Abstract
TcaR, which shares sequence homology with MarR-like transcriptional regulators, has been identified as a novel Staphylococcus aureus regulator affecting the expression of the global regulatory element SarS (SarH1), as well as that of the cell surface-associated protein SasF (N315-SA2439). Microarray analysis, confirmatory Northern blots, and genetic complementation experiments showed that TcaR upregulates sarS and thus spa transcription. In addition, it attenuates whole-length transcription of sasF, thereby producing a truncated transcript lacking the 3′ terminus, which codes for the cell wall anchor motif. Hence, in strains containing an intact tcaR gene, TcaR is likely to decrease the amount of the surface-associated protein SasF and to increase that of the surface-associated protein A. The widely used laboratory strains derived from NCTC8325 were found to be natural, truncated mutants of tcaR, harboring an inactive TcaR and therefore expressing very low levels of sarS. The data presented here identified TcaR as a further activator of sarS, and a modulator of sasF expression that has to be taken into account in studies of virulence gene expression in S. aureus.
The success of Staphylococcus aureus as an invasive pathogen is largely due to the production of a diverse array of virulence determinants. Key factors include excreted toxins, such as superantigens and exoenzymes, which are implicated in the induction of severe clinical conditions, and cell surface-associated proteins that facilitate evasion of host defenses and bacterial adhesion. Adherence of the bacterium to host factors, such as extracellular matrix and plasma components, and to artificial surfaces, such as indwelling medical devices, is supposed to be essential for host invasion and persistence. Of the 21 predicted S. aureus cell wall-associated surface proteins, 11 have been characterized and/or implicated in virulence (26). These proteins are characterized by a N-terminal signal peptide for Sec-dependent secretion, and a conserved LPXTG motif at the C terminus, which is cleaved by sortase to anchor them to the cell wall peptidoglycan (23, 26). Surface proteins have been shown to facilitate binding to host extracellular matrix proteins (16, 34), human platelets, cartilage, and fibronectin (reviewed in references 8 and 25).
The coordinated expression of these cell wall-associated virulence determinants over the growth cycle or during infection is under the control of a complex global regulatory network. Several global regulators have been shown to affect virulence factor production, including RNAIII of the agr locus (reviewed in references 3 and 25), SarA, and homologues belonging to the SarA protein family (8), the alternate sigma factor σB (13, 14), and two-component regulatory systems such as saeRS, srrAB, and arlRS (reviewed in reference 8).
The tcaRAB region has been shown to be involved in some unknown manner in teicoplanin and methicillin resistance (5). However, neither the functions of the single genes affected, their contribution to antibiotic resistance levels, nor their effect on cell wall metabolism are known. We show here that TcaR acts as a regulatory factor, which affects the transcription of sarS (synonym, sarH1), a member of the global regulatory network, and the transcription of the cell wall-anchored proteins encoded by spa and sasF.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. Bacterial cultures were grown in Luria-Bertani (LB) medium (Becton Dickinson), with shaking, at 37°C. Media were supplemented with chloramphenicol at 20 μg ml−1 or kanamycin at 50 μg ml−1 when required. Sodium salicylate (Fluka) was added to the media at a concentration of 50 μg ml−1 where indicated.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype and/or relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | NCTC8325-4, r− m+ | 18 |
| BB255 | Essentially same as NCTC8325 cured from pI524 | 4 |
| COL | mec tet | 17 |
| BB1372 | COL ΔtcaRAB::ermB | 5 |
| BB1536 | BB1372 ptcaRAB | This study |
| E. coli DH5α | F− φ80dlacZΔM15 relA1 | Invitrogen |
| Plasmids | ||
| pAW17 | S. aureus-E. coli shuttle vector ori pAMα1-ori ColE1 aac-aph; Gmr Kmr | 27 |
| pAD21 | S. aureus suicide vector derived from pAW17 by removal of pAMα1-ori | This study |
| ptcaRAB | pAW17 with a 3,962-bp insert containing the COL tcaRAB region | This study |
| pMGS100 | E. coli-S. aureus shuttle expression vector | 12 |
| ptcaA | pMGS100 with a 1,379-bp insert containing the COL tcaA gene | This study |
| ptcaRBB255 | pMGS100 with a 233-bp insert containing the truncated BB255 tcaR gene | This study |
| ptcaRCOL | pMGS100 with a 452-bp insert containing the COL tcaR gene | This study |
| psarS | pAD21 with a 390-bp insert containing an internal portion of the COL sarS gene | This study |
Gmr, gentamicin resistance; Kmr, kanamycin resistance.
