Abstract
Legionella pneumophila displays a biphasic developmental cycle in which replicating forms (RFs) differentiate postexponentially into highly infectious, cyst-like mature intracellular forms (MIFs). Using comparative protein profile analyses (MIFs versus RFs), we identified a 20-kDa protein, previously annotated as “Mip-like” protein, that was enriched in MIFs. However, this 20-kDa protein shared no similarity with Mip, a well-characterized peptidyl-prolyl isomerase of L. pneumophila, and for clarity we renamed it MagA (for “MIF-associated gene”). We monitored MagA levels across the growth cycle (in vitro and in vivo) by immunoblotting and established that MagA levels increased postexponentially in vitro (∼3-fold) and nearly 10-fold during MIF morphogenesis in HeLa cells. DNA sequence analysis of the magA locus revealed an upstream divergently transcribed gene, msrA, encoding a peptide methionine sulfoxide reductase and a shared promoter region containing direct and indirect repeat sequences as well as −10 hexamers often associated with stationary-phase regulation. While MagA has no known function, it contains a conserved CXXC motif commonly found in members of the thioredoxin reductase family and in AhpD reductases that are associated with alkylhydroperoxide reductase (AhpC), suggesting a possible role in protection from oxidative stress. MIFs from L. pneumophila strain Lp02 containing a magA deletion exhibited differences in Giménez staining, as well as an apparent increase in cytopathology to HeLa cells, but otherwise were unaltered in virulence traits. As demonstrated by this study, MagA appears to be a MIF-specific protein expressed late in intracellular growth that may serve as a useful marker of development.
Legionella pneumophila and related species reside in aquatic environments as intracellular parasites of protozoa (13). When transmitted by aerosols to susceptible human hosts, L. pneumophila infects alveolar macrophages, often producing an atypical and sometimes fatal pneumonia known as Legionnaires' disease (5, 9). In contrast to most respiratory and pneumonic diseases, Legionnaires' disease is not communicable (not transmitted from person to person), suggesting that humans are inadvertent hosts and are not part of the survival strategy or evolution of this pathogen (16, 21).
Recently, morphological and physiological evidence of a developmental cycle in which replicating L. pneumophila bacteria (replicating forms [RFs]) differentiated postexponentially into cyst-like mature intracellular forms (MIFs) in nonlymphoid cell lines or natural protozoan hosts has been presented (11, 15, 16, 18). In natural environments, MIFs likely enable L. pneumophila to survive in a dormant, highly infectious, planktonic state for long periods while between hosts. Comparative studies of MIFs with in vitro-grown stationary-phase forms revealed MIFs to be resistant to detergent lysis and the effects of antibiotics and to exhibit no measurable respiration rate (16). These studies also demonstrated that MIFs are not appreciably produced in macrophage cell culture due to early apoptotic lysis, which precludes efficient MIF formation (16). It has been suggested that inefficient production of MIFs during human infection might correlate with poor transmissibility (16).
The transition from vegetative, metabolically active RFs into MIFs must involve the coordinated regulation of many genes whose products are associated with alterations in cell wall morphology (11, 16), synthesis of storage molecules like poly-β-hydroxybutyrate (16, 24), enrichment and assembly of virulence determinants (7, 19), production of flagella (7, 8), and shifts in metabolic activities. While the MIF is exclusive to the intracellular milieu (not observed with bacteria grown in vitro), its morphogenesis likely becomes activated during the shift from exponential growth into stationary phase, a transition regulated by the stationary-phase sigma factor RpoS and the stringent response regulator RelA (1, 35).
By comparing the two-dimensional patterns of protein spots obtained from MIFs and stationary-phase bacteria as well as using pulse-chase radiolabeling experiments, Garduño et al. identified numerous protein differences and in particular a dramatic increase in the levels of a 20-kDa protein associated with MIFs purified from HeLa cells (15, 16). We identified this protein from its N-terminal amino acid sequence and renamed it MagA (for “MIF-associated gene A”) to reflect its association with the cyst-like MIF. Subsequent cloning and genetic analyses revealed that MagA contained a 20-amino-acid motif conserved among AhpD-type (thioredoxin reductase family) reductases, suggesting a possible role in protection from oxidative stress (2, 25). Deletion analysis indicated that magA is not essential for infectivity or MIF morphogenesis, but HeLa cells infected with magA mutants exhibited increased cytopathy in late stages of infection and the MIFs displayed an altered pattern of staining with Giménez stain. Thus, magA may serve as a useful genetic marker for tracking regulatory events associated with development of the highly infectious MIF of L. pneumophila.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains (L. pneumophila and Escherichia coli) and plasmids used in this study are listed in Table 1. All Legionella strains were stored at −70°C and routinely grown on buffered charcoal-yeast extract agar (BCYE) (30) for 3 to 5 days at 37°C in a humid incubator. Bacteria from primary BCYE agar plates were either used immediately for infection of HeLa cells or subcultured in BYE broth (buffered yeast extract broth minus charcoal and agar). Growth of Lp02 and its derivatives required thymidine (100 μg per ml of medium). SVir and Lp02 strains were usually grown in the presence of 100 μg of streptomycin per ml of medium. Frozen stocks of virulent L. pneumophila strains were routinely made from lysates of infected HeLa cells to which dimethyl sulfoxide was added to a final concentration of 10%.
TABLE 1.
