Abstract
RNA silencing has become a major focus of molecular and biomedical research in the last decade. This mechanism, which is conserved in most eukaryotes, has been extensively studied and is associated to various pathways implicated in the regulation of development, in the control of transposition events, heterochromatin maintenance and also playing a role in defense against viruses. Despite of its importance, the regulation of the RNA silencing machinery itself remains still poorly explored. Recently several reports in both plants and metazoans revealed that key components of RNA silencing, such as RNA-induced silencing complex component ARGONAUTE proteins, but also the endonuclease Dicer are subjected to proteasomal and autophagic pathways. Here we will review these post-translational proteolytic regulations with a special emphasis on plant research and also discuss their functional relevance.
Keywords: RNA silencing, RISC, ubiquitin, autophagy, virus, proteasome
A GLIMPSE IN THE RNA SILENCING PATHWAYS
RNA silencing involves processing of double stranded (dsRNA) by the enzyme Dicer, into small RNAs, 21–25 nucleotides in length called small interfering RNAs (siRNAs; reviewed in (Ghildiyal and Zamore, 2009; Voinnet, 2009; Krol et al., 2010). One of the two strands of each RNA fragment is then incorporated into a protein complex called RNA induced silencing complex (RISC) that invariably contains a member of the highly conserved ARGONAUTE protein (AGO) family. Once integrated into the RISC, siRNAs will base-pair to their target mRNA and induce their cleavage. The process of RNA interference (RNAi) is widely used for functional genomics and has also practical applications in therapeutics and agriculture. Most importantly, RNA silencing mediates resistance to exogenous pathogenic nucleic acids. Thus, important functions for small RNAs have emerged in the study of host-pathogen interactions and the most compelling illustration of the role of RNA silencing in defense is provided in the case of viral infections in plants, invertebrates and also more recently mammals, where populations of siRNAs are produced in infected cells directly by processing dsRNA molecules derived from the viral genome (Ding, 2010; Maillard et al., 2013). These viral-derived siRNAs are then incorporated into an antiviral RISC and turned back onto viral RNAs to trigger their degradation.
RNA silencing also regulates the expression of protein-coding genes. In this process, an important source of endogenous dsRNAs are primary transcripts of RNA-coding genes called pri-miRNAs which are processed, in the nucleus of metazoan cells, to 70-nucleotide stem-loop pre-miRNAs by the RNase III enzyme Drosha (Siomi and Siomi, 2010). After their export to the cytoplasm, pre-microRNAs are further processed via Dicer or Dicer-like (DCL) enzymes to produce miRNA duplexes. Plant genomes do not encode Drosha homologs, and all miRNA biogenesis steps at least in Arabidopsis are carried out by one of the four DCL proteins (Rogers and Chen, 2013). The microRNA (miRNA) duplex is separated, and one strand is selected as the 21-nucleotide mature miRNAs, whereas the other strand is degraded. Mature miRNAs are integrated into RISC complexes that repress the expression of one or more target mRNAs with complementary sequence by inhibiting mRNA translation or inducing their degradation. Thus miRNAs are predicted to regulate the expression of hundreds of mRNAs suggesting that they can regulate a significant proportion of the transcriptome (Leung and Sharp, 2010). Notably it has recently been shown that miRNAs are also subjected to turnover through degradation mechanisms implying both 3′–5′ and 5′–3′ exoribonucleases, adding another layer of complexity (Ramachandran and Chen, 2008; Chatterjee and Grosshans, 2009; Rüegger and Grosshans, 2012).
REGULATION OF THE RNA SILENCING MACHINERY BY AUTOPHAGY
While the biogenesis and the function of small RNAs have been extensively studied in various biological processes across many organisms, less attention was paid on the regulation of the RNA silencing machinery itself. As indicated above, AGOs are core components of the RISC (Hutvágner and Simard, 2008; Vaucheret, 2008; Voinnet, 2009). These proteins have undergone a high degree of gene duplication in metazoans and plants, counting 8 and 10 genes in humans and Arabidopsis, respectively. Genetic and biochemical analyses revealed that Arabidopsis AGO1 plays a central role in both miRNA and si-mediated RNA silencing (Mi et al., 2008; Takeda et al., 2008). Based on its key role as effectors in RNA silencing, it is expected that AGO1 protein abundance must be strictly regulated, most likely at multiple levels. Hence, either an increase or a decrease in AGO1 protein content leads to significant effects on plant development (Vaucheret et al., 2004, 2006). The most studied and best-understood mechanism controlling AGO1 homeostasis is its negative regulation by miRNA168 (Vaucheret et al., 2006; Mallory and Vaucheret, 2009). In this pathway, the miRNA miR168 represses AGO1 transcript in an AGO1-dependant manner. Besides AGO1, other elements of the RNA silencing machinery, like DCL1 or AGO2 are also regulated via specific miRNAs, respectively, miR162 and miR403 (Xie et al., 2003; Allen et al., 2005). However, it became evident that AGO1 is also regulated at the post-translational level and in particular at the level of its stability.