Sampling, RNA isolation, and transcriptional profiling.
Overnight cultures of S. aureus were diluted 1:100 into fresh prewarmed LB medium and grown to an optical density at 600 nm (OD600) of 2. The cells were harvested and snap-frozen in a dry ice-alcohol mixture. Frozen cells were then resuspended in 5 ml of ice-cold acetone-alcohol (1:1), incubated for 5 min on ice, centrifuged at 4°C, washed with 5 ml of TE buffer (10 mM Tris, 1 mM EDTA [pH 8]), and resuspended on ice in 900 μl of TE buffer. The cell suspension was transferred to 2-ml Lysing Matrix B tubes (Bio 101, Vista, Calif.) and shaken two times in an FP120 reciprocating shaker (Bio 101) at 6,000 rpm for 20 s. The cell debris was pelleted and the supernatant was used for RNA isolation by the RNeasy Midi system (Qiagen, Inc., Valencia, Calif.) according to the manufacturer's recommendations. To remove any contaminating genomic DNA, ca. 125 μg of total RNA was treated with 20 U of DNase I (Amersham Biosciences, Piscataway, N.J.) at 37°C for 30 min. The RNA was then purified with an RNeasy minicolumn (Qiagen) according to the manufacturer's cleanup protocol. Reverse transcription-PCR, cDNA fragmentation, cDNA terminal labeling, and hybridization of ca. 1.5 μg of labeled cDNA to custom-made Wyeth S. aureus GeneChips were carried out in accordance with the manufacturer's instructions (Affymetrix, Inc., Santa Clara, Calif.). GeneChip arrays were scanned by using the Agilent GeneArray laser scanner (Agilent Technologies, Palo Alto, Calif.). GeneChip scan data of biological duplicates were normalized and analyzed by using the GeneSpring gene expression software package (Silicon Genetics, Redwood City, Calif.).
RNA extraction and Northern hybridization.
Overnight cultures of S. aureus were diluted 1:100 in LB broth and pregrown for 2 h and then diluted 1:500 in LB medium and grown for 6 to 10 h. RNA was harvested at the various growth phases indicated. RNA isolation was performed as described by Cheung et al. (6), using a FastRNA kit and a Fastprep reciprocating shaker (Bio 101). Portions (10 μg) of total RNA from each sample were separated through a 1.5% agarose-0.66 M formaldehyde gel in MOPS running buffer (20 mM morpholinepropanesulfonic acid, 10 mM sodium acetate, 2 mM EDTA [pH 7.0]). Blotting of RNA onto a positively charged nylon membrane (Roche, Basel, Switzerland) was performed by using downward capillary blotting (The Source Book; FMC), with 10× SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate) as the transfer buffer. Digoxigenin-labeled DNA probes, produced by using the PCR DIG Probe synthesis kit (Roche, Basel, Switzerland), were used for the detection of specific transcripts by Northern hybridization, following the manufacturer's instructions. Primers used are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Probe or plasmid |
|---|---|---|
| Primers for construction of DIG probesa | ||
| sarSF | TTGATGAGCGTAATACTTAC | sarS |
| sarSR | GAGCTAATAATTGTTCAGCA | sarS |
| spaF | TGTAGGTATTGCATCTGTAA | spa |
| spaR | AAGTTAGGCATATTCAAGAT | spa |
| SA2439F | CAGTTATCGCTTCATCTAA | SA2439 N terminus |
| SA2439R | CATGTCCAGAACCATCATTA | SA2439 N terminus |
| SA2439ctF | GACACGTCTTCAGATGATAT | SA2439 C terminus |
| SA2439ctR | TATTGCGTCGTGATAACCAA | SA2439 C terminus |
| sdrCF | GACAATGAGTGATAGTGCAA | sdrC |
| sdrCR | CAGAATCATCGATTGTGTAA | sdrC |
| Primers for construction of recombinant plasmidsb | ||
| tcaR-f | TTTTTTCCTAGGTCATGATGCTATAGATGA | ptcaRAB |
| tcaB-r | ATTTAACCTAGGTCTATGAATAGCTCAAGT | ptcaRAB |
| HM5 | TTTTTCGGCCGGCATGGTAAAACATTTACAAGACC | ptcaRCOL |
| HM6 | ATTTTTCGCGATTTATCATTGCTATATGTTTTA | ptcaRCOL |
| HM7 | TTTTTCGGCCGGCATGAAATCTTGCCCGAAGTGCG | ptcaA |
| HM8 | ATTTTTCGCGATTTTTCTGATGTCTTGATTAAT | ptcaA |
| HM9 | ATTTTTCGCGAAGCATCGATTAACTTTTTAATTCG | ptcaRBB255 |
| sarSIAF | ATTAGAATTCCTGACTTATTTATTTCATCAG | psarS |
| sarSIAR | ATTAGGATCCGAGAAGTGATAATAGCTAGAA | psarS |
DIG, digoxigenin.