Strains and molecular DNA tools (plasmids and primers) used in this study
| Strain, plasmid, or primer | Source | Characteristic(s) or sequence | Reference |
|---|---|---|---|
| L. pneumophila | |||
| Lp1-SVir | This lab | Philadelphia-1 strain Lp1, Smr | 22 |
| Lp1-Avir | CDCa | Salt-tolerant variant of Lp1-Vir | |
| Lp02 | Ralph Isbergb | Philadelphia-1 strain, Thy auxotroph | 3 |
| 2064 | This lab | Oxford strain | 12 |
| 2064M | This lab | Salt-tolerant variant of 2064 | 12 |
| E. coli | |||
| DH5α | |||
| Plasmids | |||
| pBC | Stratagene | Cmr cloning vector | |
| pBSK | Stratagene | Apr cloning vector | |
| pBCmagA2.5 | This study | 2.5 kb, magA, msrA | |
| PBCmagA1.5 | This study | 1.5 kb, magA | |
| Primers | |||
| 24KFE (EcoRI) FOR | 5′-TAG AAT TCC GAC AAC TGT CAA-3′ | ||
| 24KRH (Hind III) REV | 5′-ATA AGC TTG TCC ATT GAG TCA A-3′ | ||
| magAKO FOR (P1) | 5′-CTG CAG ATC CTT GAC TGG AAT-3′ | ||
| magAKO SAL1 REV (P2) | 5′-GTA GTC GAC CAT ATA ACT TAC-3′ | ||
| magAKO SAL1 FOR (P3) | 5′-GAC GTC GAC TTC CCT GTC GTT-3′ | ||
| magAKO REV (P4) | 5′-GCT CTC GAG TGT TCC TTG AAA-3′ | ||
| magBAM(MAL) FOR | 5′-CGC CGA TCC ATG CAA CGT ATT AAA AAA ATC TCT ATC GCT-3′ | ||
| magBAM(MAL) REV | 5′-AGT TCG AAT TAT GAT GCA GTA CGT CCT TTA TGG GTA TC-3′ | ||
| magAF 3137 (SAC) FOR | 5′-CTC CGA GCT CAA GGA TCA CTC ACT-3′ | ||
| magAR 5836 (SAC) REV | 5′-GTA TTT CGA GAG CTC TGG ATC T-3′ |
CDC, Centers for Disease Control and Prevention Atlanta, Ga.
Tufts University School of Medicine, Boston, Mass.
Strains of E. coli were stored at −70°C in nutrient broth with 10% dimethyl sulfoxide and grown in Luria-Bertani medium supplemented with the appropriate antibiotics as indicated below.
HeLa cell infection and isolation of MIFs.
HeLa (human epithelial cell line) cells were routinely grown in either tissue culture flasks or multiwell plates (all from Falcon Plastics, Becton-Dickinson) in minimal essential medium (MEM) completed with 10% newborn calf serum and an antibiotic-antimycotic mixture (all from GIBCO Laboratories). HeLa cells in wells were infected with a suspension of phosphate-buffered saline-washed L. pneumophila at ∼108 bacteria/well (for 6-well plates) or ∼107 bacteria/well (for 24-well plates). After an overnight incubation, HeLa cells were washed twice with Earle's salt solution to eliminate free bacteria and cells, and fresh MEM medium containing 100 μg of gentamicin/ml was added for 90 min. Cells were then washed twice with Earle's salt solution to remove the gentamicin, and fresh MEM with serum but no antibiotics was added.
To produce large numbers of MIFs or intracellular RFs, HeLa cells in 75-cm2 cell culture flasks were typically infected with ∼2 × 109 bacteria per flask and bacteria were isolated from HeLa cell lysates at different times after infection by separation in continuous-density gradients of Percoll as described elsewhere (16, 17).
Standard invasion and plaque assays.
Invasion of nonphagocytic cells was determined as reported previously (16, 17). Briefly, HeLa cells were infected with a standardized inoculum, and at 3 h postinfection, monolayers were either thoroughly washed with phosphate-buffered saline and lysed (to determine the adherence index) or washed, treated with gentamicin, and lysed (to determine the invasion index). Counts of viable bacterial cells were performed in these lysates to determine the two aforementioned indexes in relation to the initial inoculum. Infectivity was also determined using a plaque assay following the method of Fernandez et al. (12) but omitting the gentamicin treatment step as previously reported (16). Briefly, monolayers of L929 (fibroblast cell line) cells in 24-well plates were incubated for 1 h with serial 10-fold dilutions of the bacterial inoculum, washed to eliminate unbound bacteria, and overlaid with cell culture medium solidified with 0.6% agarose. Four days after infection, monolayers were fixed in 4% formaldehyde and stained with crystal violet. Plaques were then enumerated, and the percent plaquing efficiency was calculated as follows: (number of plaques/number of bacteria added) × 100.
Radiolabeling of MIFs and stationary-phase bacteria and N-terminal sequencing.
MIFs were obtained from HeLa cells, and stationary-phase bacteria were harvested from growth on BCYE agar as previously described (16) and suspended in distilled water (dH2O) to an optical density at 620 nm of 0.1 in a 1-ml reaction mixture. To each suspension, 10 μCi of [35S]methionine was added, and following labeling for 1 h at 37°C, bacterial pellets were washed and suspended in sodium dodecyl sulfate (SDS) sample buffer and loaded on 7.5 to 15% gradient polyacrylamide gel electrophoresis (PAGE) gels and subsequently analyzed by autoradiography on Kodak XAO film.
Unlabeled MIF protein samples were also loaded on SDS-PAGE gels, and following electrophoresis, the proteins were transferred to Immobilon P (DuPont) membranes by transblotting (Bio-Rad) and developed as previously described (23). A prominent ∼20-kDa Coomassie-stained protein was identified and subjected to N-terminal amino acid sequencing at the University of British Columbia Biotechnology Laboratory (Vancouver, Canada). The N-terminal amino acid sequence was then used to identify the gene by BLAST-P search.
Cloning of magA and flanking sequences.
Genomic DNA was prepared from the SVir strain of L. pneumophila as previously described (22) and used as the template for PCR amplification of magA using primer pair 24KFE (EcoRI) and 24KRH (HindIII) (Table 1). The PCRs contained 2.5 μl of 10× PCR buffer with (NH4)2SO4 (MBI Fermentas, Flamborough, Ontario, Canada), 1.5 μl of 25 mM MgCl2 (MBI Fermentas), 5 μl of 1.25 mM concentrations of deoxynucleoside triphosphates (Life Technologies, Burlington, Ontario, Canada), 2 μl each of the primers (10 pmol), 1 μl of DNA template (∼10 nmol), 10.7 μl of dH2O, and 1 U of recombinant Taq DNA polymerase (MBI Fermentas). Unless otherwise indicated, the following standard PCR conditions were used: 96°C (3 min) followed by 30 to 35 cycles of 94°C (40 s), 55°C (40 s), and 76°C (1 min) and a final extension at 76°C for 10 min. The amplicon was restricted with EcoRI and HindIII and cloned into similarly restricted pBSK to create pBMAG24. Cloned magA-containing regions were verified by PCR and by Southern analysis and subjected to automated DNA sequencing at the Institute for Marine Biosciences (National Research Council of Canada) sequencing facility (Halifax, Nova Scotia).