The first evidence of selective AGO1 protein turnover was in the context of plant-viral interactions. Arabidopsis AGO1 is not only involved in the miRNA pathway, but together with AGO2 mediates antiviral defense (Alvarado and Scholthof, 2011). As a counter defense, viruses have elaborated various strategies to avoid silencing by expressing Viral Suppressors of RNA silencing (VSRs) proteins (Pumplin and Voinnet, 2013). Interestingly, it was found that certain VSRs, called P0 proteins from Poleroviruses, promote the degradation of AGO1 and thus presumably could impair RNA-based anti-viral immunity (Baumberger et al., 2007; Bortolamiol et al., 2007). This mechanism is conserved and was extended to VSRs of other viruses (Chiu et al., 2010; Fusaro et al., 2012). Interestingly, it was shown that P0 acts upstream of AGO1 loading and thus would prevent the formation of RISC (Csorba et al., 2010). This is supported by the fact that newly synthesized AGO1 after transient expression in tobacco leaves is subjected to P0-mediated destruction while endogenous AGO1 pre-assembled complex is P0-resistant. At the molecular level, viral P0 VSRs encode F-box proteins (Pazhouhandeh et al., 2006) that hijack the host SKP1-Cullin1-F-box protein (SCF) ubiquitin-protein ligase (E3) to promote ubiquitylation, which serves as a signal for degradation. This post-translational modification (PTM) regulates a broad range of physiologically and developmentally controlled processes in all eukaryotes (Ciechanover et al., 2000; Smalle and Vierstra, 2004). Because ubiquitylation of target proteins by SCF-type complexes most often leads to their proteasomal degradation, it was a surprise to find that the degradation of AGO1 by P0 was insensitive to inhibition of the proteasome (Baumberger et al., 2007). The mystery of AGO1 degradation pathway by the SCFP0 E3 ligase was, however, solved when it was reported that this process is mediated by autophagy (Derrien et al., 2012). Although recent studies already indicate a function of ubiquitylation in autophagy (McEwan and Dikic, 2011), this finding was nevertheless intriguing with respect to the presumed high selectivity of the P0-mediated ubiquitylation process, as degradation by autophagy is generally believed to be unspecific, even taking into account “selective autophagy” destroying protein aggregates and organelles.
Because viruses usually hijack host cell machineries, it was conceivable that AGO1 protein turnover by autophagy may also occur in a P0-independent context. Hence, this prediction was confirmed when it was shown that mutations affecting miRNA biogenesis and/or accumulation and thus disturbing RISC assembly, also result in AGO1 degradation by autophagy (Derrien et al., 2012). This finding, however, raises the question of which is the endogenous ubiquitin-protein ligase (E3) that promotes ubiquitylation of AGO1 in a non-viral context. Notably Arabidopsis genome encodes several classes of E3s that are the key factors defining substrate specificity and among them more than 700 hundred F-box proteins (Vierstra, 2009). One good candidate to fulfill such a function is the Arabidopsis F-box protein FBW2 (Earley et al., 2010). FBW2 was identified by a genetic suppressor screen of a null allele of SQUINT (SQN), encoding a Cyclophilin-40 chaperon, a positive regulator of AGO1 activity. While FBW2 loss-of-function mutants do not exhibit an increase in AGO1 protein level, most likely because of the miR168-dependent feedback mechanism regulating AGO1 expression (Vaucheret et al., 2006), FBW2 overexpression significantly reduces AGO1 protein content (Earley et al., 2010). Interestingly, the proteasome inhibitor MG132 was also unable to block the FBW2-mediated degradation of AGO1, a situation reminiscent to the viral SCFP0 complex. At present it remains unclear whether FBW2 mediates AGO1 destruction by autophagy similarly to viral P0 (Figure 1), with which FBW2 does not share any significant sequence similarity beside an F-box motif.