Restriction enzyme sites used for construction of recombinant plasmids are underlined.
Construction of plasmids for complementation.
Standard techniques were used for the construction of recombinant plasmids (29). All plasmids were first transformed in Escherichia coli strain DH5 α, electroporated into S. aureus RN4220 (18), and finally transduced with phage 80 α into the desired S. aureus background (4). Sequences of primers used are listed in Table 2. The 3,961-bp fragment encompassing the entire tcaRAB region from S. aureus COL was amplified with the primers tcaR-f and tcaB-r and cloned into the vector pAW17 to create ptcaRAB, which was transduced into BB1372 to give strain BB1536. The expression vector pMGS100, containing the bacA promoter, was used for the overexpression of tcaR and tcaA. Primers HM5 and HM6 were used to amplify the entire tcaR gene from COL (tcaRCOL), primers HM5 and HM9 were used to amplify the truncated tcaR gene from BB255 (tcaRBB255), and primers HM7 and HM8 were used to amplify the entire tcaA gene from COL. Inserts were then cloned into pMGS100 and the resulting plasmids ptcaRCOL, ptcaRBB255, and ptcaA were introduced into strains BB1372 and BB255.
Insertional inactivation of sarS.
Primers sarSIAF and sarSIAR (Table 2) were used to amplify a 390-bp fragment internal to the sarS gene, which was cloned into the suicide vector pAD21 creating vector psarS. psarS was then transformed into RN4220 by electroporation and transformants were selected on LB agar containing kanamycin. The insertionally inactivated sarS gene was then transduced into COL and BB1372 by using phage 80α as previously described to create strains COLsarS::pAD21 and BB1372sarS::pAD21. sarS insertional inactivation was confirmed in all strains by pulsed-field gel electrophoresis and Southern hybridization. Southern blotting and hybridization were carried out by standard techniques (29); primers sarSIAF and sarSIAR were used to amplify a digoxigenin-labeled sarS probe.
RESULTS
Genes differentially regulated in the S. aureus COL ΔtcaRAB mutant.
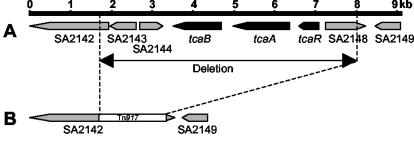
Deletion of the tcaRAB region has been shown to increase teicoplanin and decrease methicillin resistance in methicillin-resistant S. aureus (5), but the functions of the genes affected remained unknown. Microarray analysis was performed on total genomic RNA extracted from COL against that from BB1372, a strain containing a >6-kb genomic deletion encompassing the tcaRAB locus, and four additional deleted or disrupted open reading frame (ORFs) (Fig. 1). Surprisingly, comparison of the transcriptional profiles of these two strains revealed significant differences in expression for only three genes, namely, sarS (N315-SA0108), encoding staphylococcal accessory regulator S, spa (N315-SA0107), encoding protein A, and sasF (N315-SA2439), encoding a cell wall-associated protein (26). The expression values obtained for sarS were nearly sixfold higher in COL than in BB1372. Moreover, expression of spa was found to be enhanced in COL by a factor of more than 27. Both, sarS and spa were expressed abundantly in COL but expressed weakly in BB1372. In contrast, expression values obtained for sasF were >17-fold higher in BB1372 than in COL. sasF was expressed abundantly in BB1372 but expressed weakly in COL.