Construction of a magA deletion.
A stepwise PCR procedure was used to construct a magA deletion in L. pneumophila. Briefly, upstream (582 bp) and downstream (764 bp) flanking sequences of magA were amplified by PCR using primers P1 and P2 and primers P3 and P4, respectively (Table 1). The P2 and P3 primers each contained a SalI site, permitting restriction and subsequent ligation of the two amplicons creating a precise deletion of magA sequences. The ligation product was amplified with the P1 and P4 flanking primers, and following restriction with PstI and XhoI, the 1,346-bp amplicon was cloned into pBSK similarly restricted with PstI and XhoI. The Kmr determinant of plasmid p34S:km3 (29) was introduced into the SalI site, creating a 2,332-bp sequence. Allelic replacement of the magA deletion in L. pneumophila strains SVir and Lp02 was accomplished by electroporation of the 2,332-bp amplicon generated by PCR using the P1 and P4 primers. Homologous recombinants exhibiting Kmr were screened by PCR to confirm the presence and orientation of the Kmr cassette and the absence of magA sequences.
trans complementation of the magA mutant.
A 2.5-kbp DNA fragment containing magA and msrA was generated with magAF and magAR primers (Table 1) and, following restriction with SacI, was cloned into similarly restricted pBC (Stratagene) and transformed into E. coli DH5α. A 1.5-kb fragment containing magA but lacking most of the msrA gene was also cloned (HindIII/PstI) into pBC. Expression of MagA protein was verified by SDS-PAGE and immunoblotting, and the purified plasmids were introduced into L. pneumophila ΔmagA strains by electroporation.
Construction and purification of MagA as a MalE::MagA fusion protein.
By using primers MAGBAM(MAL) and MAGHIND(MAL) (Table 1), a 570-bp magA amplicon was generated via PCR and, following restriction with BamHI and HindIII, was cloned into a similarly restricted pMAL-c(MAL) vector. The pMAL-cmagA construct was electroporated into E. coli DH5α and verified by plasmid purification and restriction enzyme analysis (BamHI and HindIII).
The MalE::MagA fusion protein was purified following isopropyl-β-d-thiogalactopyranoside (IPTG)-induced overexpression from E. coli DH5α/pMAL-cmagA. Briefly, 1 mM IPTG was added to bacterial cultures grown to an optical density at 620 nm of 0.85, and after 3 h, the bacterial pellet was collected by centrifugation, washed, and suspended in 7 ml of column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 5 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride). High-speed supernatant (100,000 × g) from sonicated bacteria (30-s pulses and placed on ice between pulses) was applied to an amylose affinity column at 4°C (ca. 90 mg of total protein) according to the manufacturer's instructions (New England BioLabs, Beverly, Mass.). The fusion protein was eluted with 10 mM maltose-containing buffer, and the elution profile was monitored with a UA-5 UV monitor (ISCO, Lincoln, Nebr.). Column fractions were run on an SDS-PAGE gel and visualized with Coomassie blue stain. The pooled and concentrated fraction was dialyzed and then applied to a Sephadex G-75 (Amersham Pharmacia Biotech, Baie d'Urfé, Quebec, Canada) gel filtration column packed in a 3.1- by 90-cm column with a bed volume of ca. 560 ml (void volume of ca. 130 ml) at 4°C. The protein sample was loaded in a total volume of 25 ml of column buffer, and 6-ml fractions were collected and examined on an SDS-12% PAGE gel. Fractions containing the highest purity of the fusion protein were pooled, concentrated by ultrafiltration, and frozen at −20°C. The protein concentration was estimated by using a protein dye binding kit (Bio-Rad).
Rabbit hyperimmune serum against MalE::MagA and immunoblotting.
Two New Zealand White rabbits were immunized by subcutaneous injection (at multiple sites) with a total of 90 μg of purified MalE::MagA fusion protein emulsified in Freund's complete adjuvant. The primary injections were followed 2 weeks later by booster subcutaneous injections of a total of 90 μg of protein emulsified in Freund's incomplete adjuvant. Preimmune and immune sera were obtained from each rabbit and anti-MalE::MagA titers were determined by enzyme-linked immunosorbent assay (20), and the specificity of the hyperimmune serum towards MagA was determined on bacterial proteins blotted onto nitrocellulose membranes (34).
Microscopy.
Preparation of infected HeLa cells and MIF samples for ultrathin sectioning and staining, as well as their observation in a Philips EM300 transmission electron microscope, was performed as previously described (11, 14). Morphological changes (cytopathy) in HeLa cells infected with Lp02, Lp02ΔmagA, and Lp02ΔmagA/pBCmagA were monitored by phase-contrast microscopy (triplicate experiments) by using an inverted Nikon TMD light microscope equipped with a digital image-capturing system (Carsen Group Inc., Markham, Ontario, Canada). Image processing was performed using ImagePro version 4.0 software (Media Cybernetics, Silver Spring, Md.). Bacterial specimens stained with the Giménez stain (27) were observed in a Zeiss D7082 light microscope to score the proportion of Giménez-positive bacteria, as previously described (16).