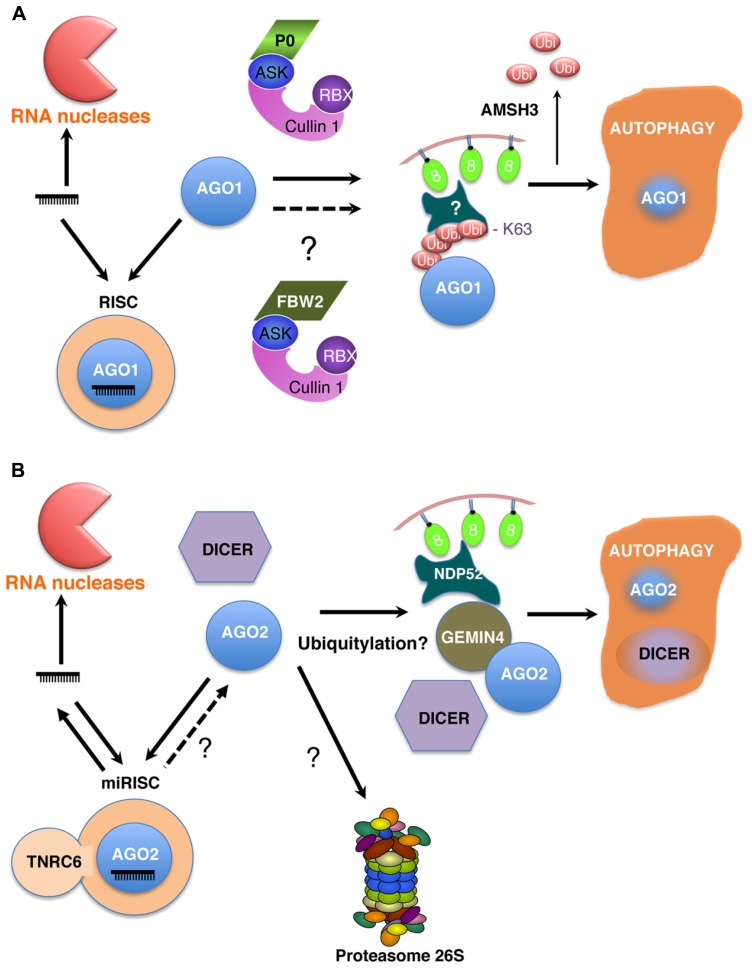
FIGURE 1.
Models for the turnover of AGO proteins in Arabidopsis (A) and mammalian cells (B). Different levels of regulation operate on the homeostasis of RISCs. First, the steady-state levels of microRNAs are regulated by degradation processes involving different ribonucleases (RNases) acting either 3′–5′ or 5′–3′. Thus microRNAs most likely are in competition for AGO binding. Recent evidences essentially from metazoans indicate that at least some microRNAs can be released from RISCs, explaining their short half-lives. However, not only microRNAs but also AGO proteins are degraded. Thus in both plants and animal cells, it is now clearly established that AGO proteins are degraded by autophagy in an RNA free form prior RISC assembly. This mechanism also co-degrades other components of the silencing machinery such as DICER in mammals (B). In Arabidopsis, the polerovirus protein P0 assembles an SCFP0 ubiquitin ligase to ubiquitylate AGO1 or an AGO1 associated protein (A). Viral P0-mediated AGO1 degradation by autophagy also requires the deubiquitylating enzyme AMSH3. However, the identity of endogenous ubiquitin ligases involved in this process have not yet been unambiguously identified. The role of ubiquitylation in the turnover of human AGO2 is at present unclear but requires GEMIN4 and NDP52 (B). Finally, whether upon guide RNA dissociation AGO2 would become accessible to autophagy and the role of the proteasome in AGOs degradation are other still open questions.
Notably AGO1 is not the only Arabidopdsis AGO so tightly regulated at the post-translational level. Thus, at least in transient expression assays in tobacco leaves, P0 is also able to mediate the degradation of AGO2, AGO4-6, and AGO9 (Baumberger et al., 2007). Whether those AGOs are targeted by the endogenous SCFFBW2 is presently unknown, thought some of them have already been identified as ubiquitylated by proteomic approaches (Maor et al., 2007; Kim et al., 2013).