FIG. 1.
Schematic representation of the tcaRAB locus. (A) Map of the chromosomal region surrounding the tcaRAB operon. The nomenclature of the ORFs corresponds to the published S. aureus N315 genome sequence. (B) Depiction of the Tn917 induced deletion that occurred in BB1372.
Confirmation of microarray results by Northern analysis.
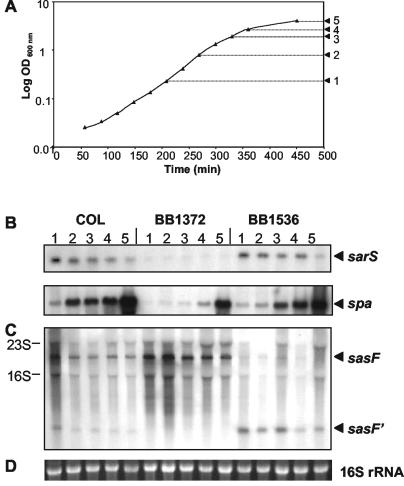
The effect of the BB1372 deletion on the transcription of ORFs identified by microarray analysis was monitored by Northern blots over the growth cycle. RNA from COL, the deletion mutant BB1372, and its ptcaRAB-complemented strain BB1536, was probed with sarS, spa, and sasF (Fig. 2). All Northern blot analyses confirmed the microarray findings. The Northern analysis revealed that sarS transcription was almost completely abolished in BB1372 and was restored to wild-type levels by complementation with ptcaRAB (Fig. 2B). Transcripts of spa followed the same trend, with spa transcription in BB1372 being reduced during early growth stages up to the end of exponential growth. Levels of spa transcription were also increased again upon complementation of the deletion mutant with ptcaRAB. sasF transcription was found to be significantly altered in the deletion mutant. In COL, two weak transcripts, one of ∼1,900 nucleotides, corresponding to the whole length of the sasF ORF, and a smaller transcript, sasF′ of ∼400 nucleotides, were detected (Fig. 2C). In BB1372, the smaller transcript disappeared, while there was a clear increase in the abundance of the larger transcript. Complementation with ptcaRAB resulted in the detection once again of the smaller transcript, accompanied by a strong downregulation of the larger transcript. sasF codes for a cell wall-sorted protein with a N-terminal signal sequence and a C-terminal sortase motif, directing the protein to the cell wall pentaglycine bridge. The probe used in this northern covered the 5′ end of the ORF. When probed with a 3′-specific probe, the small transcript was not detected in the wild-type COL. However, the upregulation of the larger transcript in BB1372 was still present (data not shown).
FIG. 2.
Transcription of sarS, spa, and sasF during growth. (A) Growth curve of S. aureus strain COL. The arrows indicate the OD600 points at which RNA was isolated. (B) Northern blot analyses of sarS and spa. Samples 1 to 5 correspond to RNA samples isolated at the OD600 values indicated in panel A. (C) Transcription of sasF. Specific transcripts are indicated on the right; two additional hybridizing bands, which corresponded to the sizes of the 23S rRNA and 16S rRNA, are indicated on the left. (D) As an indication of RNA loading, the 16S rRNA band from the ethidium bromide-stained gel is shown.
Identification of tcaR as the effector.
Complementation experiments (Fig. 2) demonstrated that it was the tcaRAB locus and not the other ORFs affected by the deletion that was responsible for affecting the regulation of all three ORFs identified by the microarray analysis.