RESULTS
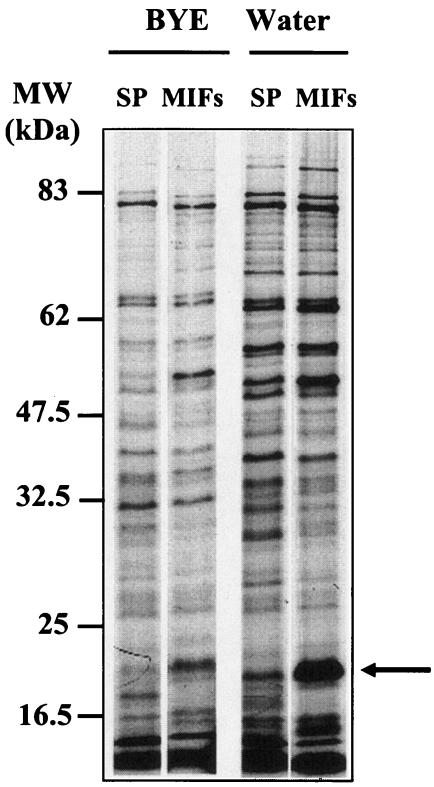
In previous studies, the proteomes of MIFs obtained from HeLa cells and stationary-phase bacteria were analyzed by two-dimensional gel electrophoresis, and several proteins that were enriched in extracts prepared from MIFs, including a 20-kDa protein subsequently named MagA, were identified (15, 16). To determine whether MagA and several other proteins were indeed selectively synthesized during MIF morphogenesis, MIFs isolated from infected HeLa cells and stationary-phase agar-grown bacteria were suspended in dH2O or diluted BYE broth and pulse radiolabeled with [35S]methionine. As shown in Fig. 1, several proteins were preferentially labeled (“Water”) in the MIF population, including the 20-kDa MagA protein (indicated by the arrow). This protein was also present in stationary-phase bacteria but was not as extensively radiolabeled. While MIFs and stationary-phase bacteria incorporated less radiolabel in diluted BYE broth than in water suspension, the patterns of labeled protein bands were similar to those obtained with water-suspended bacteria. The ability of MIFs to incorporate radiolabel in vitro suggests that the maturation process most likely continues postcellularly, as noted for cysts of other bacteria (32).
FIG. 1.
Expression of MagA in L. pneumophila SVir grown in BYE to stationary phase (SP) or in HeLa cells to produce MIFs. Stationary-phase bacteria or MIFs were resuspended in fresh BYE or in double-distilled water and radiolabeled as described in the text. Samples of bacterial pellets (equivalent amounts of protein) were run in SDS-PAGE (12% acrylamide) gels and subjected to autoradiography. The arrow indicates the position of MagA. Molecular size indicators are shown at left.
N-terminal sequence of MagA.
The prominent MagA protein obtained from MIFs was subjected to N-terminal sequencing, and a BLAST search of its N-terminal sequence (MQRIKKISIANAQGKAK) revealed that MagA shared identity with a 24-kDa Mip-like protein previously reported by Miyamoto et al. (28) and with no assigned function. Since this protein is unrelated to the 24-kDa Mip protein, a well-characterized prolyl cis-trans isomerase (10), we chose to rename the protein MagA (for “MIF-associated gene”) to reflect its abundance in MIFs.
Postreplicative upregulation of MagA expression in vitro and in vivo.
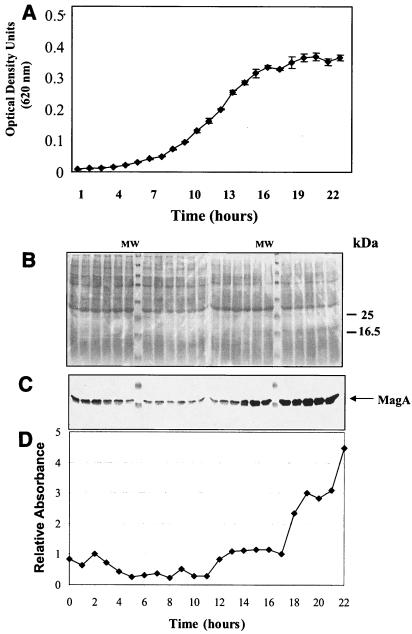
Using monospecific polyclonal antibody raised to the purified MagA::MalE fusion protein, we monitored the expression of MagA during the growth cycle of Lp1-SVir in broth culture by immunoblotting (Fig. 2). The lowest levels of MagA were observed for bacteria harvested during exponential phase, and MagA levels increased in bacteria harvested from the postexponential phase and peaked during stationary phase (Fig. 2C). Densitometry scans indicated a three- to fourfold increase in MagA levels between exponential and postexponential bacteria (Fig. 2D).
FIG. 2.
Expression of MagA along the growth curves of Lp1-SVir. (A) Growth curve. (B) SDS-PAGE of samples taken along the growth curve shown in panel A. (C) Immunoblot of material shown in panel B. (D) Densitometry scan of material shown in panel C. Bacterial cell density was measured using 1:10 dilutions of BYE cultures grown at 37°C with shaking. Each point in panel A represents the mean ± standard deviation of triplicate measurements (from three independent cultures, n = 3). Culture samples for immunoblotting were taken every hour and pooled and centrifuged, and bacterial pellets were frozen until the end of the experiment, when all samples were processed simultaneously. Equal amounts of protein were loaded in each lane.
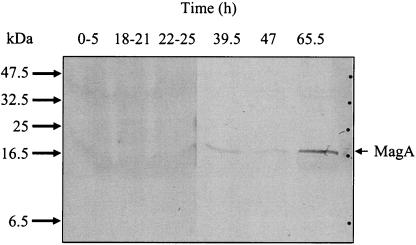
When the Lp1-SVir strain was grown in HeLa cells, we also observed a close correlation between MagA expression and entrance into stationary phase in vivo (Fig. 3). To obtain sufficient quantities of Lp1-SVir for early time points, bacteria harvested from several samples were pooled. MagA was not detected in the early exponential phases of growth (<39 h postinoculation), and the highest levels appeared at ∼65 h postinfection (Fig. 3), corresponding with MIF maturation. Thus, upregulation of MagA expression constitutes a robust marker of the postreplicative phase in vitro and in vivo.
FIG. 3.