Is autophagy-mediated regulation of AGO proteins specific to the green linage? The answer is no and several findings suggest that the fate of the animal RNA silencing machinery shares some striking similarities with plants. Previous studies already reported that in mammals, AGO proteins are regulated at the post-translational level. For instance, in human cells, AGO2 (the only mammalian AGO producing RNA cleavage) is both hydroxylated and phosphorylated (Qi et al., 2008; Zeng et al., 2008). In particular hydroxylation was shown to influence both AGO2 subcellular localisation and stability, although the biological significance of this modification is still unclear.
Significant molecular insights on the post-translational control of metazoan AGO proteins emerged only recently. First it has been shown that the molecular chaperone HSP90 is required for the stability of mammalian AGO1 and AGO2 (Johnston et al., 2010). Thus inhibition of HSP90 function by geldanamycin triggered the degradation of both AGOs, an effect that could be alleviated, at least partially, by the proteasome inhibitor MG132. Interestingly, HSP90 does not bind AGO2 complexes that contain miRNAs and was therefore proposed to act upstream of RISC action indicating already that it is RNA-free AGO2 that is degraded in this pathway (Johnston et al., 2010). However, the first ubiquitin E3 ligase proposed to control mammalian AGO2 stability was the mouse Trim domain containing protein mLIN41 (Rybak et al., 2009). This protein is preferentially expressed in several stem cell niches and participates in the control of stem cell maintenance. mLIN41 physically interacts with AGO2 through its coiled-coil domain and promotes AGO2 ubiquitylation in vitro and in vivo through its RING and B-Box domains, all located in the Trim domain. Moreover, the ectopic overexpression of mLin41 reduced the level of endogenous Ago2 in embryonic carcinoma cells and this effect was attenuated by inhibition of the proteasome with MG132. However, more recent studies put into question the control of AGO2 stability by mLIN41 (Chang et al., 2012; Chen et al., 2012). In particular, it was shown that mLin41 promotes neuronal progenitor cell maintenance through FGF signaling by ubiquitylation of Shc SH2-binding protein 1 (SHCBP1), but not via the regulation of AGO2 stability (Chen et al., 2012).
While the turnover of AGO2 by the mLIN41-proteasome pathway will need further investigations, the degradation of Argonautes proteins by the autophagy pathway turned out to be conserved across kingdoms (Figure 1). Hence it was shown that both DICER and AGO2 levels increased in HeLa cells treated with chemical inhibitors known to block autophagy and in siRNA-depleted cells for different component of the autophagy pathway, such as ATG5, ATG6, ATG7 or NDP52 (Gibbings et al., 2012). Of particular interest was NDP52, a known autophagy receptor, which confers some cargo selectivity typically by recognizing conjugated ubiquitin (Rogov et al., 2014). At present the mechanism by how NDP52 recognizes AGO2 and DICER is unclear, but GEMIN4, a component of the multi-protein SMN (survival of motor neuron) complex, is required in this process eventually by interacting with both NDP52 and AGO2. Whether AGO2 ubiquitylation is a prerequisite to be directed to autophagy is unknown. In contrast DICER might be recruited by a mechanism independent of GEMIN4. Moreover similar to plant AGO1 decay (Csorba et al., 2010), mammalian AGO2 autophagy-mediated degradation occurs upstream of the formation of miRISC (Gibbings et al., 2012).
This novel paradigm of the post-translational control of the RNA silencing machinery exhibits nevertheless some variations. Hence in Caenorhabditis elegans, the Ago homologs ALG-1 and ALG-2 accumulate into aggregates in autophagy mutants only under certain stress conditions and the role of selective autophagy in their regulation under normal physiological conditions is presently unclear (Zhang and Zhang, 2013). Instead, AIN-1, a homolog of mammalian GW182/TNRC6 that interacts with AGO and mediates silencing, is clearly degraded by autophagy (Zhang and Zhang, 2013). AIN-1 colocalizes with SQST-1, the homolog of mammalian p62 that acts as a receptor for autophagic degradation of ubiquitylated protein aggregates and also directly interacts with Atg8/LC3 contributing to cargo specificity. This mechanism seems also to involve EPG-7 a scaffold protein linking cargo-receptor complexes with the autophagic assembly machinery (Lin et al., 2013). The putative role of ubiquitylation in the mechanism of AIN-1 destruction will nevertheless need further investigations.