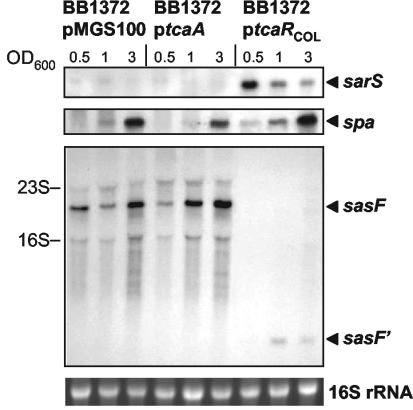
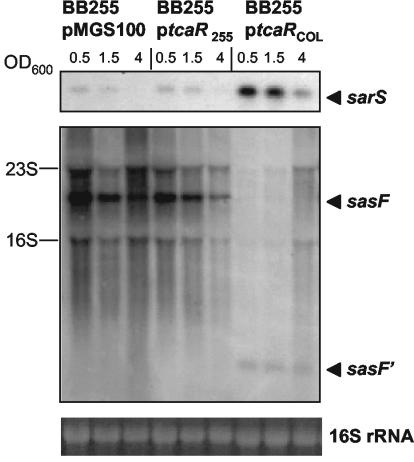
In order to determine which of the tca genes facilitated the transcriptional regulation of the three ORFs, BB1372 was complemented with plasmids constructed to overexpress either tcaR (ptcaRCOL), or tcaA (ptcaA). Northern blots showed that complementation of the deletion mutant with ptcaR increased transcription levels of sarS and those of spa (Fig. 3) and restored the wild-type sasF transcriptional profile, i.e., it accentuated the production of the truncated sasF′ to the detriment of sasF. Complementation of BB1372 with the empty pMGS100 vector or ptcaA had no effects on the transcription levels of sarS, spa, or sasF (Fig. 3).
FIG. 3.
Complementation of the COL ΔtcaRAB phenotype by tcaR. Northern blots of BB1372 complemented with the empty plasmid vector pMGS100, ptcaA, or ptcaRCOL against the sarS, spa, and sasF probes. Total RNA was harvested at growth stages corresponding to the OD600 values indicated above the lanes. As an indication of RNA loading, the 16S rRNA band from the ethidium bromide stained gel is shown.
NCTC8325 derivatives are tcaR mutants.
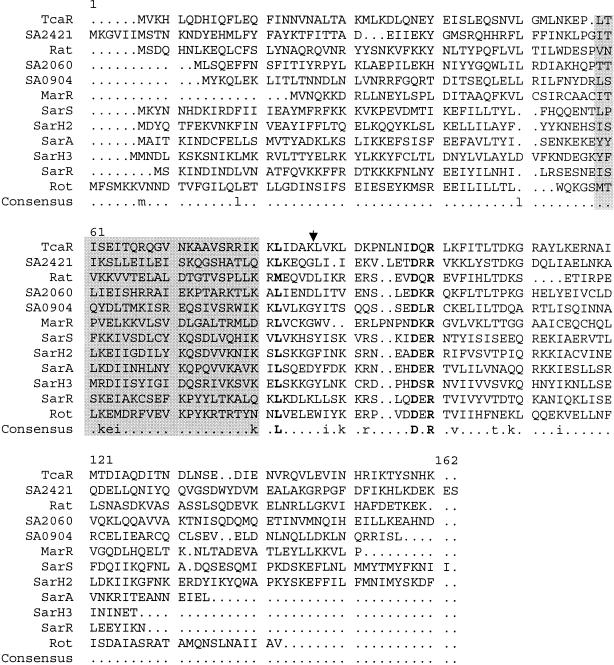
TcaR shares amino acid sequence homology with members of the MarR family of DNA-binding transcriptional regulators, as does N315-SA0641, the protein published recently as Rat, Mgr, and NorR, and three further hypothetical S. aureus proteins containing the helix-turn-helix MarR domain (Fig. 4). TcaR also shares similarity, but to a lesser degree with the SarA family of proteins (Fig. 4). Comparison of all available TcaR amino acid sequences from S. aureus genome sequences revealed that the TcaR in the NCTC8325 derivative 8325-4 is truncated by a stop codon at position 79. Sequencing of the tcaR gene of BB255, RN4220, and RN6390 revealed the same mutation to be present in all strains analyzed, suggesting that the mutation originated from NCTC8325, which is the common ancestor for all of these strains. An arrowhead above the COL TcaR sequence in Fig. 4 marks the position of the stop codon in NCTC8325-4. Comparison of the TcaR amino acid sequence with those of other MarR-like proteins indicated that TcaR, although truncated after the predicted helix-turn-helix motif, is missing a number of the universally conserved residues highlighted in Fig. 4 and thus may be inactive.
FIG. 4.