Immunoblot of MagA expression by L. pneumophila Lp1-SVir grown intracellularly in HeLa cells. Samples of infected HeLa cells were lysed in double-distilled water, dilution plated on BCYE to determine the number of CFU per milliliter, pelleted by centrifugation, and frozen at −20°C until further processing. Loading of SDS-PAGE gels was normalized to an equivalent of 5 million CFU per lane. For early times, samples had to be pooled (due to low initial counts of CFU per milliliter); thus, the lanes labeled 0-5 and 18-21 contain three samples collected between 0 and 5 h or between 18 and 21 h postinfection, respectively. The first three lanes from the left look slightly darker due to a longer developing time of immunostaining and higher background of HeLa cell proteins. Molecular size indicators are shown at left.
Genetic analysis of the magA locus.
The magA gene shared 99% identity with the previously published DNA sequence (28) and 100% nucleotide identity with the sequence posted in the Legionella genome website at Columbia University for the Philadelphia-1 strain of L. pneumophila (http://genome3.cpmc.columbia.edu/∼legion/). The magA gene sequence ends in an inverted repeat of 10 bp typical of rho-independent terminators (22), suggesting that magA is not contained in an operon. Upstream of magA, and in the opposite orientation, is msrA, whose predicted product shares similarity with peptide methionine sulfide reductases that are associated with detoxification of oxidized methionine residues in proteins (33). BLAST searches indicated that orthologs of magA are present in the annotated genomes of “Microbulbifer degradans,” Ralstonia metallidurans, Pseudomonas sp., Vibrio vulnificus, Streptococcus pneumoniae, and Neisseria meningitidis, with protein homologies of 68, 61, 58, 57, 49, and 48%, respectively (data not shown). Downstream of magA were several genes whose predicted products included a series of transposases and the AbiD phage protein-encoding gene (4). Brassinga et al. have previously reported that the magA locus is contained within a 65-kb pathogenicity island (LpPI-1) that is unique to the Philadelphia-1 strain of L. pneumophila (4).
MagA is predicted to be a cytoplasmic protein of 184 amino acids with a pI of 7.73. A cytoplasmic location was confirmed by immunogold electron microscopy (data not presented). While MagA has no known function, we identified a EQIALVSAGYNGCNYCASAH motif (bold type indicates conserved amino acids) and additional regions of amino acid similarity with a diverse group of proteins annotated as orthologs of AhpD reductases that typically supply reducing equivalents to alkylhydroperoxide reductase AhpC (25). However, the location of the CXXC motif of AhpD proteins with demonstrated AhpC reductase activity is in the carboxyl end of the protein rather than central, as found in MagA. Moreover, the conserved motif (EX5SAX4CXXCX3H of the MagA group) differs slightly from the AhpD group (EX6SAX3CXXCX3H) and therefore may not be functional in this capacity. With the exception of similarity in this conserved region, the AhpD of L. pneumophila (whose gene is contiguous with and downstream of ahpC) exhibits little similarity to MagA.
MagA regulatory sequences.
The promoter region spanning the intergenic region of msrA and magA contained direct and indirect repeat motifs that might be associated with regulation, one of which (a 6-bp inverted repeat) is located within the −35 and −10 promoter region of magA (Fig. 4). Upstream of msrA is a consensus −10 RpoS-binding sequence (TATACT) (26), and for magA, a −10 hexamer of GATACT as well as a TATAAAA (RpoD) hexamer were found. Located between the two divergent promoters were two 12-bp direct repeats generated by repeated EcoRI restriction sites. Our attempts to clone this intergenic sequence were complicated by deletion and rearrangement of this DNA, suggesting that it may be a potential hot spot for recombination.
FIG. 4.
Promoter region of msrA and magA. SD, Shine-Dalgarno ribosome binding site. Arrows and italics indicate direct (DR) and indirect (IR) repeat sequences. The −10 and −35 hexamers are indicated in bold type and exhibit similarity to promoters recognized by RpoS.
Phenotypic characterization of a magA deletion mutant.
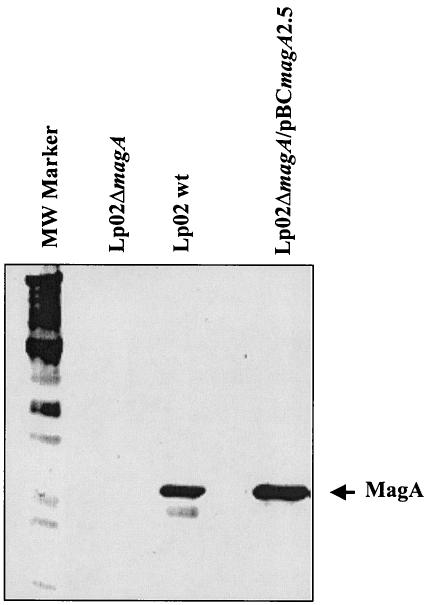
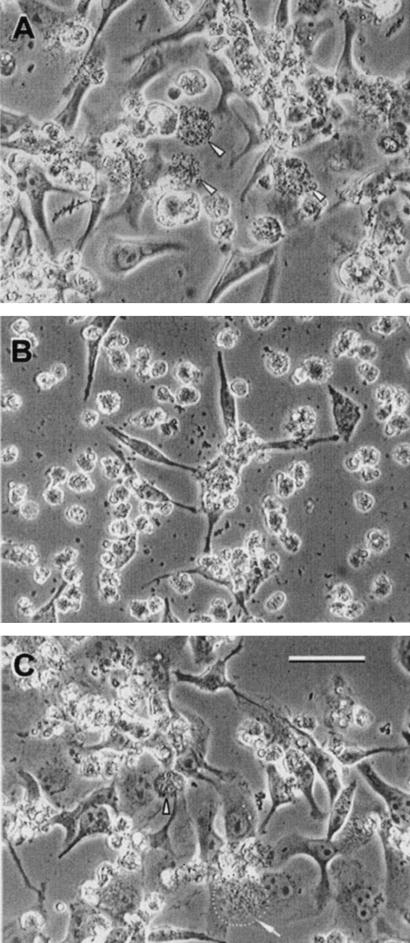
A magA deletion was created by insertion of a Kmr cassette in L. pneumophila Lp02 (Lp02ΔmagA) and the loss of MagA was verified by immunoblotting as depicted in Fig. 5. MagA production was restored by introduction of pBCmagA2.5 into this mutant (Fig. 5). The magA mutant, when compared with WT Lp02, was not defective in its growth characteristics in vitro, as evaluated by both the time taken to form similar numbers of visible colonies on BCYE plates and its growth rate in BYE broth (data not shown). Moreover, there was no loss in viability for stationary-phase bacteria (magA mutant or parent Lp02) suspended in dH2O after 3 days (>100% survival), indicating that the fitness of the mutant in water was preserved. With respect to infectivity, the magA mutant was invasive and able to grow intracellularly, as demonstrated in attachment-invasion assays with HeLa cells (data not shown) and in plaque assays with L929 cells (Table 2). However, we noticed that the magA mutant caused HeLa cells to round out and detach from the polystyrene substratum (Fig. 6). trans complementation of the magA mutant (either pBCmagA1.5 or pBCmagA2.5) restored vesicle formation and HeLa cell morphology (Fig. 6). In spite of the apparent cytopathic effects produced by the magA mutant on HeLa cells, the yield of bacteria per monolayer (three independent experiments) was not reduced compared to what was seen with the parental Lp02 strain (data not presented). The observation that HeLa cells infected with the magA mutant were unable to produce MIF-laden vesicles prompted us to examine whether MagA might function in MIF morphogenesis.
