretion apparatus. J. Bacteriol. 183:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, H. M., K. C. Wang, Y. L. Liu, H. Y. Yew, L. Y. Chen, W. M. Leu, D. C. Chen, and N. T. Hu. 2000. Association of the cytoplasmic membrane protein XpsN with the outer membrane protein XpsD in the type II protein secretion apparatus of Xanthomonas campestris pv. campestris. J. Bacteriol. 182:1549-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindeberg, M., G. P. Salmond, and A. Collmer. 1996. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologues reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol. Microbiol. 20:175-190. [DOI] [PubMed] [Google Scholar]
- 13.Manoil, C., and J. Bailey. 1997. A simple screen for permissive sites in proteins: analysis of Escherichia coli Lac permease. J. Mol. Biol. 267:250-263. [DOI] [PubMed] [Google Scholar]
- 14.Nouwen, N., H. Stahlberg, A. P. Pugsley, and A. Engel. 2000. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 19:2229-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Possot, O. M., M. Gerard-Vincent, and A. P. Pugsley. 1999. Membrane association and multimerization of secreton component PulC. J. Bacteriol. 181:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Possot, O. M., G. Vignon, N. Bomchil, F. Ebel, and A. P. Pugsley. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 182:2142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postle, K., and R. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 18.Robert, V., F. Hayes, A. Lazdunski, and G. P. Michel. 2002. Identification of XcpZ domains required for assembly of the secreton of Pseudomonas aeruginosa. J. Bacteriol. 184:1779-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russel, M. 1994. Phage assembly: a paradigm for bacterial virulence factor export? Science 265:612-614. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 22.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 23.Tsai, R. T., W. M. Leu, L. Y. Chen, and N. T. Hu. 2002. A reversibly dissociable ternary complex formed by XpsL, XpsM and XpsN of the Xanthomonas campestris pv. campestris type II secretion apparatus. Biochem. J. 367:865-871. [DOI] [PMC free article] [PubMed] [Google Scholar]