Amino acid sequence alignment of TcaR against MarR from E. coli, SarA, and identified SarA homologues from S. aureus and other homologous protein sequences identified from the S. aureus N315 genome by BLASTP analysis. The sequences of SarS (SarH1) and SarH2, which both contain two domains with homology to SarA, were truncated so that only the N-terminal domain of each was included in the alignment. The region containing the predicted helix-turn-helix motif of TcaR (Network Protein Sequence analysis [9]) and several of the other homologues is highlighted in gray. Strongly conserved residues are indicated in the consensus line and universally conserved residues are in bold type. The arrowhead represents the position (amino acid 79) at which the TcaR protein in NCTC8325-4 is truncated.
To determine whether the truncated TcaR protein from BB255 (TcaRBB255) retained activity, Northern analyses with probes for sarS and sasF were performed on RNA extracted from BB255 and its derivatives, complemented either with the empty expression vector pMGS100, plasmid ptcaRBB255, overexpressing the truncated TcaR from BB255, or ptcaRCOL, overexpressing the intact TcaR from COL. The expression of sarS and sasF in BB255 containing the empty pMGS100 vector was analogous to that observed in the COL ΔtcaRAB deletion mutant, i.e., weak expression of sarS and strong expression of the large sasF transcript with no detection of the smaller sasF′ transcript. Complementation with the truncated ptcaRBB255 had no effect on the transcription of these genes. However, complementation with ptcaRCOL significantly upregulated sarS transcription and altered sasF transcription, mirroring the transcription profile of these genes in the wild-type COL background (Fig. 5).
FIG. 5.
Transcription of sarS and sasF in BB255 and BB255 complemented with either the empty plasmid vector pMGS100, the tcaR gene from B255 (ptcaRBB255), or the tcaR gene from COL (ptcaRCOL). Total RNA was harvested at growth stages corresponding to the OD600 values shown. As an indication of RNA loading the 16S rRNA band from the ethidium bromide-stained gel is shown.
Disruption of sarS.
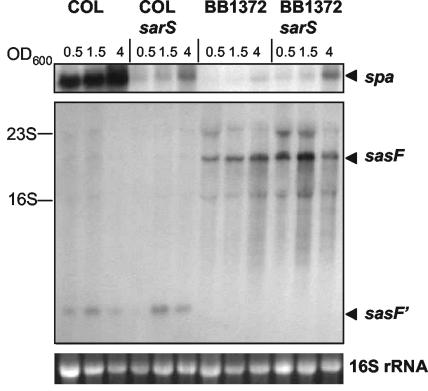
To test whether TcaR influences the expression of spa and sasF directly, or indirectly as a result of altered sarS expression, the sarS gene was disrupted by insertional inactivation. The transcription profiles of spa and sasF for COL, the COL sarS mutant (COLsarS::pAD21), BB1372, and the BB1372 sarS mutant (BB1372sarS::pAD21) were compared by Northern analysis (Fig. 6). Levels of spa transcription were significantly decreased in both sarS mutant strains, confirming previous reports that SarS is one of the key factors required for induction of spa expression (7, 32). Thus, the low spa transcription level in BB1372 will be, at least in part, a direct result of decreased sarS expression. Whether TcaR can also directly influence spa expression remains to be determined. The transcription profile of sasF was not affected by inactivation of sarS, indicating that sarS is not involved in the regulation of this gene locus.
FIG. 6.
Effect of sarS mutation on the expression of spa and sasF. Northern analysis of COL and BB1372 and their corresponding sarS mutants, COLsarS::pAD21 and BB1372sarS::pAD21. Total RNA was harvested at growth stages corresponding to the OD600 values shown. As an indication of RNA loading the 16S rRNA band from the ethidium bromide-stained gel is shown.
Salicylic acid does not affect the regulatory function of TcaR.
Salicylic acid is known to bind to and inactivate MarR in Escherichia coli, altering the expression of genes under MarR control (1). In addition, it has been shown to significantly influence virulence factor expression in S. aureus (19). No effects of salicylate could be detected in our system when we analyzed the intensity and pattern of the spa, sarS, and sasF transcripts in COL and BB1372 grown in the presence or absence of sodium salicylate, suggesting that salicylate is unlikely to bind and affect the activity of TcaR (data not shown).