FIG. 5.
Western blot with MagA-specific antibody showing the absence of MagA in the Lp02ΔmagA mutant and restoration of MagA production in the complemented mutant. MW, molecular weight.
TABLE 2.
Comparison of parent strain Lp02 and magA mutant (ΔmagA) grown on BCYE agar plates or in HeLa cells (to form MIFs) with respect to plaque formation efficiency in L929 cells
| Strain | Agar grown
|
MIFsa
|
||||
|---|---|---|---|---|---|---|
| Inoculum per 105 L929 cells (107)b | No. of plaques (106)b | % Plaquing efficiency | Inoculum per 105 L929 cells (104) | No. of plaques (103) | % Plaquing efficiency | |
| Lp02 | 4.4 ± 2.6 | 2.2 ± 1.6 | 5.0 | 1.9 ± 1.3 | 0.34 ± 0.0 | 1.8 |
| ΔmagA | 5.4 ± 3.0 | 1.9 ± 0.05 | 3.5 | 1.3 ± 0.3 | 0.2 ± 0.15 | 1.5 |
MIFs from infected HeLa cells were isolated in a Percoll density gradient, washed in dH2O to eliminate residual Percoll, and used as inoculum to infect L929 cells.
Results are means ± standard deviations for duplicate experiments.
FIG. 6.
Phase-contrast light micrographs of HeLa cells infected with the L. pneumophila parent strain Lp02 (A), Lp02ΔmagA mutant (B), or the complemented magA mutant, Lp02ΔmagA/pBCmagA (C). Notice the reduction in attached HeLa cells as well as the increased number of small round cells in the monolayers infected by the magA mutant (B). Also apparent is the presence of numerous MIF-laden vesicles (indicated by arrowheads) in monolayers infected with the parent strain (A) and complemented mutant (C). These vesicles were almost completely absent in panel B. Bar, 25 μm (micrographs are the same scale).
The cell envelope ultrastructure of MIFs obtained from the magA mutant, as analyzed by transmission electron microscopy, was indistinguishable from that of wild-type MIFs, which has been described previously in great detail (11, 16). However, MIFs of the magA mutant exhibited alterations in their Giménez staining (Table 3), suggesting changes in organization or composition of the cell surface. It should be noted that a Gim(−) phenotype refers to bacteria showing a grayish, green, or blue-red shade after Giménez staining, as opposed to the Gim(+) phenotype which refers to bright red-colored bacteria after the Giménez staining (16). The efficiencies of plaque formation were very similar between the MIFs of the parent strain and the magA mutant, indicating that the lack of MagA did not impair the ability of the magA mutant to grow intracellularly (Table 2).
TABLE 3.
Comparison of MIFs (grown in HeLa cells)a of the parent strain Lp02, the magA mutant (ΔmagA), and the trans-complemented magA mutant (ΔmagA/pBCmagA) with respect to Giménez staining in four independent experiments
| Expt. no. | Lp02
|
Lp02ΔmagA
|
Lp02ΔmagA/pBCmagA
|
|||
|---|---|---|---|---|---|---|
| Total no. of bacteria counted | % Gim(+) bacteria | Total no. of bacteria counted | % Gim(+) bacteria | Total no. of bacteria counted | % Gim(+) bacteria | |
| 1 | 244 | 77.8 | 292 | 16.1 | 190 | 61.0 |
| 2 | 101 | 71.0 | 146 | 21.2 | 161 | 75.8 |
| 3 | 129 | 88.4 | 250 | 48.8 | 165 | 76.4 |
| 4 | 125 | 87.2 | 179 | 28.5 | 189 | 61.0 |
MIFs from infected HeLa cells were isolated in a Percoll density gradient, washed in dH2O to eliminate residual Percoll, and spread on a glass slide that was Giménez stained and observed using bright-field light microscopy. The number of red-stained bacteria was scored against the total number of bacteria found in 10 random fields.