SOME PERSPECTIVES
It is clear that more work is required to better understand post-translational regulations of AGOs in both plants and metazoans. Moreover in plants, it will also be important to characterize the different protein complexes containing AGOs and their subcellular locations. We already know that plant AGO1 is present in both low and high molecular protein complexes that co-fractionate with small RNAs (Baumberger and Baulcombe, 2005; Qi et al., 2005; Csorba et al., 2010). Whether these multi-protein complexes resemble those identified in mammals (Filipowicz et al., 2008) remains, however, to be established. Moreover, evidence of two distinct cellular pools of AGO1 (siRNA versus miRNA loaded AGO1) RISCs was also recently established (Schott et al., 2012). In addition, in Arabidopsis, at least a fraction of AGO1 is also associated to membranes and isoprenoid biosynthesis which is important for membrane protein localization and trafficking, is required for miRNA function (Brodersen et al., 2012). Mammalian AGO2 was already known to bind to cellular membranes, most likely as a component of RISC (Cikaluk et al., 1999). AGO proteins are therefore present in cells as various pools representing likely different functional states. How are these different AGO protein pools regulated at the post-translational level and what is the impact of these regulations on RNAi function are major questions that will have to be solved.
Concerning the process of AGOs degradation by autophagy, an important issue will be to clarify the role of ubiquitylation. In plants, immunoprecipitation assays revealed an enrichment of polyubiquitin conjugates of AGO1 and/or an AGO1-associated protein and MLN-4924, a drug that inhibits the activity of cullin-RING ubiquitin ligases, impaired P0-dependant AGO1 degradation in Arabidopsis (Derrien et al., 2012). In mammals, AGO2 or one of its associated protein was also found ubiquitylated in cells treated with siRNAs to deplete autophagy activity (Gibbings et al., 2012). Notably, ubiquitin contains seven internal lysine residues and all can serve as conjugation sites to build up poly-ubiquitin chains that depending on their topologies can direct the substrate to the 26S proteasome or to the autophagy pathway (Grabbe et al., 2011; McEwan and Dikic, 2011). Therefore future experiments should reveal the identity of endogenous ubiquitin E3 ligases involved in this process, where and how they recognize AGOs or other associated proteins, the topology of the polyubiquitin chains that are generated and how these chains will be selected by the autophagy pathway.
Notably, at present we cannot rule out the possibility that the 26S proteasome also plays important functions in controlling the homeostasis of the RNA silencing machinery, as both proteolytic pathways may coexist, eventually in different cell types or specific developmental contexts. For instance, several studies incriminate the proteasome in controlling the stability of Drosophila and mammalian AGO effector proteins (Johnston et al., 2010; Smibert et al., 2013). Also in plants, the silencing suppressor protein P25 of Potato virus X (PVX) triggers AGO1 destabilization by the proteasome (Chiu et al., 2010). The mechanism by which this is achieved is unknown, but P25 might recruit a still unknown endogenous ubiquitin ligase complex to achieve such a function. Moreover, the Double-stranded RNA Binding protein (DRB4) that interacts with DCL4, one of the four Dicer-like proteins present in Arabidopsis, is also degraded by the proteasome after being recognized by the APC/C (anaphase promoting complex or cyclosome), a master ubiquitin protein ligase that usually targets cell cycle regulatory proteins (Marrocco et al., 2012). Thus to understand the contribution of proteasomal degradation versus the autophagy pathway in fine-tuning components of the RNA silencing machinery needs further investigations, both in metazoans and plants.
Finally, the most interesting question is what could be the physiological function(s) of these proteolytic pathways? The current model indicates that the stability of AGO proteins depends on miRNA biogenesis and thus unloaded AGOs are unstable (Derrien et al., 2012; Martinez and Gregory, 2013; Smibert et al., 2013). If AGO proteins are degraded essentially prior RISC assembly (Csorba et al., 2010; Johnston et al., 2010; Gibbings et al., 2012), the key regulatory step would be at the level of small RNA production that would compete for binding of available AGOs. In such a scenario, the P0 proteins from poleroviruses would destroy AGO1 at an early step to prevent viral siRNAs produced during infection to be incorporated into novel RISCs and this would compromise antiviral RNA silencing. However, what is the fate of AGO proteins once part of small RNA programmed RISCs? In mammals the half-life of Ago2 bound to small RNAs seems rather stable, at least under normal grow conditions (Johnston et al., 2010). Similarly, a half-life of 2–3 days of AGO1 RISCs was estimated in plants (Csorba et al., 2010). However, recent findings revealed that target RNAs could destabilize the interaction between human Ago2 and their corresponding guide RNAs, indicating that at least some RISCs can be unloaded (De et al., 2013). Such a dynamic loading and unloading mechanism might not only allow reprogramming of Ago2 by novel guide RNAs, but might also expose the protein to cellular degradation machineries such as the autophagy pathway. If this holds true, what would be the functional relevance of this degradation on RISC homeostasis and reprogramming?