DISCUSSION
Regulation and fine-tuning of virulence factors is one of the most important tasks for the successful establishment of an infection and propagation of S. aureus. Many of the virulence genes are controlled simultaneously by different regulators, and there is an increasing amount of evidence suggesting that marked differences exist in the genetic setup of these regulators between different strain lineages. Here we add an additional factor to this network of regulators, TcaR, which we showed to activate the expression of the global regulatory locus, sarS (sarH1).
To date, the majority of the studies of virulence gene regulation have been carried out with strains derived from NCTC8325 (25). The widely used laboratory strain derivatives of NCTC8325, such as 8325-4, BB255, RN4220, and RN6390, were found here to contain a truncated and thus an inactive TcaR; a phenotype that could be complemented by the addition of the COL tcaR gene in trans. This observation is of particular interest since all information previously published concerning sarS function has been obtained from NCTC8325 derivatives (8, 30, 32). Moreover, NCTC8325 derivatives are additionally known to be rsbU mutants, rendering these strains phenotypically σB defective. Expression of sarS was also demonstrated to be influenced by σB activity (4a, 32). Hence, the influence of either TcaR and/or σB on the regulatory circuit proposed for sarS and spa transcription in wild-type TcaR+ and RsbU+ strains, such as the strain COL used here, is likely to differ from that described for NCTC8325 derived strains.
TcaR is one of several MarR-like transcriptional regulators identified to play a role in the regulation of virulence determinants in S. aureus. Others include the sarA family of regulators and the gene products of N315-SA0641, named Rat (also known as NorR and Mgr) (15, 22, 33), and N315-SA1583, designated Rot (24). Rat, influences the production of a variety of virulence factors, such as type 8 capsular polysaccharide, protein A, alpha-toxin, nuclease, lipase, protease, and coagulase (22). Rat also affects autolysis, possibly by regulating genes influencing autolytic activities such as abcA, scdA, and sspA (15) and is known to regulate the multidrug efflux pump NorA (33). Rot was shown to act as a global regulator with the capacity to both positively and negatively influence a large range of target genes, including sarS and spa, which were found to be upregulated (28).
TcaR, although related on the amino acid level to Rot, Rat, and the SarA family, which belong to the winged-helix family of DNA-binding proteins (2, 20, 21, 28, 31), appears to have a very narrow spectrum of action. Unlike SarA, which was found to affect at least 120 genes (10), Rot, which potentially regulates up to 146 genes (28), and Rat, which appears to influence the regulation of at least 10 genes (15, 22, 33) in S. aureus, TcaR apparently directly affects the transcription of only two genes. While acting as an activator of sarS and thus spa expression, TcaR repressed the production of a further exoprotein, SasF. SasF has been described as a probable adhesin, but as yet no function has been ascribed to it (26). TcaR is the first regulator described for sasF. The mode of TcaR mediated repression of sasF transcription appears to be unusual. TcaR appears to somehow cause the abortive transcription or the transcriptional processing of sasF. The shortened transcript corresponds to the 5′ end of the ORF and is thus lacking the C-terminal sortase motif required for cell wall anchoring. It remains to be determined whether the sasF′ transcript fulfils a specific function and, if translated, is targeted to the cytoplasm or excreted.
The conditions controlling expression of tcaR are not yet known. tcaR transcription appeared to be low but consistent over all growth phases (22a). Complementation of the deletion mutant with tcaR did not affect teicoplanin nor methicillin resistance, nor did tcaR overexpression enhance tcaAB transcription (unpublished results), suggesting that TcaR is not involved in these resistance mechanisms.
The significantly lower levels of sarS in tcaR mutants, such as BB1372 and 8325 derivatives, indicate that TcaR is, besides SarT (30), a major activator of sarS. Our results confirmed moreover the earlier findings of Cheung et al. and Tegmark et al. (7, 32) that spa transcription depends on SarS activation. Additional Northern blot experiments, to determine whether TcaR influenced sarT expression, indicated that sarT is not an intermediate in the TcaR-dependent pathway of sarS regulation (results not shown).
MarR-type regulators have been shown to act as repressors of transcription, as positive regulators, or as both positive and negative regulators (11). SarA family proteins have also been shown to act as activators and repressors of transcription (8). TcaR also appears to be a bifunctional regulator, with two very specific targets.