DISCUSSION
In natural environments, L. pneumophila cycles between an intracellular RF in protozoa and an extracellular resilient cyst-like planktonic form or MIF (16, 18). In this study, we investigated the likelihood that the transition from RFs to MIFs involves many genes whose expression is developmentally controlled. Using a combination of comparative proteomics and pulse-chase radiolabeling of stationary-phase bacteria (in vitro) and MIFs obtained from infected HeLa cells, we found MagA to be the most abundant of several proteins produced during MIF morphogenesis and to a lesser extent in vitro in the transition from exponential to stationary phase. The structural gene (magA) is located in a pathogenicity island (LpPI-1) that is unique to the Philadelphia-1 strain of L. pneumophila serogroup 1 (4) and shares a divergent promoter region with msrA that encodes peptide methionine sulfoxide reductase, one of several such genes in the LpPI-1 (4). Deletion of magA had little effect on in vitro growth or virulence as assessed by plaque formation in L929 cells or growth in HeLa cells. However, several phenotypic traits associated with loss of function of MagA and restored by trans complementation were noted, including the following: (i) cyst-like MIFs obtained from HeLa cells infected with the magA mutant poorly retained the Giménez stain, a hallmark of the MIF phenotype (16), perhaps indicating a role for MagA in cell envelope morphogenesis; (ii) HeLa cells infected with the magA mutant did not form large bacteria-laden endosomal vesicles following disintegration of the host cells (Fig. 6); and (iii) magA mutant-infected HeLa cells displayed increased cytopathy (rounding up and detachment from polystyrene surface). The latter observations, together with a noted increase in the number of free bacteria in the HeLa cell culture, would suggest that the magA mutant hastens vesicle lysis and early release of bacteria. Thus, MagA, a putative redox active protein, may function to suppress events associated with early exit from host cells, a potential competitive advantage in natural environments.
The MagA protein was originally described by Miyamoto et al. (28) as a macrophage-induced L. pneumophila protein that was not observed during in vitro growth on agar or in broth culture. They further distinguished this protein from the well-characterized Mip protein, a prolyl cis-trans isomerase (10). However, annotation as a 24-kDa Mip-like protein has led to the perpetuation of an inaccurate description in the annotations of orthologs in other microbial genomes. Our studies confirm the work of Miyamoto and colleagues by indicating that MagA is not Mip and extend their work by demonstrating that magA is both cell cycle regulated in vitro and developmentally regulated in vivo. A combination of pulse radiolabeling and immunoblot tracking, together with previously performed two-dimensional gel analyses, showed that MagA appears postexponentially in vitro, coinciding with the onset of stationary phase, and late in intracellular infection of HeLa cells during post-stationary-phase MIF morphogenesis (15, 16). In contrast to the findings of Miyamoto et al., we detected low-level expression of MagA in exponentially growing bacteria in vitro but not early in infection of HeLa cells. The inability to detect MagA early in infection of HeLa cells is not due to low bacterial numbers at the early time points, since equal CFU were analyzed at each time point (pooled bacteria from several HeLa cell monolayers were used for the early time points). We suggest that magA expression may be more tightly controlled in vivo.
Several groups have shown that postexponential gene expression in L. pneumophila is generally under the control of the stationary-phase sigma factor RpoS and the stringent response regulator RelA (1, 7, 19, 35). Indeed, preliminary studies with an rpoS mutant of L. pneumophila Lp02 (obtained from Michele Swanson, University of Michigan) suggest that magA expression as well as MIF morphogenesis may be regulated in part by RpoS (unpublished data). Consistent with this possibility, the −10 hexamers in the shared promoter region for magA and msrA show sequence similarity with consensus RpoS promoter motifs. Since MagA levels are significantly higher in MIFs than in stationary-phase bacteria (16), other developmentally controlled regulatory factors, perhaps binding to direct and indirect repeat sequences noted in the intergenic promoter region, enhance gene expression in concert with RpoS.
MagA was originally annotated as a gene of unknown function (28); however, BLAST searches against complete and incomplete microbial genomes revealed a broad distribution among bacteria from Ralstonia spp. to the pseudomonads. These proteins contain a motif resembling that found in the alkylhydroperoxide reductase (AhpD) (6, 25). However, the motif found in MagA differs slightly from that found in AhpD (EX5SAX4CSSCX3H versus EX6SAX3CXXCX3H) and phylogenetic analyses suggest that these differences define two lineages. Indeed, MagA shows little similarity to the AhpD of L. pneumophila, whose gene is located adjacent to the ahpC alkylhydroperoxide reductase gene in the genome sequence (see Columbia University website mentioned above). These analyses do not necessarily exclude MagA from a role in providing reducing equivalents to AhpC as part of an oxygen defense system. Interestingly, our analysis reveals that L. pneumophila contains two copies of ahpC, one whose expression is up-regulated in macrophages and is in a locus containing an ortholog of glutaredoxin and the Fe-superoxide dismutase (31) and the other which is in a locus containing ahpD and the CuZn-superoxide dismutase. The lack of identity in amino acid sequence between the two AhpC proteins indicates that these genes are of different origins and potentially might be activated by different redox systems. At least three different redox systems have been described for activation of alkylhydroperoxide reductases (6, 25), and further study is required to resolve a role for MagA, if any, in protection from oxidative stress.
The genetic diversity among strains of L. pneumophila has only recently been explored, and like so many other pathogens, the legionellae also appear to have acquired genes from other organisms (4). It is noteworthy that the abundance and duplication of genes associated with protection from oxidative stress in L. pneumophila play critical roles in protecting the bacteria from the deleterious products produced by phagocytic hosts, whether amoebae or macrophages. An abundance of redox active proteins, including MagA in MIFs, might also serve as a source of reducing equivalents, perhaps similar to glutathione or thioredoxin, that are required in order to maintain a low redox potential during planktonic periods.
In summary, L. pneumophila displays a developmental cycle when it is intracellular in host cells and MagA, a putative reductase related to AhpD reductases, is an early marker of the transition from RFs to MIFs. MIFs are the most infectious form of L. pneumophila, capable of surviving for extended periods while between hosts. Interestingly, MIFs are not efficiently produced in macrophages, suggesting that MIFs are important in transmission of Legionnaires' disease (16). Further studies of magA and other developmentally regulated genes should advance our understanding of the developmental process and its underlying regulation.
Acknowledgments
We thank Kim Jefferies for assisting in the studies of MagA expression in HeLa cells.
This work was supported by grant MOP-14443 from the Canadian Institute for Health Research to P.S.H. and R.A.G. A.K.C.B. is a recipient of a postdoctoral fellowship from the Canadian Institutes for Health Research.