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
“This work has been published under the framework of the LABEX: ANR-10-LABX-0036_NETRNA and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program” and from the European Research Council under the European Union’s Seventh Framework Program (FP7/2007-2013)/ERC grant agreement n° [338904].
REFERENCES
- Allen E., Xie Z., Gustafson A. M., Carrington J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221 10.1016/j.cell.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Alvarado V. Y., Scholthof H. B. (2011). AGO2: a new argonaute compromising plant virus accumulation. Front. Plant Sci. 2:112 10.3389/fpls.2011.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D. C. (2005). Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 102 11928–11933 10.1073/pnas.0505461102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Tsai C.-H., Lie M., Havecker E., Baulcombe D. C. (2007). The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17 1609–1614 10.1016/j.cub.2007.08.039 [DOI] [PubMed] [Google Scholar]
- Bortolamiol D., Pazhouhandeh M., Marrocco K., Genschik P., Ziegler-Graff V. (2007). The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17 1615–1621 10.1016/j.cub.2007.07.061 [DOI] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Schaller H., Khafif M., Schott G., Bendahmane A., et al. (2012). Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 1778–1783 10.1073/pnas.1112500109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.-M., Martinez N. J., Thornton J. E., Hagan J. P., Nguyen K. D., Gregory R. I. (2012). Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 3 923 10.1038/ncomms1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S., Grosshans H. (2009). Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461 546–549 10.1038/nature08349 [DOI] [PubMed] [Google Scholar]
- Chen J., Lai F., Niswander L. (2012). The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 26 803–815 10.1101/gad.187641.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu M.-H., Chen I.-H., Baulcombe D. C., Tsai C.-H. (2010). The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 11 641–649 10.1111/j.1364-3703.2010.00634.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A., Orian A., Schwartz A. L. (2000). Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 22 442–451 [DOI] [PubMed] [Google Scholar]
- Cikaluk D. E., Tahbaz N., Hendricks L. C., DiMattia G. E., Hansen D., Pilgrim D., et al. (1999). GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell 10 3357–3372 10.1091/mbc.10.10.3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T., Lózsa R., Hutvágner G., Burgyán J. (2010). Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 62 463–472 10.1111/j.1365-313X.2010.04163.x [DOI] [PubMed] [Google Scholar]
- De N., Young L., Lau P.-W., Meisner N.-C., Morrissey D. V, MacRae I. J. (2013). Highly complementary target RNAs promote release of guide RNAs from human Argonaute2. Mol. Cell 50 344–355 10.1016/j.molcel.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien B., Baumberger N., Schepetilnikov M., Viotti C., De Cillia J., Ziegler-Graff V., et al. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U.S.A. 109 15942–15946 10.1073/pnas.1209487109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.-W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10 632–644 10.1038/nri2824 [DOI] [PubMed] [Google Scholar]
- Earley K., Smith M., Weber R., Gregory B., Poethig R. (2010). An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1 15 10.1186/1758-907X-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9 102–114 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- Fusaro A. F., Correa R. L., Nakasugi K., Jackson C., Kawchuk L., Vaslin M. F. S., et al. (2012). The Enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology 426 178–187 10.1016/j.virol.2012.01.026 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M., Zamore P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10 94–108 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D., Mostowy S., Jay F., Schwab Y., Cossart P., Voinnet O. (2012). Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat. Cell Biol. 14 1314–1321 10.1038/ncb2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabbe C., Husnjak K., Dikic I. (2011). The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 12 295–307 10.1038/nrm3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G., Simard M. J. (2008). Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9 22–32 10.1038/nrm2321 [DOI] [PubMed] [Google Scholar]
- Johnston M., Geoffroy M.-C., Sobala A., Hay R., Hutvágner G. (2010). HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol. Biol. Cell 21 1462–1469 10.1091/mbc.E09-10-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-Y., Scalf M., Smith L. M., Vierstra R. D. (2013). Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25 1523–1540 10.1105/tpc.112.108613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11 597–610 10.1038/nrg2843 [DOI] [PubMed] [Google Scholar]