Acknowledgments
We thank S. Projan, P. Dunman, E. Murphy, and Wyeth for providing facilities and support to M.B. for generating the microarray data and S. Fujimoto, Gunma University School of Medicine, for kindly providing pMGS100.
This study was supported by the Swiss National Science Foundation grant NRP49-63201 to B.B.-B. and grant 560030 of the Forschungskredit der Universität Zürich to M.B.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181: 4669-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8: 710-714. [DOI] [PubMed] [Google Scholar]
- 3.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291: 159-170. [DOI] [PubMed] [Google Scholar]
- 4.Berger-Bächi, B. 1983. Increase in transduction efficiency of Tn551 mediated by the methicillin resistance marker. J. Bacteriol. 154: 533-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Bischoff, M., P. Dunman, J. Kormanec, E. Murphy, B. Berger-Bächi, and S. Projan. Microarray-based analysis of the σB-regulon of Staphylococcus aureus. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 5.Brandenberger, M., M. Tschierske, P. Giachino, A. Wada, and B. Berger-Bächi. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta 1523: 135-139. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222: 511-514. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein a synthesis in Staphylococcus aureus. Infect. Immun. 69: 2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., and G. Y. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7: D1825-D1842. [DOI] [PubMed] [Google Scholar]
- 9.Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25: 147-150. [DOI] [PubMed] [Google Scholar]
- 10.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183: 7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egland, P. G., and C. S. Harwood. 1999. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J. Bacteriol. 181: 2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimoto, S., and Y. Ike. 2001. pAM401-based shuttle vectors that enable overexpression of promoterless genes and one-step purification of tag fusion proteins directly from Enterococcus faecalis. Appl. Environ. Microbiol. 67: 1262-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giachino, P., S. Engelmann, and M. Bischoff. 2001. sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183: 1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184: 5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingavale, S. S., W. van Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48: 1451-1466. [DOI] [PubMed] [Google Scholar]
- 16.Josefsson, E., K. W. McCrea, D. Ni Eidhin, D. O'Connell, J. Cox, M. Hook, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144: 3387-3395. [DOI] [PubMed] [Google Scholar]
- 17.Kornblum, J., B. Kreiswirth, S. J. Projan, H. Ross, and R. P. Novick. 1990. Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 18.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. N. Maslow, A. McGeer, D. E. Low, and R. P. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259: 227-230. [DOI] [PubMed] [Google Scholar]
- 19.Kupferwasser, L. I., M. R. Yeaman, C. C. Nast, D. Kupferwasser, Y. Q. Xiong, M. Palma, A. L. Cheung, and A. S. Bayer. 2003. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J. Clin. Investig. 112: 222-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, R., A. C. Manna, S. Dai, A. L. Cheung, and G. Zhang. 2003. Crystal structure of the SarS protein from Staphylococcus aureus. J. Bacteriol. 185: 4219-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, Y. F., A. Manna, R. G. Li, W. E. Martin, R. C. Murphy, A. L. Cheung, and G. Y. Zhang. 2001. Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98: 6877-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185: 3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Maki, H., N. McCallum, M. Bischoff, A. Wada, and B. Berger-Bächi. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 23.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalyzed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40: 1049-1057. [DOI] [PubMed] [Google Scholar]
- 24.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182: 3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48: 1429-1449. [DOI] [PubMed] [Google Scholar]
- 26.Roche, F. M., R. Massey, S. J. Peacock, N. P. J. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149: 643-654. [DOI] [PubMed] [Google Scholar]
- 27.Rossi, J., M. Bischoff, A. Wada, and B. Berger-Bächi. 2003. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob. Agents Chemother. 47: 2558-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by rot. J. Bacteriol. 185: 610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2003. SarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 71: 5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schumacher, M. A., B. K. Hurlburt, and R. G. Brennan. 2001. Crystal structures of SarA, a pleiotropic regulator of virulence genes in S. aureus. Nature 409: 215-219. [DOI] [PubMed] [Google Scholar]
- 32.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37: 398-409. [DOI] [PubMed] [Google Scholar]
- 33.Truong-Bolduc, Q. C., X. M. Zhang, and D. C. Hooper. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 185: 3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tung, H. S., B. Guss, U. Hellman, L. Persson, K. Rubin, and C. Ryden. 2000. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem. J. 345: 611-619. [PMC free article] [PubMed] [Google Scholar]