REFERENCES
- 1.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 2.Baker, L. M., A. Raudonikiene, P. S. Hoffman, and L. B. Poole. 2001. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 183:1961-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 4.Brassinga, A. K. C., M. F. Hiltz, G. R. Sisson, M. G. Morash, N. Hill, E. Garduno, P. H. Edelstein, R. A. Garduno, and P. S. Hoffman. 2003. A 65-kilobase pathogenicity island is unique to Philadelphia-1 strains of Legionella pneumophila. J. Bacteriol. 185:4630-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner, D. J., A. G. Steigerwalt, R. E. Weaver, J. E. McDade, J. C. Feeley, and M. Mandel. 1978. Classification of the Legionnaires' disease bacterium: an interim report. Curr. Microbiol. 1:71-75. [Google Scholar]
- 6.Bryk, R., C. D. Lima, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073-1077. [DOI] [PubMed] [Google Scholar]
- 7.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelstein, P. H., and R. D. Meyer. 1984. Legionnaires' disease. A review. Chest 85:114-120. [DOI] [PubMed] [Google Scholar]
- 10.Engleberg, N. C., C. Cater, D. R. Weber, N. P. Cianciotto, and B. I. Eisenstein. 1989. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect. Immun. 57:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faulkner, G., and R. A. Garduño. 2002. Ultrastructural analysis of differentiation in Legionella pneumophila. J. Bacteriol. 184:7025-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez, R. C., S. H. S. Lee, D. Haldane, R. Sumarah, and K. R. Rozee. 1989. Plaque assay for virulent Legionella pneumophila. J. Clin. Microbiol. 27:1961-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garduño, R. A., G. Faulkner, M. A. Trevors, N. Vats, and P. S. Hoffman. 1998. Immunolocalization of Hsp60 in Legionella pneumophila. J. Bacteriol. 180:505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garduño, R. A., E. Garduño, M. Hiltz, D. Allan, and P. S. Hoffman. 2002. Morphological and physiological evidence for a developmental cycle in Legionella pneumophila, p. 82-85. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Lück (ed.), Legionella. ASM Press, Washington, D.C.
- 16.Garduño, R. A., E. Garduño, M. Hiltz, and P. S. Hoffman. 2002. Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 70:6273-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garduño, R. A., F. D. Quinn, and P. S. Hoffman. 1998. HeLa cells as a model to study the invasiveness and biology of Legionella pneumophila. Can. J. Microbiol. 44:430-440. [PubMed] [Google Scholar]
- 18.Greub, G., and D. Raoult. 2003. Morphology of Legionella pneumophila according to their location within Hartmanella vermiformis. Res. Microbiol. 154:619-621. [DOI] [PubMed] [Google Scholar]
- 19.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 20.Helsel, L. O., W. F. Bibb, C. A. Butler, P. S. Hoffman, and R. M. McKinney. 1988. Recognition of a genus-wide antigen of Legionella by a monoclonal antibody. Curr. Microbiol. 16:201-208. [Google Scholar]
- 21.Hoffman, P. S. 1997. Invasion of eukaryotic cells by Legionella pneumophila: a common strategy for all hosts? Can. J. Infect. Dis. 8:139-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffman, P. S., C. A. Butler, and F. D. Quinn. 1989. Cloning and temperature-dependent expression in Escherichia coli of a Legionella pneumophila gene coding for a genus-common 60-kilodalton antigen. Infect. Immun. 57:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman, P. S., J. H. Seyer, and C. A. Butler. 1992. Molecular characterization of the 28- and 31-kilodalton subunits of the Legionella pneumophila porin. J. Bacteriol. 174:908-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James, B. W., W. S. Mauchline, P. J. Dennis, C. W. Keevil, and R. Wait. 1999. Poly-3-hydroxybutyrate in Legionella pneumophila, an energy source for survival in low-nutrient environments. Appl. Environ. Microbiol. 65:822-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koshkin, A., C. M. Nunn, S. Djordjevic, and P. R. Ortiz de Montellano. 2003. The mechanism of Mycobacterium tuberculosis alkylhydroperoxidase AhpD as defined by mutagenesis, crystallography, and kinetics. J. Biol. Chem. 278:29502-29508. [DOI] [PubMed] [Google Scholar]
- 26.Lacour, S., A. Kolb, and P. Landini. 2003. Nucleotides from −16 to −12 determine specific promoter recognition by bacterial σS-RNA polymerase. J. Biol. Chem. 278:37160-37168. [DOI] [PubMed] [Google Scholar]
- 27.McDade, J. E. 1979. Primary isolation using guinea pigs and embryonated eggs, p. 69-75. In G. L. Jones and G. A. Hébert (ed.), “Legionnaires',” the disease, the bacterium, and methodology. HEW publication no. (CDC)79-8375. U.S. Department of Health, Education, and Welfare, Atlanta, Ga.
- 28.Miyamoto, H., S. Yoshida, H. Taniguchi, M. H. Qin, H. Fujio, and Y. Mizuguchi. 1993. Protein profiles of Legionella pneumophila Philadelphia-1 grown in macrophages and characterization of a gene encoding a novel 24-kDa Legionella protein. Microb. Pathog. 15:469-484. [DOI] [PubMed] [Google Scholar]
- 29.Oka, A., H. Sugsaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn 903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 30.Pascule, A. W., J. C. Feeley, R. J. Gibson, L. G. Cordes, R. L. Meyerowitz, C. M. Patton, G. W. Gorman, C. L. Carmack, J. W. Ezzell, and J. N. Dowling. 1980. Pittsburgh pneumonia agent: direct isolation from human lung tissue. J. Infect. Dis. 141:727-732. [DOI] [PubMed] [Google Scholar]
- 31.Rankin, S., Z. Li, and R. R. Isberg. 2002. Macrophage-induced genes of Legionella pneumophila: protection from reactive intermediates and solute imbalance during intracellular growth. Infect. Immun. 70:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadoff, H. L. 1975. Encystment and germination in Azotobacter vinelandii. Bacteriol. Rev. 39:516-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor, A. B., D. M. Benglis, Jr., S. Dhandayuthapani, and P. J. Hart. 2003. Structure of Mycobacterium tuberculosis methionine sulfoxide reductase A in complex with protein-bound methionine. J. Bacteriol. 185:4119-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]