- Leung A. K. L., Sharp P. A. (2010). MicroRNA functions in stress responses. Mol. Cell 40 205–215 10.1016/j.molcel.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Yang P., Huang X., Zhang H., Lu Q., Zhang H. (2013). The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J. Cell Biol. 201 113–129 10.1083/jcb.201209098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard P. V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S. W., et al. (2013). Antiviral RNA interference in mammalian cells. Science 342 235–238 10.1126/science.1241930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A. C., Vaucheret H. (2009). ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 10 521–526 10.1038/embor.2009.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor R., Jones A., Nühse T. S., Studholme D. J., Peck S. C., Shirasu K. (2007). Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell Proteomics 6 601–610 10.1074/mcp.M600408-MCP200 [DOI] [PubMed] [Google Scholar]
- Marrocco K., Criqui M.-C., Zervudacki J., Schott G., Eisler H., Parnet A., et al. (2012). APC/C-mediated degradation of dsRNA-binding protein 4 (DRB4) involved in RNA silencing. PLoS ONE 7:e35173 10.1371/journal.pone.0035173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N. J., Gregory R. I. (2013). Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA 19 605–612 10.1261/rna.036434.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D. G., Dikic I. (2011). The three musketeers of autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 21 195–201 10.1016/j.tcb.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 133 116–127 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M., Dieterle M., Marrocco K., Lechner E., Berry B., Brault V., et al. (2006). F-box-like domain in the Polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. U.S.A. 103 1994–1999 10.1073/pnas.0510784103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Voinnet O. (2013). RNA silencing suppression by plant pathogens: defense, counter-defense and counter-counter-defense. Nat. Rev. Microbiol. 11 745–760 10.1038/nrmicro3120 [DOI] [PubMed] [Google Scholar]
- Qi H. H., Ongusaha P. P., Myllyharju J., Cheng D., Pakkanen O., Shi Y., et al. (2008). Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 455 421–424 10.1038/nature07186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Denli A. M., Hannon G. J. (2005). Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19 421–428 10.1016/j.molcel.2005.06.014 [DOI] [PubMed] [Google Scholar]
- Ramachandran V., Chen X. (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321 1490–1492 10.1126/science.1163728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25 2383–2399 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., Kirkin V. (2014). Interactions between autophagy receptors and Ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53 167–178 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Rüegger S., Grosshans H. (2012). MicroRNA turnover: when, how, and why. Trends Biochem. Sci. 37 436–446 10.1016/j.tibs.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Hadian K., Smirnova L., Wulczyn E. A., Michel G., et al. (2009). The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 11 1411–1420 10.1038/ncb1987 [DOI] [PubMed] [Google Scholar]
- Schott G., Mari-Ordonez A., Himber C., Alioua A., Voinnet O., Dunoyer P. (2012). Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J. 31 2553–2565 10.1038/emboj.2012.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Siomi M. C. (2010). Post-transcriptional regulation of microRNA biogenesis in animals. Mol. Cell 38 323–332 10.1016/j.molcel.2010.03.013 [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R. D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590 10.1146/annurev.arplant.55.031903.141801 [DOI] [PubMed] [Google Scholar]
- Smibert P., Yang J.-S., Azzam G., Liu J.-L., Lai E. C. (2013). Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol. 20 789–795 10.1038/nsmb.2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49 493–500 10.1093/pcp/pcn043 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13 350–358 10.1016/j.tplants.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Mallory A. C., Bartel D. P. (2006). AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22 129–136 10.1016/j.molcel.2006.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P., Bartel D. P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197 10.1101/gad.1201404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10 385–397 10.1038/nrm2688 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136 669–687 10.1016/j.cell.2009.01.046 [DOI] [PubMed] [Google Scholar]
- Xie Z., Kasschau K. D., Carrington J. C. (2003). Negative feedback regulation of Dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13 784–789 10.1016/S0960-9822(03)00281-1 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Sankala H., Zhang X., Graves P. R. (2008). Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J. 413 429–436 10.1042/BJ20080599 [DOI] [PubMed] [Google Scholar]
- Zhang P., Zhang H. (2013). Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 14 568–576 10.1038/embor.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]